Abstract
Objectives
To evaluate the automated two-point Dixon screening sequence for the detection and estimated quantification of hepatic iron and fat compared with standard sequences as a reference.
Methods
One hundred and two patients with suspected diffuse liver disease were included in this prospective study. The following MRI protocol was used: 3D-T1-weighted opposed- and in-phase gradient echo with two-point Dixon reconstruction and dual-ratio signal discrimination algorithm (“screening” sequence); fat-saturated, multi-gradient-echo sequence with 12 echoes; gradient-echo T1 FLASH opposed- and in-phase. Bland–Altman plots were generated and correlation coefficients were calculated to compare the sequences.
Results
The screening sequence diagnosed fat in 33, iron in 35 and a combination of both in 4 patients. Correlation between R2* values of the screening sequence and the standard relaxometry was excellent (r = 0.988). A slightly lower correlation (r = 0.978) was found between the fat fraction of the screening sequence and the standard sequence. Bland–Altman revealed systematically lower R2* values obtained from the screening sequence and higher fat fraction values obtained with the standard sequence with a rather high variability in agreement.
Conclusions
The screening sequence is a promising method with fast diagnosis of the predominant liver disease. It is capable of estimating the amount of hepatic fat and iron comparable to standard methods.
Key points
• MRI plays a major role in the clarification of diffuse liver disease.
• The screening sequence was introduced for the assessment of diffuse liver disease.
• It is a fast and automated algorithm for the evaluation of hepatic iron and fat.
• It is capable of estimating the amount of hepatic fat and iron.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Diffuse liver diseases such as non-alcoholic fatty liver disease (NAFLD) and chronic liver iron overload have become a global burden and major problem in daily clinical routine. NAFLD is the most common cause of chronic liver disease in Western countries and a known risk factor for the development of the metabolic syndrome [1, 2]. Hepatic iron overload (HIO) can exist as a separate disease in hereditary haemochromatosis or hereditary anaemia, or may occur as secondary iron deposition in many diffuse liver diseases, especially in patients with NAFLD [3–5]. HIO has the potential to accelerate liver damage which can lead to cirrhosis and hepatocellular carcinoma [6].
Magnetic resonance imaging (MRI) is a widely available noninvasive tool for the detection of lipids and paramagnetic metal ions, and therefore plays a major role in the clarification of diffuse liver disease. Various methods like chemical shift imaging, susceptibility weighted imaging or spectroscopy are applied to detect and quantify parenchymal changes. Liver iron is mainly evaluated by the signal intensity ratio technique and by R2 and R2* relaxometry techniques [7]. Fatty liver disease can be evaluated by standard in- and opposed-phase imaging, magnetic resonance spectroscopy, conventional fat-suppressed imaging or complex chemical shift-based water–fat separation [8]. Most of the MRI techniques that are used for the evaluation of liver fat and iron require post-processing with special software. Recently, a promising method based on a routine 3D opposed- and in-phase T1-weighted gradient-echo acquisition with two-point Dixon reconstruction has been described [9–11]. This algorithm, which is performed during image reconstruction, automatically calculates signal intensity ratios from four sets of images: in-phase, opposed-phase, fat-only and water-only. Furthermore a large volume of interest within the liver is selected from the two-point Dixon data by using an automated sampling algorithm [12] and the liver is automatically classified into four different categories (normal, fat, iron or combined disease) together with an additional estimation of the R2* value (related to iron content) and/or fat (in % fat fraction) for categories other than “normal”. The main idea behind this two-point Dixon technique thereby is to have a fast liver classification method which can be used to quickly screen for hepatic iron overload or fatty liver disease. Hence, in the following this method is referred to as the “screening” sequence or algorithm.
The aim of this prospective study was to evaluate the automated two-point Dixon screening method for the detection of hepatic iron and fat, and the estimated quantification of the R2* and fat. Therefore the screening method was compared with standard R2* relaxometry regarding iron and T2*- and T1-corrected chemical shift-based sequences regarding fat estimation.
