Abstract
Purpose
To compare a faster, new, high-resolution accelerated 3D-fast-spin-echo (3D-FSE) acquisition sequence (CS-SPACE) to traditional 2D and high-resolution 3D sequences for knee 3-T magnetic resonance imaging (MRI).
Materials and methods
Twenty patients received knee MRIs that included routine 2D (T1, PD ± FS, T2-FS; 0.5 × 0.5 × 3 mm3; ∼10 min), traditional 3D FSE (SPACE-PD-FS; 0.5 × 0.5 × 0.5 mm3; ∼7.5 min), and accelerated 3D-FSE prototype (CS-SPACE-PD-FS; 0.5 × 0.5 × 0.5 mm3; ∼5 min) acquisitions on a 3-T MRI system (Siemens MAGNETOM Skyra). Three musculoskeletal radiologists (MSKRs) prospectively and independently reviewed the studies with graded surveys comparing image and diagnostic quality. Tissue-specific signal-to-noise ratios (SNR) and contrast-to-noise ratios (CNR) were also compared.
Results
MSKR-perceived diagnostic quality of cartilage was significantly higher for CS-SPACE than for SPACE and 2D sequences (p < 0.001). Assessment of diagnostic quality of menisci and synovial fluid was higher for CS-SPACE than for SPACE (p < 0.001). CS-SPACE was not significantly different from SPACE but had lower assessments than 2D sequences for evaluation of bones, ligaments, muscles, and fat (p ≤ 0.004). 3D sequences had higher spatial resolution, but lower overall assessed contrast (p < 0.001). Overall image quality from CS-SPACE was assessed as higher than SPACE (p = 0.007), but lower than 2D sequences (p < 0.001). Compared to SPACE, CS-SPACE had higher fluid SNR and CNR against all other tissues (all p < 0.001).
Conclusions
The CS-SPACE prototype allows for faster isotropic acquisitions of knee MRIs over currently used protocols. High fluid-to-cartilage CNR and higher spatial resolution over routine 2D sequences may present a valuable role for CS-SPACE in the evaluation of cartilage and menisci.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
3D fast spin-echo (FSE) MRI offers several potential improvements upon the 2D MRI traditionally used in musculoskeletal imaging, including improved spatial resolution and the potential for multiplanar reconstructions (MPRs) [1–5]. However, incorporation of currently available 3D FSE sequences into practice has been limited by longer scan and review times and uncertain diagnostic benefit when compared to routine 2D acquisitions [6–8].
Standard 2D MRI protocols most often employ FSE acquisitions repeated in multiple planes with high in-plane resolution, but comparatively thick slices that predispose to partial volume averaging. Newer 3D FSE acquisitions allow for an isotropic resolution with equally high in-plane resolution and slice thickness, which reduces partial volume averaging when compared to 2D techniques and allows for multiplanar reformations (MPRs) in any imaging plane [4, 9, 10].
Although some studies have shown diagnostic efficacy of 3D protocols for common internal derangements of the knee [3, 5, 11–13], evidence has not clearly demonstrated that 3D FSE sequences can replace standard 2D MRI protocols for routine diagnostic knee MRI [6–8, 10]. However, the improved spatial resolution and MPRs provide a potential role for 3D FSE sequences, particularly for imaging of cartilage [2, 14–18]. As a result, institutions that currently incorporate 3D FSE sequences into the knee MRI protocol often do so in addition to the standard 2D MRI sequences [16].
The use of accelerated 3D acquisitions with MPR can decrease motion artifacts and increase scanner availability compared with non-accelerated 3D protocols with MPR and multi-planar 2D MRI protocols. Over the years, many ultrafast techniques have evolved to reduce acquisition times [19–23], including recent advanced techniques that under-sample the data and process it by removing resultant artifacts to reconstruct a full image [23]. Compressed sensing is a specific type of acceleration technique that undersamples k-space, followed by a nonlinear iterative reconstruction in order to preserve image quality and prevent aliasing artifacts [24], and has been successfully applied to multiple MRI modalities [25]. It also inspired various extensions including reconstruction techniques using multiple coil arrays and parallel imaging techniques [26–29].
