Abstract
Objectives
To investigate optimised isotropic 3D turbo spin echo (TSE) and gradient echo (GRE)-based pulse sequences for visualisation of articular cartilage lesions within the knee joint.
Methods
Optimisation of experimental imaging sequences was completed using healthy volunteers (n=16) with a 3-Tesla (3T) MRI scanner. Imaging of patients with knee cartilage abnormalities (n=57) was then performed. Acquired sequences included 3D proton density-weighted (PDW) TSE (SPACE) with and without fat-suppression (FS), and T2*W GRE (TrueFISP) sequences, with acquisition times of 6:51, 6:32 and 5:35 min, respectively.
Results
One hundred sixty-one confirmed cartilage lesions were detected and categorised (Grade II n=90, Grade III n=71). The highest sensitivity and specificity for detecting cartilage lesions were obtained with TrueFISP with values of 84.7% and 92%, respectively. Cartilage SNR mean for PDW SPACE-FS was the highest at 72.2. TrueFISP attained the highest CNR means for joint fluid/cartilage (101.5) and joint fluid/ligament (156.5), and the lowest CNR for cartilage/meniscus (48.5). Significant differences were identified across the three sequences for all anatomical structures with respect to SNR and CNR findings (p-value <0.05).
Conclusion
Isotropic TrueFISP at 3T, optimised for acquisition time, accurately detects cartilage defects, although it demonstrated the lowest contrast between cartilage and meniscus.
Key points
• Cartilage is better visualised with 3D TrueFISP than 3D SPACE sequences.
• 3D TrueFISP is a reliable sequence for detecting low- and high-grade cartilage defects.
• 3D TrueFISP at 3T provides excellent contrast between cartilage and joint fluid.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Magnetic resonance imaging (MRI) is considered the gold standard technique for evaluating and monitoring cartilage pathology and the effect of pharmacological and surgical therapies [1–5]. Recent advances in MRI technology have led to the increasing clinical use of 3-Tesla (3T) MRI [6], which has created opportunities to enhance MR image quality by improvements in signal-to-noise ratio (SNR), contrast-to-noise ratio (CNR) and spatial resolution [7]. Whilst the majority of published material has focused on applications of 3T systems for neuroimaging, there is increasing interest in musculoskeletal (MSK) imaging at 3T [6]. Recent studies involving the knee joint have demonstrated that three-dimensional (3D) sequences could compliment the more routinely acquired two-dimensional (2D) sequences, but they currently cannot replace 2D techniques due to long acquisition times, ranging from 9 to 11 min, and poorer image quality [8]. 3D sequences are described as suffering from more image blurring and noise compared to 2D [8, 9]. While a key advantage of 3D acquisitions is their inherently high spatial resolution, the major potential of isotropic 3D MRI is that it permits image reformatting in any plane (including the conventional axial, sagittal and coronal planes). If high-quality 3D MRI datasets could be obtained, without a significant acquisition time penalty, then it might be possible to replace the typically acquired 2D multi-planar sequences with a single 3D sequence that can delineate pathology with an even greater fidelity than conventional 2D sequences [10]. In this study, we have assessed the performance of novel optimised 3D gradient echo (True-FISP) and turbo spin echo based (SPACE) pulse sequences for visualisation of articular cartilage lesions within the knee joint.
Materials and methods
Full institutional ethical approval was attained prior to commencing patient recruitment. Optimisation of the 3D sequences was undertaken in order to shorten the acquisition times. The strategies adopted for sequence optimisation involved scan parameter manipulation (TR, bandwidth, ETL and matrix size). Optimising the 3D sequences included scanning 16 healthy volunteers (female n=9, male n=7; age range 22–58 years; mean age 34 years). The healthy volunteers were medical or paramedical personnel, with no history of attending a clinician and/or physiotherapist for evaluation and/or treatment of knee joint diseases.
Following the experimental phases, the work then focused on applying the optimised 3D sequences on patients and evaluating the findings. Fifty-seven consecutive adult patients (male n=31, female n=26; age range 21–67 years; mean age 41 years) with suspected knee cartilage abnormalities were recruited. Patients included in this study presented with a clinical indication of a cartilage abnormality, e.g. pain, swelling and stiffness (n=36); injury (n=14); osteoarthritis using Kellgren and Lawrence grading system (n=7; grade 2 = 4, grade 3 = 3).
All participants were prospectively scanned over a 3-month period from April to June 2015. Written informed consent was obtained from all the participants before the examinations. Participants with any contraindication according to the MR safety-screening questionnaire for the 3T scanner were excluded [11].