Materials and methods
Patients
This prospective study was approved by our institutional review board (Innsbruck Medical University). Written informed consent was obtained from each patient. The authors who were employees of Siemens Healthcare had no control of all data for the duration of the study.
The study included 109 patients that were referred to our department for the evaluation of diffuse liver disease between May 2013 and April 2014. All patients had suspected HIO or diffuse liver disease on the basis of clinical criteria or because of elevated serum ferritin (>300 μg/L in male patients and >200 μg/L in female patients), transferrin saturation (>45 % in male patients and >50 % in female patients) or liver enzymes. General contraindications to MRI were used as exclusion criteria. No patients were excluded from our study for these reasons.
MR imaging
MRI was performed on a 1.5-Tesla system (Magnetom Avanto, Siemens, Germany) in the supine position using an anterior six-element body matrix coil and 12 elements of the inbuilt spine matrix coil. The liver MRI protocol included the following sequences: transverse 3D T1-weighted opposed- and in-phase spoiled gradient echo (TR = 7.5 ms, TE = 2.38/4.76 ms, flip angle 10°, slice thickness 4 mm, typical image size 320 × 164 pixels, 56 images from a data matrix of 320 × 164 × 40 samples, FOV 380 × 309 mm, undersampling with CAIPIRINHA (“Controlled Aliasing in Parallel Imaging Results in Higher Acceleration”) [13], acquisition time 22 s) with two-point Dixon reconstruction and dual-ratio signal discrimination algorithm (work in progress package 718B, Siemens Healthcare) [12]; transverse 2D fat-saturated spoiled multi-gradient echo (12 echoes, TR = 200 ms, TE = 0.99 ms + n × 1.41 ms, flip angle 20°, matrix 128 × 128 pixels, FOV 360 × 360 mm, slice thickness 10 mm, acquisition time 17 s) in five different slice positions; 2D transverse T1-weighted opposed- and in-phase spoiled gradient echo (TR = 103 ms, TE = 2.37/5.05 ms, flip angle 70°, matrix 256 × 192 pixels, FOV 300 × 400 mm, slice thickness 5 mm, multi breath hold acquisition time 9 s/slice). In the following the multi-gradient-echo sequence and the opposed- and in-phase sequence will be called “standard sequences”.
MR data analysis
One radiologist (BH, with more than 5 years of experience in liver imaging) reviewed the obtained images. For all patients the classification (normal, fat, iron or combined) automatically provided by the screening sequence was recorded. According to this classification patients were divided into four groups. For patients with a pathologic classification, the dual-ratio discrimination prototype also provides estimated values of R2* and fat fraction of a full liver volume sampled by an automatic algorithm [12]. In short, the magnitude images of both echoes were used to calculate an R2* map using log-linear fitting. The fat–water separation algorithm was described elsewhere [14]. Briefly, the phase map due to hardware imperfection such as gradient delay, eddy currents and concomitant field and accumulated from B0 heterogeneity during the time between the two echoes was calculated using a region-growing algorithm [14], and removed from the original data; then the resultant fat and water quantities were computed. In patients classified as normal, no estimated values are offered by the screening sequence prototype.
The data of the standard multi-gradient-echo sequence and the standard in- and opposed-phase sequence were analysed using ImageJ (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA) by a single reader who was unaware of the clinical results. R2* maps were calculated for the multi-gradient-echo sequence by pixel-wise fitting with a monoexponential decay function using a custom-written ImageJ plugin. Proton density fat fraction (FF) was calculated by correcting the in-phase and opposed-phase signals for T2* decay using a global mean T2* value as obtained from the above T2* maps and by correcting for T1 bias [15, 16]. The reader placed three circular regions of interest (ROIs) in the liver parenchyma of one transversal section (two in the right lobe and one in the left lobe) in corresponding locations of the T2* (R2*) and FF maps. ROIs had a diameter of 10–13 mm (area 0.8–1.3 cm2). Focal liver lesions or artefacts and biliary or vascular structures were avoided. The mean R2* and FF values within all ROIs were recorded. An average fat fraction of more than 5.6 % was determined to be abnormal and indicate hepatic steatosis [17]. For the multi-gradient-echo sequence, R2* was assessed as pathologic according to the reference values of the literature with a threshold of more than 70 s−1 for abnormal iron deposition [18–20].