In this study, we investigate a new, accelerated 3D FSE sequence prototype that applies a compressed sensing technique of incoherent k-space undersampling and a nonlinear SENSE-type (SENSitivity Encoding) reconstruction [29, 30], called CS-SPACE, to an existing commercially available 3D FSE sequence (SPACE) on a 3-T MRI system. The CS-SPACE sequence reduces acquisition time in the knee from about 7.5 min for the SPACE sequence to about 5 min with the same isotropic resolution. The goal of this study is to evaluate image quality and MSKR-assessed tissue-specific diagnostic value of knee MRI on a 3-T system with a CS-SPACE acquisition as compared to traditional SPACE and routine 2D sequences.
Methods
Study design
IRB approval was obtained for this HIPAA-compliant prospective study. Patients aged 18 to 89 scheduled to undergo knee MRI on our 3-T Siemens system were screened and consented consecutively. Patients undergoing contrast-enhanced studies were excluded. Patients with prior surgery were included in the eligible population.
Study population
Between February and August of 2014, 20 of 23 screened patients scheduled for knee MRI consented to participation, comprising the study group (eight males, mean age 45 years; 12 females, mean age 47 years). Six patients had known prior surgeries of the involved knee. Five patients went on to have knee arthroscopies at our institution before December 2014.
Imaging protocol
All MRI acquisitions were performed on a 3-T MRI scanner (MAGNETOM Skyra, Siemens, Erlangen Germany) with a dedicated 15-element transmit/receive knee coil (QED, Mayfield Village, OH, USA). Our institutional protocol for routine knee MRI on this system includes the following 2D turbo spin-echo (TSE) sequences: axial fat-suppressed (FS) T2-weighted sequence, coronal FS proton density, coronal non-fat-suppressed (NFS) T1-weighted, sagittal NFS proton density, and sagittal FS T2-weighted TSE (Table 1). In addition to our routine 2D knee sequence, a sagittal FS SPACE (“Sampling Perfection with Application-optimized Contrast using different flip angle Evolutions”) and an accelerated sagittal FS SPACE sequence called CS-SPACE utilizing incoherent undersampling and a nonlinear SENSE-type reconstruction [29, 30], were acquired (Table 1). Obtained sequences were approximate proton density (PD) acquisitions. Post-acquisition coronal and sagittal MPRs for these sequences were made using the scanner software. Radial reconstructions for selected cases were performed on original software designed in MatLab (MathWorks, Natick, MA, USA).
Imaging analysis
Three fellowship-trained musculoskeletal radiologists (MSKRs), with 8, 5, and 1 years of experience independently reviewed the 20 exams. The MSKRs were each asked to complete an assessment as they reviewed the exams, in which they evaluated aspects of perceived image quality (contrast, resolution, noise, artifact, and overall quality) as well as perceived ability to diagnose pathology in specific tissues (bone, cartilage, ligaments/tendons, menisci, synovial fluid, muscle, fat, and blood vessels) on a scale from 1 to 6 (1 = very poor: extremely limited diagnostic value, 2 = poor: substantially limited diagnostic value, 3 = pretty bad: somewhat limited diagnostic value, 4 = okay: acceptable for majority of diagnoses, 5 = pretty good: good for the majority of diagnoses, 6 = great: optimal diagnostic value). Specific artifacts and pathology were noted by reviewers in comments.