MR imaging protocol
Knee images were acquired using a 3T MR system (MAGNETOM Skyra Siemens Healthcare GmbH, Erlangen, Germany) with a compatible 15-channel knee coil (TxRx, Quality Electrodynamics, Mayfield, Ohio, USA). The standard 2D sequences of the knee joint and the three different 3D sequences were acquired from consenting participants to determine the diagnostic performance characteristics of these sequences. 3D PDW Sampling Perfection with Application optimised Contrast using different flip angle Evolutions (SPACE), 3D PDW SPACE with fat saturation (FS) technique, and 3D gradient echo TrueFISP incorporating spectral attenuated inversion recovery (SPAIR) fat suppression technique were applied. Table 1 summarises the parameters for the MR pulse sequences evaluated.
Post-processing
All 3D-source data sets were reconstructed in the coronal, axial and sagittal planes, corresponding to the acquisition planes for the 2D sequences, with 0.5-mm slice thickness using Syngo software (Syngo, Siemens Medical Solutions, Erlangen, Germany). Images reconstructed in the coronal plane were planned from axial images, with the plane of reconstruction angled parallel to the medial and lateral femoral epicondyles at the attachment points of the collateral ligaments. Reconstructed axial images were planned from coronal images by aligning the reconstruction plane parallel to the medial and lateral femoral epicondyles. Sagittal reconstructions were planned from axial images with the reconstruction plane adjusted parallel to the lateral femoral epicondyle, and therefore to the anterior cruciate ligament.
Image analysis
Qualitative assessment
Ambient light levels of below 50 lux were maintained during the image evaluation sessions [12] and images were displayed on a PACS monitor with 5 megapixel resolution (Barco®).
Healthy volunteers: The evaluation was performed by two MSK radiologists with 9 and 10 years of experience, respectively, and specialists in MSK MR imaging.
Qualitative assessment included evaluation of the visualisation of anatomical structures: menisci, cartilages and ligaments [13] using an ordinal 5-point Likert-scale. Each anatomical structure was rated based on the appearance as follows: anatomical structure is not visible (poor); anatomical structure only partially visible (below average); anatomical structure fully visible, with substantial blurring of borders (fair); anatomical structure fully visible, with slight blurring of borders (good) and; excellent delineation of anatomical structure (excellent) [14].
Patients: Qualitative evaluation was undertaken based on the independent, subjective assessment of the degree of visibility of abnormalities involving the knee joint. Evaluation was performed by two MSK radiologists with 10 and 16 years of experience, respectively, and specialists in MSK MR imaging. The radiologists first independently reviewed the 2D MR images. To reduce a potential learning bias, radiologists independently reviewed the 3D images 14–21 days later [13, 15]. At the time of each image review session, the radiologists were blinded to the sequence parameters, patient’s clinical history and clinical findings.
Six articular cartilage surfaces were evaluated as part of this study: medial and lateral femoral cartilage, medial and lateral tibial cartilage, retropatellar cartilage, and femoral trochlear cartilage [16]. For each sequence type, the two radiologists graded cartilage abnormalities at the above six regions using an ordinal 3-point Likert-scale. The abnormalities were rated based on the MR appearance of the pathology as follows: normal cartilage surface (Grade I); low-grade cartilage defect <50% of total cartilage thickness (grade II); high-grade cartilage defects of more than 50% (Grade III) [16].
Quantitative assessment
Quantitative measurements were evaluated by calculating numerical indices of MR image quality for the representative images through the knee joint including: SNR and CNR for the 3D MR pulse sequences using sagittal plane [15]. Regions of interest (ROIs) for determining the signal intensities were drawn within the following tissue types: menisci, cartilage, ligaments, bones, muscles, fat and joint fluid in the knee joint [8, 13]. The CNR values were calculated for the following anatomical structures: cartilage/meniscus, joint fluid/meniscus, joint fluid/cartilage and joint fluid/ligament.
The SNR within the anatomical regions was calculated as follows: SNR = the mean signal intensity (SI) of the regions/SD of the noise in the background [13]. The CNR of the regions was calculated as follows: CNR = (the mean SI of the region – the mean SI of the normal structure adjacent to the region)/SD of the background noise [13]. The SD of noise was measured by placing a 5 mm2 circular ROI within the image background, avoiding artefact if present [13].