Statistical analysis
Analysis of the data was performed using Excel software (Microsoft Corporation, Redmond, Washington, USA). Mean values together with standard deviations were used to describe the obtained variables. Quantitative results of the sequences were compared by means of Bland–Altman plots as well as correlation and regression analysis. The agreement of classification into “normal” or “pathologic” for either iron overload or steatosis between standard and screening sequences was assessed by calculating the portion of overall agreement (OA), specific agreement indices “percent of positive agreement” (PA), “percent of negative agreement” (PN) as described by Cicchetti and Feinstein and the Cohen’s kappa coefficient, which was calculated following Watkins et al. [21, 22]. For ROC analysis the pROC package for the R Project for Statistical Computing (R Development Core Team (2006), Vienna, Austria, http://www.R-project.org, version 2.13.1) was used [23].
Results
All MRI examinations were performed successfully. Seven patients were excluded because of unsuccessful liver sampling of the screening Dixon sequence, where no estimated values were calculated. A total of 102 patients (72 males and 30 females; mean age 47.9; age range 10–102) were included in the study.
Results of the screening sequence
The screening sequence labelled 33 patients as category fat, 35 patients as iron and 4 patients as combined. Furthermore 30 patients were classified as normal. Results of the used sequences are summarized in Tables 1 and 2. Examples of patient images are shown in Figs. 1 and 2.
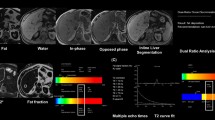
A 38-year-old male patient with suspicion of hepatic iron overload due to genetic C282Y homozygosity. Standard chemical shift sequence shows an increase of signal from in-phase (a) to opposed-phase (b). The screening sequence (c) results in the classification “iron” and estimates an R2* of 127.9 s−1. With standard multi-echo relaxometry (d) R2* was 175.9 s−1
A 66-year-old male patient with suspicion of hepatic iron overload due to pathologic laboratory tests. Standard chemical shift sequences show a decrease of signal from in-phase (a) to opposed-phase (b). The screening sequence (c) results in the classification “fat” and estimates a fat fraction of 15.9 %. With standard multi-echo relaxometry (d) R2* was 68.13 s−1 which confirms the absence of pathologic iron overload. The T2*- and T1-corrected fat fraction was calculated as 14.01 %
Comparison between screening sequence classification and screening sequence value estimates
For 2/33 patients who were classified by the screening sequence as category “fat” the estimated fat percentage given by the screening sequence was below 5.6 % (4.9 % and 5.1 %). For 11/35 patients who were classified as “iron” the estimated R2* values were below 70 s−1 (mean 45.4 s−1; range 29.5–60.1 s−1) and for patients classified as “combined disease” 4/4 estimated R2* values were below 70 s−1 (mean 14.3 s−1; range 5.8–15.6 s−1) and 3/4 FF estimates were below 5.6 % (mean 5.2 %; range 5.0–5.4 %)
Comparison of screening sequence classification and standard sequences
In patients classified as normal by the screening sequence we found 8/30 patients with relatively high R2* values (mean 78.9 s−1; range 70.1–111.1 s−1) when using the standard relaxometry method and 3/30 patients with an FF above 5.6 % (mean 6.42; range 6.19–6.83 %).
The group classified with iron overload by the screening sequence revealed 6/35 patients which had combined disease by the reference method (FF above 5.6 %, mean 9.5 %; range 7.0–17.1 %) and 2/35 patients with relatively low R2* (mean 61.9 s−1; range 58.9–64.9 s−1). In the fat-classified group 1/33 patients showed an FF below 5.6 % (5.5 %), all other patients showed an FF above 5.6 % (mean 17.3 %; range 5.7–31.0 %) but 7/33 patients had combined disease by the reference method (R2* in excess of 70 s−1, mean 80.0 s−1; range 71.1–100.8 s−1).