Separately, tissue-specific regions-of-interest (ROIs) were measured to obtain tissue specific signal intensities and signal-to-noise ratios (SNR) for air, bone, cartilage, menisci, ligaments, fat, muscle, and fluid. Routine ROIs on a sagittal sequence, respectively, were the anterior air at the level of the ACL insertion, medial femoral condyle in the first full slice from the intercondylar notch for bone, patellar cartilage (lateral femoral condylar cartilage was also measured for internal control and followed the same pattern as patellar cartilage), the body of the medial meniscus, the distal insertion of the PCL, Hoffa’s fat pad, the gastrocnemius muscle medial head, and the intercondylar notch fluid. ROIs were kept consistent between sequences in the same study, and involved areas without pathology. Measurements were also used to calculate tissue-specific contrast-to-noise ratio (CNR) for fluid against menisci, cartilage, ligament, bone, fat, muscle, and cartilage against bone. ROIs were obtained in consistent areas of anatomy and with consistent sizes between sequences. SNR was calculated by dividing the average signal value of ROI measured tissue signal (μtissue) by the standard deviation of the tissue ROI (σtissue). Standard deviation of the tissue was used instead of background as SNR calculated by the background standard deviation is artificially elevated due to relative clamping. Tissue-specific CNR was calculated by the following equation: |(μ tissue1 − μ tissue2)/√ (σ tissue1 2 + σ tissue2 2)|.
Vendor role
The associated vendor (Siemens, Erlangen, Germany) had no role in the IRB approval, study design, data acquisition, or analysis in this study. The involved MSKRs had no vested interest or conflict of interest. The vendors created the accelerated acquisition technique CS-SPACE, and installed it on one of the institutional scanners for this study. The accelerated sequence has not been used by the institution outside of the purposes of this study. The vendor chose the name of the sequence (CS-SPACE), and helped edit the paper only for accuracy regarding the description of the sequence.
Statistics
Confidence intervals were calculated for MSKR-assessed image and diagnostic quality based upon the 1 to 6 evaluation scale. MSKR evaluations of assessed image and diagnostic quality were compared using analysis of variance (ANOVA). Individual averages were compared using Student’s t test. Similar calculations were performed to compare differences in measured tissue-specific signal intensity, SNR, and CNR.
Results
Twenty knee MRI examinations were performed with CS-SPACE, SPACE, and routine 2D acquisitions. For perceived tissue-specific diagnostic quality as assessed by three MSKRs (Table 2), CS-SPACE had higher assessed diagnostic value for evaluation of cartilage than SPACE and 2D sequences (Fig. 1, p < 0.001). Assessment values of CS-SPACE were higher than SPACE and not statistically different from 2D sequences for evaluation of menisci (p < 0.001 and p = 0.347, respectively) and synovial fluid (p < 0.001 and p = 0.321, respectively). CS-SPACE was not assessed significantly different from SPACE and had lower assessment values than 2D sequences for evaluation of bones (p = 0.167 and p < 0.001, respectively), ligaments (p = 0.095 and p < 0.001 respectively), muscles (p = 0.901 and p < 0.001, respectively), and fat (p = 0.057 and p = 0.004, respectively). Both CS-SPACE and SPACE had significantly higher assessment values than 2D sequences for evaluation of blood vessels (p < 0.001).
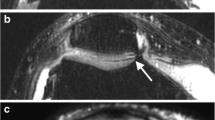
Full-thickness cartilage defect in the lateral trochlea (arrows) is best demonstrated on an accelerated FS CS-SPACE acquisition (a), which demonstrates higher fluid signal and higher fluid-to-cartilage contrast than FS SPACE (b) despite performing the acquisition 1.5 times faster. The higher fluid signal more closely resembles a 2D FS T2 sequence (c) while having increased out-of-plane resolution. Thicker slices of 3D sequences can be created (d) in any optimized plane that creates an image with less noise and higher contrast, more closely resembling 2D sequences
For assessed image quality (Table 3), the overall image quality from CS-SPACE had higher assessed values than SPACE (p = 0.007), but lower than traditional 2D sequences (p < 0.001). CS-SPACE and SPACE had higher assessed spatial resolution (p < 0.001), but lower assessed contrast (p < 0.001) when compared to 2D sequences. CS-SPACE had more assessed noise than SPACE (p = 0.004) and 2D sequences (p < 0.001).