Reference standard
In line with previous studies, the conventional 2D MR images of the knee joint were used as the reference standard and the three different 3D sequences were compared to the 2D technique in order to calculate the sensitivity and specificity [16]. The optimum reference standard across all 2D images was determined by consensus decision between the two radiologists. Whilst arthroscopy is acknowledged as the gold standard reference [8], the majority of the patients (87%) included in this study did not clinically require arthroscopic assessment.
Statistical analysis
All statistical analyses were carried out using SPSS (IBM SPSS Statistics for Macintosh, Version 23.0, IBM Corp., Armonk, NY, USA), and Excel (Microsoft Excel for Macintosh, Version 14.5). Statistical significance was assumed for p <0.05.
Qualitative data
Sensitivity and specificity for the 3D sequences were calculated for all patients; the conventional 2D technique was used as a reference standard. Cartilage lesions were classified as either negative (grade I) or positive for an abnormality, with low-grade cartilage defect (grade II), and with high-grade cartilage defects (grade III). The sensitivity and specificity were calculated for grade II and grade III cartilage lesions for all six of the articular surfaces. Sensitivity was calculated as the proportion of times the 3D technique agreed with the reference standard [8]. Specificity was calculated as the proportion of times patients were correctly classified as not having the specific abnormality. Sensitivity and specificity were also calculated for grade II and grade III cartilage lesions combined (vs. grade I).
Kappa (κ) statistics determined the inter-observer agreement during MR image scoring and analysis [17].
Quantitative data
For each patient, SNR and CNR were calculated for the 3D sequences. Repeated measures analysis of variance (ANOVA) was used to test for a difference in SNR and CNR across sequence types [18]. If the SNR and CNR were found to differ across sequences, pairwise post hoc tests were carried out (paired t-test with Bonferroni correction) to identify pairs of sequences with differing SNR and CNR.
Results
Qualitative findings
Healthy volunteers
The anatomical structure visualisation findings for the 3D sequences are summarised in Table 2. The 3D TrueFISP sequence performed better than tested 3D SPACE sequences for visualising cartilage and meniscus.
Patients
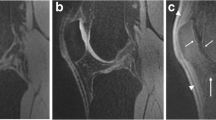
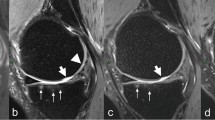
In total, 161 cartilage lesions were confirmed, distributed as follows: Grade II n=90, Grade III n=71. The 3D TrueFISP sequence was identified as having the highest sensitivity for detecting cartilage lesions (84.7%), followed by PDW SPACE-FS and PDW SPACE with 82%, and 79%, respectively. The sensitivity of the 3D sequences was found to be lower for detecting low-grade cartilage defects (Fig. 1), with sensitivity of 77.7%, 72.7% and 67.7% for TrueFISP, PDW SPACE-FS and PDW SPACE, respectively for grade II. In contrast, the sensitivity of 3D sequences was higher with high-grade cartilage defects (Fig. 2), with 94.3%, 93.6% and 93% for PDW SPACE-FS, TrueFISP and PDW SPACE, respectively.
The specificity was higher with TrueFISP (92%), followed by PDW SPACE and PDW SPACE-FS with 82% and 80%, respectively. Similar to sensitivity values, the specificity of the 3D sequences was higher with high-grade cartilage abnormalities, with 98%, 96.5% and 96% for TrueFISP, PDW SPACE and PDW SPACE-FS. The specificity of the TrueFISP sequence was considered high for low-grade abnormalities (92.8%), but low for PDW SPACE-FS and PDW SPACE with values of 85.5% and 85.3%, respectively.
The κ values for inter-rater agreement in evaluation of cartilage lesions are listed in Table 3, and indicated excellent agreement (0.81–1) between both readers for all 3D sequence types.
Quantitative findings
For most anatomical structures, the mean SNR was higher for PDW SPACE-FS and TrueFISP sequences. As seen in Table 4, PDW SPACE-FS and TrueFISP SNR mean values for cartilage, joint fluid and muscle were higher than those for PDW SPACE. However, the PDW SPACE mean SNR scores for fat and bone marrow were substantially greater than for other sequences (Table 4 and Fig. 3). Meniscus and ligaments showed very low SNR of 5.6 and 5.5 for meniscus, and 8.5 and 9.5 for ligaments with PDW SPACE-FS and PDW SPACE, respectively. However, TrueFISP produced higher SNR for the meniscus (20.5) and ligaments (14).