In the group classified as “combined disease” only 1/4 patients was also classified as “combined disease” by the reference method (R2* = 99.8 s−1, FF = 7.5 %), 2/4 were classified by the reference method as “normal” (R2* = 65.8 s−1 and 32.4 s−1; FF = 4.5 % and 2.8 %) and 1/4 as “iron” only (R2* = 71 s−1, FF = 4.3 %)
By individual disease type, using R2* from the multi-echo sequence as a reference 15/102 patients were therefore classified false negative, 4/102 as false positive, 48/102 as true negative and 35/102 as true positive (OA 81.37 %, 95 % CI 57.8–100 %; PA 78.65 %, 95 % CI 55.6–100 %; PN 83.48 %, 95 % CI 59.5–100 %; kappa 0.502). With the FF from the standard sequence as a reference 9/102 were false negative, 4/102 were false positive, 56/102 were true negative and 33/102 were true positive (OA 87.3 %, 95 % CI 62.4–100 %; PA 83.5 %, 95 % CI 59.5–100 %; PN 89.6 %, 95 % CI 64.3–100 %; kappa 0.59).
Using ROC analysis (Fig. 3) the optimal FF cut-off between screening Dixon classification “fat” and our reference method was found to be 7.18 %, with a specificity of 100 % (95 % CI 1.0–1.0) and sensitivity of 86.5 % (95 % CI 75.68–97.3). The optimal R2* cut-off value between the multi-echo sequence and screening DIXON classification “iron” was found to be 75.485 s−1, with a specificity of 90 % (95 % CI 76.67–100.0) and sensitivity of 84.62 % (95 % CI 71.79–94.87). With these optimal cut-off values between screening Dixon classification and our reference methods the above numbers for R2* change to 10/102 false negative classifications, 6/102 false positive, 53/102 true negative and 33/102 true positive (OA 84.31 %, 95 % CI 60.1–100 %; PA 80.49 %, 95 % CI 57.1–100 %; PN 86.89 %, 95 % CI 62.2–100 %; kappa 0.51). For FF the values change to 5/102 false negative classifications, 4/102 false positive, 60/102 true negative and 33/102 true positive (OA 91.18 %, 95 % CI 65.6–100 %; PA 88.0 %, 95 % CI 63.0–100 %; PN 93.0 %, 95 % CI 67.0–100 %; kappa 0.672).
ROC curves to determine the optimal R2* (left) and FF (right) cut-off between screening Dixon classification “iron” or “fat” and the standard chemical shift sequence or multi-echo sequence, respectively. The values given beside the filled circles represent the optimal cut-off values and in brackets the specificity and sensitivity, respectively. The obtained cut-off values are somewhat higher than the values taken from the literature
Correlation of the used sequences
The correlation coefficient between R2* values of the screening sequence and the standard multi-echo relaxometry was 0.988. The slope of the corresponding linear regression line was 1.064 with an intercept of −48.19 (Fig. 4). Thereby the slope was found to be not significantly different from 1 (p = 0.9673), whereas the intercept was significantly different from 0 (p < 0.001). The correlation coefficient for the FF between the screening sequence and the standard chemical shift sequence was 0.978. The slope of the corresponding linear regression line was 1.1 with an intercept of −2.4 (Fig. 5). Thereby the slope was found to be not significantly different from 1 (p = 0.3886), whereas the intercept was significantly different from 0 (p < 0.001).
The Bland–Altman plot for the comparison of the two R2* estimation methods shows systematic lower R2* values (mean difference 36.13 s−1; standard deviation of difference 22.62 s−1) obtained from the screening sequence as compared to the multi-echo relaxometry with the difference between both methods tending to be lower for very high R2* values (Fig. 6).
The Bland–Altman plot for the comparison of the two fat estimation methods shows slightly higher FF values for the used 2D opposed/in-phase sequence (mean difference 0.724 %; standard deviation of difference 1.78 %) (Fig. 7).
Discussion
In this prospective study we assessed a recently introduced MRI screening sequence regarding the evaluation of hepatic steatosis and siderosis [9, 12]. Therefore we compared this method with standard MRI methods (R2* relaxometry and T2*-corrected chemical shift imaging) with respect to diagnosis and quantitative accuracy.