For calculated tissue-specific signal characteristics (Tables 4 and 5), CS-SPACE had higher fluid signal intensity (p < 0.001) and SNR (p < 0.001) when compared to traditional SPACE. CNR of fluid against cartilage, menisci, ligaments, bone, fat, and muscle was resultantly higher for CS-SPACE when compared to SPACE (all p < 0.001). When compared to traditional SPACE, CS-SPACE had lower cartilage signal intensity (p < 0.001) and SNR (p < 0.001). This resulted in increased contrast between cartilage and fluid for CS-SPACE compared to SPACE, but decreased contrast between cartilage and bone (p < 0.001). There was no significant difference between CS-SPACE and SPACE for SNR in bone (p = 0.475) and fat (p = 0.074). There was decreased SNR in CS-SPACE sequences when compared to SPACE in menisci (p < 0.001), ligaments (p < 0.001), and muscles (p = 0.017).
CS-SPACE demonstrated no significant difference in fluid SNR when compared to T2 sequences (p = 0.475), but had significantly lower SNR than PD sequences (p = 0.019). There was no significant difference between CS-SPACE and 2D sequences in CNR of fluid against cartilage, ligaments, and bone. CS-SPACE demonstrated no significant difference in CNR of fluid against menisci, fat, and muscle when compared to T2 sequences. Proton density sequences had higher SNR in bone, ligaments, muscle, and fat when compared to CS-SPACE and SPACE (p < 0.001).
The high assessed contrast in CS-SPACE in addition to increased spatial resolution when compared to 2D sequences resulted in higher MSKR-perceived diagnostic value for cartilage pathology in the knee, including femoral cartilage (Fig. 1) and patellar cartilage (Fig. 2). There was similarly higher MSKR-perceived diagnostic value for CS-SPACE in the diagnosis of meniscal tears when compared to SPACE (Fig. 3). In specific instances, improved spatial resolution relative to 2D sequences also was perceived as valuable for the diagnosis of more subtle meniscal tears (Fig. 4). Notably, this study did not directly measure diagnostic accuracy.
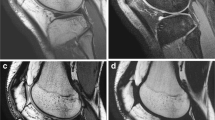
Focal patellar near-full-thickness cartilage defect (arrows) is well demonstrated on an accelerated FS CS-SPACE sequence (a), demonstrating high contrast between fluid, cartilage, and bone. Relatively lower fluid signal and higher cartilage signal on FS SPACE sequences (b) results in lower overall fluid-cartilage contrast. A 2D STIR sequence (c) in this patient with ACL reconstruction hardware demonstrates high cartilage-fluid contrast, but lower out-of-plane resolution
Medial meniscus posterior horn horizontal tear (arrows) is well demonstrated on an accelerated FS CS-SPACE sequence (a) related to high fluid-meniscus contrast. A FS SPACE sequence (b) by comparison demonstrates lower fluid signal and lower fluid-meniscus contrast, making the tear less conspicuous than other sequences, with the macerated portion of the meniscus being better defined on the FS CS-SPACE sequences (b). The higher contrast seen on FS CS-SPACE sequence more closely resembles those seen in the 2D acquisition (c)
Medial meniscus posterior horn inner margin tear (arrows) is volume-averaged and inconspicuous on 2D FS T2 sequences while scrolling through the tear on sagittal plane (a-1 through a-3), which cover a span of nearly 10 mm over three slices. Similar volume averaging occurs in the axial plane (a-4). Other 2D PD sequences not presented have a similar resolution and meniscal appearance as T2 sequences. 3D isotropic sequences demonstrate the inner margin tear more conspicuously as users scroll through images on the sagittal plane (c and d, images 1 through 3), which cover a span of 1.5 mm over three slices, or on the axial plane (b-4 and c-4)
While routine planes were reconstructed for the purposes of assessment on this evaluation, nonstandard planes can be constructed with post-acquisition processing, if desired. 3D isotropic sequences have been shown to improve diagnostic performance for evaluation of cartilage when including radial reformats for the evaluation of cartilage [18], and can be applied to the accelerated CS-SPACE sequence (Fig. 5). Similar radial reconstructions could be applied to the menisci for characterization of tear types as well as evaluation of meniscal extrusion (Fig. 6).