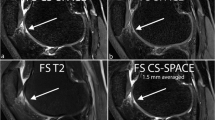
Sagittal 3D MR images acquired at 3T for a 28-year-old female. TrueFISP image demonstrated the highest contrast between cartilage and joint fluid across all images (a). PDW SPACE-FS image showed blurring and noise effects, with high contrast between cartilage and joint fluid (c), while there is low contrast between cartilage and joint fluid with PDW SPACE (b)
Similar to SNR values, PDW SPACE-FS and TrueFISP CNR means were higher than for PDW SPACE, and this includes the following groups: cartilage/meniscus, joint fluid/meniscus, joint fluid/cartilage and joint fluid/ligament (Table 5). TrueFISP CNR means were higher than PDW SPACE-FS for the groups that did not include meniscus, while PDW SPACE-FS CNR means were the highest for groups that included the meniscus.
Discussion
In this study, we found that the optimised 3D isotropic TrueFISP and SPACE-FS sequences can be reliably used for delineation of knee cartilage pathology within practical acquisition times and with acceptable image quality. The high contrast between cartilage and joint fluid associated with these sequences improved the detection of high-grade and low-grade cartilage defects.
Previous studies have evaluated the advanced 15-channel knee RF coil and its use for knee MR imaging at 3 Tesla [8, 16, 19, 20]. These prior studies found that long acquisition times and image blurring were the principal limiting factors for the routine application of 3D pulse sequences. In an effort to address these limitations we performed acquisition time reduction optimisation involving healthy volunteers and experimental phantom before instigating this clinical study.
A 3D TSE-based sequence of the knee was previously investigated by Notohamiprodjo et al. [8]. In this study, image quality was graded as excellent but at the cost of a relatively prolonged acquisition time (10:44 min), a length which may not be practical in the clinical setting. Prolonged acquisition times increase the risk of motion artefact and may not be tolerated by some patient cohorts, for example those with acute pain [1, 8, 13]. In clinical practice, it is important to achieve a balance between scan time and image quality. The question thus remains, despite previous studies, as to whether 3D sequences can be optimised to such a degree that they could replace conventional multi-planar 2D sequences for knee joint imaging.
Based on qualitative image evaluation, the investigated sequences (True-FISP, PDW SPACE-FS and PDW SPACE) demonstrated high sensitivity and specificity for detecting cartilage lesions, at substantially reduced acquisition times of 5:35, 6:32 and 6:51 min, respectively, when compared to previous studies [8, 16]. The 3D TrueFISP was identified as being the most sensitive and specific technique for detecting cartilage defects of any grade, and these findings are consistent with previous literature [8]. While 3D sequences demonstrated high sensitivity and specificity for high-grade cartilage defects, the performance of the 3D sequences was lower for low-grade abnormalities. The detection of low-grade abnormalities was primarily compromised by image blurring and reduced contrast between cartilage and adjacent joint fluid on these 3D sequences, which were more severe with PDW SPACE images.
Based on quantitative image evaluation, mean SNR and CNR values for TrueFISP and PDW SPACE-FS were higher than for the PDW SPACE sequence especially for articular cartilage and joint fluid, findings that are in line with previous studies [8, 16, 21, 22]. High SNR and CNR values have been shown to improve the detection sensitivity for subtle cartilage lesions, and similarly so in this study. The TrueFISP CNR means for joint fluid/cartilage and joint fluid/ligament were higher than for PDW SPACE-FS (Fig. 3). Prior literature found that the TrueFISP pulse sequence provides higher contrast between cartilage and joint fluid compared to other 3D sequences such as SPACE, Fast-Low Angle Shot (FLASH) and Double-Echo Steady-State (DESS), as was determined in this study [19, 22, 23]. Gradient-based sequences showed lower values than TSE-based sequences for contrast between cartilage and meniscus, both in this study and in previous work [23]. However, the clinical performance of the applied TrueFISP sequence was higher than that of other sequences tested.
In this study, TrueFISP was found to be the most reliable sequence for detecting cartilage abnormalities, which is similar to previous research opinion [21, 22]. Previous literature has reported a lower sensitivity using the TrueFISP sequence compared to other 3D sequences due the effects of banding artefact [16]. Whilst this artefact was noted in the image periphery for 70% of our cases (n=40), it did not obscure critical anatomy. The improved performance of the TrueFISP sequence in our study may relate to sequence optimisation, which involved the removal of the parallel imaging option from this 3D sequence. The optimised 3D TrueFISP sequence may therefore replace multiple or at least one of the conventional 2D sequences (e.g., axial PDW with fat saturation) to increase diagnostic confidence.