The main idea behind the screening sequence is to provide a quick assessment of the liver regarding the presence of diffuse liver disease. This information should enable the radiologist or technologist to decide if a further evaluation of the liver with additional, more accurate sequences is necessary. The evaluation of the data of the screening sequence is performed automatically at the end of the measurement and the results are immediately available to the radiological staff. Whenever the screening protocol delivers a pathologic classification an additional multi-echo or spectroscopic acquisition for more accurate evaluation can be performed subsequently. These techniques provide a more accurate quantitative assessment than the dual-echo imaging of the screening sequence [15, 24–27]. Nevertheless our results show an excellent correlation (r = 0.988) concerning the estimation of R2* between the screening sequence and the multi-echo sequence in patients classified as “iron” by the screening sequence. Simultaneously an excellent correlation (r = 0.978) was found between the FF estimates of the screening sequence and the standard sequence. In 7/109 (6 %) patients liver sampling failed; in each case this was due to a failure of the automated liver segmentation algorithm used for the screening sequence in patients with very high hepatic iron content. It has to be pointed out that the used sequence has a “work in progress” status and the obtained results will in the future lead to improvements of the underlying algorithms.
In several patients we observed a discrepancy between the classification of the screening sequence and the estimated R2* or FF values from the screening sequence. This has to do with the fact that classification of the screening sequence is not based on the estimated R2* and FF values but instead on a dual-ratio algorithm [12] and the value estimates are calculated separately.
Concerning the detection of pathologic increased hepatic iron our results show promising results for the screening method. With respect to the presence of iron, only 4/102 patients were classified as false positive and 15/102 as false negative when compared to the reference method. It again has to be noted that the classification done by the screening sequence is not based on a direct quantification of R2* and FF values but instead is based on the evaluation of in- and opposed-phase as well as fat-only/water-only ratios and therefore does not rely on R2* or FF thresholds [12]. Therefore the observed false negative patients strongly depend on the chosen threshold for pathologic iron overload in the reference method (R2* was between 70.10 s−1 and 111.1 s−1 in the 15 false negative patients) and the internal dual-ratio discrimination used by the screening sequence. It is important to recognize that the accepted “normal” values for R2* and their clinical implications are still open. The “normal” liver iron concentration is between 10 μmol/g (0.558 mg/g) and 36 μmol/g (2.010 mg/g) of dry weight, high iron overload is usually considered to be in excess of 80 μmol/g (4.467 mg/g) of dry weight [28]. On the basis of calibration curves given by Hankins et al. and Wood et al. [18, 29] in our study therefore an R2* threshold of 70 s−1 was considered as pathologic which would have resulted in 15 false negative patients when using the screening algorithm only. Using ROC analysis only a slightly increased optimal cut-off of 75.485 s−1 was obtained which leads to less false negative patients (10/102) whereas at the same time a slight increase of false positive results (6/102) was observed and in the end no significant improvement of the agreement between our reference method and the screening method was obtained. Actually there is no full consensus about the ideal cut-off value, especially when dealing with clinical relevance and treatment decisions. Further larger studies and collaboration with clinicians are necessary especially when dealing with hepatic iron and the question of when R2* is identified as pathologic.
In our study the Bland–Altman plot shows systematic lower R2* values obtained from the screening sequence. This can be explained by the different sequence parameters, where the screening sequence is using a dual-echo method to estimate R2* and our multi-echo sequence is based on fitting of 12 echoes. Despite the observed systematic differences in R2* values the obtained Cohen’s kappa coefficients indicate a moderate agreement between both methods with regard to classification of patients as “normal” or “pathologic”.
We found 9/102 false negative results for the detection of fatty liver disease under the assumption that the standard T2*- and T1-corrected chemical shift sequence is an acceptable reference. Only 3 patients with the screening classification “normal” showed an FF in the standard sequence above our threshold with an FF range of 6.19–6.83 % (mean 6.42 %). In addition 7 patients classified by the screening sequence as “iron” had an FF above our threshold with values of up to 17.1 % which should have been classified as “combined disease”. Using the threshold found by ROC analysis (7.18 %) the number of false negative patients decreased to 5 without change of false positive results leading to a clear improvement of agreement between our reference method and the screening method. Although for fatty liver disease the used cut-off values are also open for discussion, our results indicate that the algorithm used for liver classification in the “work in progress” screening sequence eventually incorporates a too high cut-off value between normal and fatty liver and might have to be adapted in future work.