FS CS-SPACE radial multiplanar reformation centered on the medial femoral condyle of the same patient in Fig. 1 demonstrates a lateral trochlear full-thickness cartilage defect (large arrow) as well as a more subtle medial trochlear cartilage defect (small arrow)
Discussion
In this study, we demonstrate the successful clinical application of a compressed sensing acceleration technique to a traditional 3D FSE sequence in the knee. We compare the accelerated 3D FSE sequence CS-SPACE to a traditional 3D FSE (SPACE) and traditional 2D FSE sequences. Interestingly, MSKRs assessed the novel accelerated sequence, CS-SPACE, to have higher perceived image quality than SPACE and higher perceived diagnostic value for evaluation of knee cartilage when compared to SPACE and traditional 2D sequences.
The 1.5-fold acceleration of the high-resolution isotropic 3D FSE acquisition decreased scan time from nearly 7.5 min to 5 min (Table 1). This compares to a total of 10 min of scanning for the 2D multi-planar acquisitions routinely used at this institution. The CS-SPACE sequence results in identical resolution as the traditional SPACE, and can be reconstructed in any imaging plane, precluding the need for multiple planar acquisitions that increase scan time.
CS-SPACE unexpectedly demonstrated some favorable differences in signal characteristics when compared to the longer SPACE acquisition, particularly in fluid. Specifically, CS-SPACE demonstrated higher signal intensity in fluid, more closely following 2D T2 TSE signal characteristics and resulting in increasing fluid-cartilage and fluid-meniscus contrast. Importantly, some of the assessed differences in contrast may be attributable to specific protocol differences that may affect contrast not directly related to the acceleration. Such differences may in part relate to vendor optimization of the commercially available SPACE acquisitions. The prototype-accelerated technique, on the other hand, has not been fully optimized. For example, on review, the authors noted differences in CS-SPACE and SPACE for the applied k-space reordering technique; specifically, conventional SPACE used radial reordering and CS-SPACE used linear reordering. Some of these optimizations applied to SPACE may not be needed for the accelerated technique, particularly those related to further acquisition time reduction and motion artifact, such as radial k-space reordering.
Despite high assessed diagnostic quality performance of CS-SPACE in cartilage and menisci, 2D sequences demonstrated higher tissue SNR as well as improved assessed diagnostic quality in bone, ligaments, muscles, and fat than both SPACE and CS-SPACE. This high assessed diagnostic performance of 2D sequences in the ligaments and other tissues coincides with the high diagnostic performance of 2D MRI for these tissues established in the literature [3, 5, 6, 11]. This is not an unexpected finding, as there is typically a trade-off between SNR and spatial resolution, which is lower in 2D as compared to 3D sequences. This could be partially compensated for in isotropic 3D acquisitions by averaging thicker slices from the isotropic data. While increasing volume averaging, thicker slices would result in improved SNR and contrast and result in an image that more closely approximates the 2D image (Fig. 1). This optimized averaging can be created in any imaging plane desired on 3D acquisitions.
Limitations of this evaluation include the small sample size and indirect assessment of perceived diagnostic value without surgical correlation. The purpose of our work was to compare the imaging characteristics between the currently available sequences and a novel accelerated 3D sequence. A few of the patients did go to surgery, with the findings at surgery correlating with those on imaging review. However, larger studies would need to be performed that directly measure diagnostic accuracy with the gold standard of surgery in order to establish a true diagnostic benefit. Simultaneous assessment of 2D, SPACE, and CS-SPACE sequences by individual readers was done in order to allow readers to directly compare sequences, which could introduce bias in the interpretation, particularly as it applies to diagnostic assessment. We did not evaluate non-fat-suppressed 3D sequences in this study, which could potentially change diagnostic confidence in assessing fat, muscle, or bone/marrow. Additionally, the improved signal characteristics of the accelerated 3D sequence relative to the non-accelerated sequence was unexpected. Further investigations should be done to better understand the factors that may improve signal characteristics in the application of acceleration techniques, including differences in sequence optimizations when compared to standard 3D techniques. Although the compressed sensing acceleration technique improves scanning time, it does not affect post-processing time or high image counts inherent to 3D isotropic techniques, which can affect work flow in clinical practice.