It is acknowledged that arthroscopic correlation was not possible due to the small number of patients who were referred for both knee MRI and arthroscopy. Arthroscopies could not be mandated for all study patients due to ethical and clinical reasons. Limiting the study to just those patients who had 3D imaging and arthroscopy would have significantly reduced the potential patient cohort and possibly introduced a bias in terms of patient selection (younger patients with significant abnormalities are more likely to have arthroscopy). A study by Thorlund et al. [24] showed that arthroscopy has insignificant benefit for middle-aged and older patients. A previous study has employed a similar 2D sequence reference standard when evaluating cartilage defects and knee joint abnormalities [16]. A potential limitation of our study is that, despite the radiologists being blinded to the sequence parameters, each MR sequence generated images with signal intensity characteristics that were recognisable to the reviewing radiologists. Furthermore, a single observer measured the numerical values of SNR and CNR by drawing ROIs; however, this is in line with previous studies [25, 26].
A technical limitation noted in this study was that despite the 0.5 mm3 voxel size and 0.5 mm reconstruction interval, image blurring was more pronounced with 3D reformatted images than with the original 3D image data sets, which could affect lesion visualisation [27]. In a previous study at 3T, Kijwoski et al. [9] using a 3D isotropic TSE-based sequence, found image blurring reduced the ability to detect lateral meniscal root tears. We similarly found that image blurring was more obvious on 3D PDW SPACE sequences, which could potentially reduce sensitivity to cartilage abnormalities.
Conclusion
This study investigated three optimised 3D isotropic sequences for evaluating articular cartilage defects. 3D isotropic TrueFISP and PDW SPACE-FS images demonstrated high sensitivity and specificity for detecting cartilage lesions within practical imaging times. The 3D isotropic TrueFISP and PDW SPACE-FS pulse sequences in particular demonstrated high contrast between cartilage and joint fluid, which aided in the detection of subtle cartilage abnormalities. These findings support the reliability of the 3D technique for evaluating cartilage abnormalities within the knee joint. Although optimised 3D TrueFISP rendered excellent images and was associated with high sensitivity and specificity values for both low- and high-grade cartilage defects, it demonstrated the lowest contrast between cartilage and meniscus. Further research is recommended to establish whether the removal of parallel imaging from the TrueFISP sequence, as performed in this study, reduces the severity of banding artefact.
References
Crema MD, Nogueira-Barbosa MH, Roemer FW et al (2013) Three-dimensional turbo spin-echo magnetic resonance imaging (MRI) and semiquantitative assessment of knee osteoarthritis: comparison with two-dimensional routine MRI. Osteoarthritis Cartilage 21:428–433
Milewski MD, Smitaman E, Moukaddam H et al (2012) Comparison of 3D vs. 2D fast spin echo imaging for evaluation of articular cartilage in the knee on a 3T system scientific research. Eur J Radiol 81:1637–1643
Crema MD, Roemer FW, Marra MD et al (2011) Articular cartilage in the knee: current MR imaging techniques and applications in clinical practice and research. Radiographics 31:37–62
Hafezi-Nejad N, Guermazi A, Roemer FW et al (2016) prediction of medial tibiofemoral compartment joint space loss progression using volumetric cartilage measurments: data from the FNIH OA biomakers consortium. Eur Radiol 27:464–473
Alizai H, Roemer FW, Hayashi D, Crema MD, Felson DT, Guermazi A (2015) An update on risk factors for cartilage loss in knee osteoarthritis assessed using MRI-based semiquantitative grading methods. Eur Radiol 25:883–893
Shapiro L, Staroswiecki E, Gold G (2010) Magnetic resonance imaging of the knee: optimizing 3 Tesla imaging. Semin Roentgenol 45:238–249
Magee T, Williams D (2006) 3.0-T MRI of meniscal tears. AJR Am J Roentgenol 187:371–375
Notohamiprodjo M, Horng A, Kuschel B et al (2012) 3D-imaging of the knee with an optimized 3D-FSE-sequence and a 15-channel knee-coil. Eur J Radiol 81:3441–3449