The proton density fat fraction is widely used in the literature and previous studies that used chemical shift-based methods showed excellent agreement with spectroscopy for quantification of hepatic fat [24, 25, 30–33]. It is well known that the T2* decay and T1 effects can affect fat quantification, especially when evaluating the liver for NAFLD where concomitant iron deposition may occur. Therefore it is essential to correct for all known confounding factors [8]. In our study T2* correction was used on the chemical shift sequence together with T1 correction to compensate for the used high flip angle which otherwise would probably lead to systematically higher FF values due to T1 bias.
Many MRI-based techniques for the evaluation of hepatic fat or iron rely on manual selection of region of interest or volume of interest. In particular, the multi-echo approach where ROIs are selected in a post-processing procedure is time consuming because of the manual placement in numerous sites throughout the liver. In our study we compared both types of measurements; the ROI-based method was used for the conventional sequences and a whole liver algorithm was used in the screening sequence. McCarville et al. compared small ROI placement and a whole liver method in 41 patients with iron overload [34]. They found a strong correlation between liver iron content and R2* measurement by small ROI and whole liver ROI method, although they found slightly greater interobserver variability when using the small ROI technique. Other study groups also used a global whole liver method with promising measurements and lower interobserver variability [35]. Currently there is no full consensus on the best approach for the type of measurement.
The study by Bashir et al. found that the screening sequence was accurate in determining the presence of deposition disease (93.1 %) [9]. Their work did not focus on patients with suspicion of diffuse liver disease, the population was mixed and only 8 patients had iron deposition. They found a positive predictive value of 80 % for the screening algorithm indicating that it may result in some false positive results, but no patient with proven fat or iron deposition was misdiagnosed as unaffected. In our study for the detection of iron we had only 4/102 and for the detection of fat only 1/102 false positive patients. Nevertheless we had 15/102 false negative results for iron and 9/102 false negative results for fat which very likely depends on the chosen thresholds of 70 s−1 for the standard R2* relaxometry and 5.6 % for standard T2*- and T1-corrected fat estimation.
We could find no study assessing the screening algorithm in patients where diffuse liver disease was suspected. Our patient collective was selected in order to provide a large number of cases of diffuse liver disease on the basis of clinical criteria. This clarifies our high number of pathologic results in the screening algorithm with 33 patients having steatosis, 35 patients with iron overload and 4 patients with combined disease.
This study has limitations that must be addressed. For the evaluation of hepatic fat fractions from our opposed- and in-phase gradient echo sequence only a single-peak fat assumption was used which limits the accuracy of our fat estimates. More accurate fat quantification techniques would use multi-echo sequences together with multi-peak fat spectral modelling, taking the spectral complexity of fat into account [8, 33]. We used a global T2* value for correcting in- and opposed-phase acquisition which could lead to errors in the FF calculation if T2* is strongly heterogeneous in the liver. However, such strong heterogeneity was rarely seen in our data.
A further limitation of our study is that none of our patients had liver biopsy for confirmation. This aspect is may be only of minor importance as it is known that biopsy is prone to sampling errors. Additionally we did not use MR spectroscopy, which is widely accepted as a gold standard in evaluating liver fat but is still generally available only at academic centres [8].
In conclusion the screening sequence is a promising method in a number of ways. First it can provide a fast diagnosis of the predominant pathologic liver deposition. Second the algorithm is capable of estimating the amount of hepatic fat or iron comparable to standard, widely available methods.