In conclusion, this study demonstrates a feasible application of accelerated compressed sensing 3D scanning using incoherent undersampling and iterative reconstruction to the existing high-resolution 3D FSE SPACE sequence. In addition to comparable assessed diagnostic performance in several tissues, the accelerated CS-SPACE sequence unexpectedly demonstrated some favorable signal characteristics in fluid. High fluid-to-cartilage CNR and higher spatial resolution over routine 2D sequences may suggest a valuable role for accelerated 3D FSE sequences, such as CS-SPACE, in the evaluation of cartilage and menisci with potential for MPRs. Acceleration techniques can save valuable MRI scanning time while maintaining diagnostic value, particularly for 3D isotropic sequences that can be acquired in addition to 2D sequences in the knee for targeted roles.
References
Gold GE, Busse RF, Beehler C, et al. Isotropic MRI of the knee with 3D fast spin-echo extended echo-train acquisition (XETA): initial experience. AJR Am J Roentgenol. 2007;188(5):1287–93.
Gold GE, Fuller SE, Hargreaves BA, Stevens KJ, Beaulieu CF. Driven equilibrium magnetic resonance imaging of articular cartilage: initial clinical experience. J Magn Reson Imaging. 2005;21(4):476–81.
Kijowski R, Davis KW, Woods MA, et al. Knee joint: comprehensive assessment with 3D isotropic resolution fast spin-echo MR imaging—diagnostic performance compared with that of conventional MR imaging at 3.0 T. Radiology. 2009;252(2):486–95.
Yao L, Pitts JT, Thomasson D. Isotropic 3D fast spin-echo with proton-density-like contrast: a comprehensive approach to musculoskeletal MRI. AJR Am J Roentgenol. 2007;188(2):W199–201.
Ristow O, Steinbach L, Sabo G, et al. Isotropic 3D fast spin-echo imaging versus standard 2D imaging at 3.0 T of the knee—image quality and diagnostic performance. Eur Radiol. 2009;19(5):1263–72.
Subhas N, Kao A, Freire M, Polster JM, Obuchowski NA, Winalski CS. MRI of the knee ligaments and menisci: comparison of isotropic-resolution 3D and conventional 2D fast spin-echo sequences at 3 T. AJR Am J Roentgenol. 2011;197(2):442–50.
Kijowski R, Davis KW, Blankenbaker DG, Woods MA, Del Rio AM, De Smet AA. Evaluation of the menisci of the knee joint using three-dimensional isotropic resolution fast spin-echo imaging: diagnostic performance in 250 patients with surgical correlation. Skeletal Radiol. 2011.
Van Dyck P, Gielen JL, Vanhoenacker FM, et al. Diagnostic performance of 3D SPACE for comprehensive knee joint assessment at 3 T. Insights Imaging. 2012;3(6):603–10.
Kijowski R, Gold GE. Routine 3D magnetic resonance imaging of joints. J Magn Reson Imaging. 2011;33(4):758–71.
Naraghi A, White LM. Three-dimensional MRI of the musculoskeletal system. AJR Am J Roentgenol. 2012;199(3):W283–93.
Jung JY, Yoon YC, Kwon JW, Ahn JH, Choe BK. Diagnosis of internal derangement of the knee at 3.0-T MR imaging: 3D isotropic intermediate-weighted versus 2D sequences. Radiology. 2009;253(3):780–7.
Jung JY, Yoon YC, Kim HR, Choe BK, Wang JH, Jung JY. Knee derangements: comparison of isotropic 3D fast spin-echo, isotropic 3D balanced fast field-echo, and conventional 2D fast spin-echo MR imaging. Radiology. 2013;268(3):802–13.
Lefevre N, Naouri JF, Bohu Y, Klouche S, Herman S. Partial tears of the anterior cruciate ligament: diagnostic performance of isotropic three-dimensional fast spin-echo (3D-FSE-cube) MRI. Eur J Orthop Surg Traumatol. 2014;24(1):85–91.