Kijowski R, Davis KW, Blankenbaker DG, Woods MA, Del Rio AM, De Smet AA (2012) Evaluation of the menisci of the knee joint using three-dimensional isotropic resolution fast spin-echo imaging: diagnostic performance in 250 patients with surgical correlation. Skeletal Radiol 41:169–178
Jung JY, Yoon YC, Kim HR, Choe BK, Wang JH, Jung JY (2013) Knee derangements: comparison of isotropic 3D fast spin-echo, isotropic 3D balanced fast field-echo, and conventional 2D fast spin-echo MR imaging. Radiology 268:802–813
Kanal E, Barkovich AJ, Bell C (2007) ACR gudiance document for safe MR practice: 2007. AJR Am J RoentgeonI 188:1447–1474
Brennan PC, McEntee M, Evanoff M, Phillips P, O'Connor WT, Manning DJ (2007) Ambient lighting: effect of illumination on soft-copy viewing of radiographs of the wrist. AJR Am J Roentgenol 188:W177–W180
Tokuda O, Harada Y, Shiraishi G et al (2012) MRI of the anatomical structures of the knee: the proton density-weighted fast spin-echo sequence vs the proton density-weighted fast-recovery fast spin-echo sequence. Br J Radiol 85:e686–e693
Sutter R, Hodek R, Fucentesen SF, Nittka M, Pfirrmann CW (2013) Total knee arthroplasty MRI featurig slice-encoding for metal artifact correction: reduction of artifacts for STIR and proton density-weighted sequences. AJR Am J Roentgeonl 201:1315–1324
Ristow O, Steinbach L, Sabo G et al (2009) Isotropic 3D fast spin-echo imaging versus standard 2D imaging at 3.0 T of the knee--image quality and diagnostic performance. Eur Radiol 19:1263–1272
Van Dyck P, Vanhevel F, Vanhoenacker FM et al (2015) Morphological MR imaging of the articular cartilage of the knee at 3 T-comparison of standard and novel 3D sequences. Insights Imag 6:285–293
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33:159–174
Vasey MW, Thayer JF (1987) The continuing problem of false positives in repeated measures ANOVA in psychophysiology: a multivariate solution. Psychophysiology 24:479–486
Moriya S, Miki Y, Kanagaki M, Matsuno Y, Miyati T (2014) 90 degrees -flip-angle three-dimensional double-echo steady-state (3D-DESS) magnetic resonance imaging of the knee: isovoxel cartilage imaging at 3T. Eur J Radiol 83:1429–1432
Rehnitz C, Kupfer J, Streich NA et al (2014) Comparison of biochemical cartilage imaging techniques at 3 T MRI. Osteoarthr Cartilage 22:1732–1742
Duc SR, Pfirrmann CW, Koch PP, Zanetti M, Hodler J (2008) Internal knee derangement assessed with 3-minute three-dimensional isovoxel true FISP MR sequence: preliminary study. Radiology 246:526–535
Duc SR, Pfirrmann CW, Schmid MR et al (2007) Articular cartilage defects detected with 3D water-excitation true FISP: prospective comparison with sequences commonly used for knee imaging. Radiology 245:216–223
Friedrich KM, Reiter G, Kaiser B et al (2011) High-resolution cartilage imaging of the knee at 3T: basic evaluation of modern isotropic 3D MR-sequences. Eur J Radiol 78:398–405
Thorlund JB, Juhl CB, Roos EM, Lohmander LS (2015) Arthroscopic surgery for degenerative knee: systematic review and meta-analysis of benefits and harms. Br J Sports Med 49:1229–1235
Cui Y, Dyvorne H, Besa C, Cooper N, Taouli B (2015) IVIM diffusion-weighted imaging of the liver at 3.0T: comparison with 1.5T. Eur J Radiol Open 2:123–128
Chen CA, Lu W, John CT et al (2009) Multiecho IDEAL gradient-echo water-fat separation for rapid assessment of cartilage volume at 1.5 T: initial experience. Radiology 252:561–567
Jung JY, Jee WH, Park MY, Lee SY, Kim JM (2012) Meniscal tear configurations: categorization with 3D isotropic turbo spin-echo MRI compared with conventional MRI at 3 T. AJR Am J Roentgenol 198:W173–W180
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Prof Louise Rainford.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Funding
The authors state that this work has not received any funding.
Statistics and biometry
One of the authors has significant statistical expertise and is employed as a statistician in a third level academic institution.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• prospective
• experimental
• performed at one institution
Rights and permissions
About this article
Cite this article
Abdulaal, O., Rainford, L., MacMahon, P. et al. 3T MRI of the knee with optimised isotropic 3D sequences: Accurate delineation of intra-articular pathology without prolonged acquisition times. Eur Radiol 27, 4563–4570 (2017). https://doi.org/10.1007/s00330-017-4816-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-017-4816-x