Al saleh H, Hernandez L, Lee KS, Rosas HG, Block WF, Kijowski R. Rapid isotropic resolution cartilage assessment using radial alternating repetition time balanced steady-state free-precession imaging. J Magn Reson Imaging. 2014;40(4):796–803.
Chen CA, Kijowski R, Shapiro LM, et al. Cartilage morphology at 3.0T: assessment of three-dimensional magnetic resonance imaging techniques. J Magn Reson Imaging. 2010;32(1):173–83.
Kijowski R, Blankenbaker DG, Woods M, Del Rio AM, De Smet AA, Reeder SB. Clinical usefulness of adding 3D cartilage imaging sequences to a routine knee MR protocol. AJR Am J Roentgenol. 2011;196(1):159–67.
Welsch GH, Zak L, Mamisch TC, et al. Advanced morphological 3D magnetic resonance observation of cartilage repair tissue (MOCART) scoring using a new isotropic 3D proton-density, turbo spin-echo sequence with variable flip angle distribution (PD-SPACE) compared to an isotropic 3D steady-state free precession sequence (true-FISP) and standard 2D sequences. J Magn Reson Imaging. 2011;33(1):180–8.
Gustas CN, Blankenbaker DG, Rio AM, Winalski CS, Kijowski R. Evaluation of the articular cartilage of the knee joint using an isotropic resolution 3D fast spin-echo sequence with conventional and radial reformatted images. AJR Am J Roentgenol. 2015;205(2):371–9.
Tsao J. Ultrafast imaging: principles, pitfalls, solutions, and applications. J Magn Reson Imaging. 2010;32(2):252–66.
Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magn Reson Med. 1997;38(4):591–603.
Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952–62.
Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. 2002;47(6):1202–10.
Tsao J, Kozerke S. MRI temporal acceleration techniques. J Magn Reson Imaging. 2012;36(3):543–60.
Donoho DL. Compressed sensing. IEEE Trans Inf Theory. 2006;52(4):1289–306.
Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–95.
Hamilton LH, Fabregat JA, Moratal D, et al. “PINOT”: time-resolved parallel magnetic resonance imaging with a reduced dynamic field of view. Magn Reson Med. 2011;65(4):1062–74.
Jung H, Sung K, Nayak KS, Kim EY, Ye JC. K-t FOCUSS: a general compressed sensing framework for high resolution dynamic MRI. Magn Reson Med. 2009;61(1):103–16.
Otazo R, Kim D, Axel L, Sodickson DK. Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med. 2010;64(3):767–76.
Stalder AF, Schmidt M, Quick HH, et al. Highly undersampled contrast-enhanced MRA with iterative reconstruction: integration in a clinical setting. Magn Reson Med. 2014.
Li G, Zaitsev M, Buchert M, et al. Improving the robustness of 3D turbo spin-echo imaging to involuntary motion. MAGMA. 2015;28(4):329–45.
Acknowledgments
We would like to acknowledge Bruce Spottiswoode, PhD, and Ke Cheng Liu, PhD, from Siemens for their role in applying the CS-SPACE sequence and setting up the sequence on the institutional MRI scanner. We would also like to acknowledge Dipl-Inf Christoph Forman from Siemens Healthcare GmbH and Jens Wetzl, MSc from the Department of Computer Science of Friedrich-Alexander-University Erlangen-Nuremburg, Germany for their important role in developing the CS-SPACE reconstruction framework.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This research study was not sponsored financially by any institution. Dr. Raithel is employed by Siemens Healthcare GmbH (Erlangen, Germany). Her contribution to the paper was limited to development of the accelerated sequence and elucidation of the technical factors and descriptive language of the MRI sequences. The study design, data acquisition, data analysis, and interpretation were performed entirely by the remaining authors, who declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Altahawi, F.F., Blount, K.J., Morley, N.P. et al. Comparing an accelerated 3D fast spin-echo sequence (CS-SPACE) for knee 3-T magnetic resonance imaging with traditional 3D fast spin-echo (SPACE) and routine 2D sequences. Skeletal Radiol 46, 7–15 (2017). https://doi.org/10.1007/s00256-016-2490-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00256-016-2490-8