Abbreviations
- FF:
-
fat fraction
- HIO:
-
hepatic iron overload
- NAFLD:
-
non-alcoholic fatty liver disease
- MRI:
-
magnetic resonance imaging
References
Harrison SA, Neuschwander-Tetri BA (2004) Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Clin Liver Dis 8:861–879, ix
Clark JM, Diehl AM (2003) Defining nonalcoholic fatty liver disease: implications for epidemiologic studies. Gastroenterology 124:248–250
Moirand R, Mortaji AM, Loreal O, Paillard F, Brissot P, Deugnier Y (1997) A new syndrome of liver iron overload with normal transferrin saturation. Lancet 349:95–97
Bonkovsky HL, Jawaid Q, Tortorelli K et al (1999) Non-alcoholic steatohepatitis and iron: increased prevalence of mutations of the HFE gene in non-alcoholic steatohepatitis. J Hepatol 31:421–429
Henninger B, Kremser C, Rauch S et al (2013) Evaluation of liver fat in the presence of iron with MRI using T2* correction: a clinical approach. Eur Radiol 23:1643–1649
Kew MC (2009) Hepatic iron overload and hepatocellular carcinoma. Cancer Lett 286:38–43
Hernando D, Levin YS, Sirlin CB, Reeder SB (2014) Quantification of liver iron with MRI: state of the art and remaining challenges. J Magn Reson Imaging 40:1003–1021
Reeder SB, Sirlin CB (2010) Quantification of liver fat with magnetic resonance imaging. Magn Reson Imaging Clin N Am 18:337–357, ix
Bashir MR, Zhong X, Dale BM, Gupta RT, Boll DT, Merkle EM (2013) Automated patient-tailored screening of the liver for diffuse steatosis and iron overload using MRI. AJR Am J Roentgenol 201:583–588
Bashir MR, Merkle EM, Smith AD, Boll DT (2012) Hepatic MR imaging for in vivo differentiation of steatosis, iron deposition and combined storage disorder: single-ratio in/opposed phase analysis vs. dual-ratio Dixon discrimination. Eur J Radiol 81:e101–e109
Boll DT, Marin D, Redmon GM, Zink SI, Merkle EM (2010) Pilot study assessing differentiation of steatosis hepatis, hepatic iron overload, and combined disease using two-point dixon MRI at 3 T: in vitro and in vivo results of a 2D decomposition technique. AJR Am J Roentgenol 194:964–971
Bashir MR, Dale BM, Merkle EM, Boll DT (2012) Automated liver sampling using a gradient dual-echo Dixon-based technique. Magn Reson Med 67:1469–1477
Breuer FA, Blaimer M, Heidemann RM, Mueller MF, Griswold MA, Jakob PM (2005) Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi-slice imaging. Magn Reson Med 53:684–691
Jellus V (2010) Phase correction method. US20100201364 A1
Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB (2007) Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med 58:354–364
Yang IY, Cui Y, Wiens CN, Wade TP, Friesen-Waldner LJ, McKenzie CA (2014) Fat fraction bias correction using T1 estimates and flip angle mapping. J Magn Reson Imaging 39:217–223
Szczepaniak LS, Babcock EE, Schick F et al (1999) Measurement of intracellular triglyceride stores by H spectroscopy: validation in vivo. Am J Physiol 276:E977–E989
Hankins JS, McCarville MB, Loeffler RB et al (2009) R2* magnetic resonance imaging of the liver in patients with iron overload. Blood 113:4853–4855
St Pierre TG, Clark PR, Chua-anusorn W et al (2005) Noninvasive measurement and imaging of liver iron concentrations using proton magnetic resonance. Blood 105:855–861
Henninger B, Kremser C, Rauch S et al (2012) Evaluation of MR imaging with T1 and T2* mapping for the determination of hepatic iron overload. Eur Radiol 22:2478–2486
Cicchetti DV, Feinstein AR (1990) High agreement but low kappa: II. Resolving the paradoxes. J Clin Epidemiol 43:551–558
Watkins MW, Pacheco M (2000) Interobserver agreement in behavioral research: importance and calculation. J Behav Educ 10:205–2012
Robin X, Turck N, Hainard A et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma 12:77
Meisamy S, Hines CD, Hamilton G et al (2011) Quantification of hepatic steatosis with T1-independent, T2-corrected MR imaging with spectral modeling of fat: blinded comparison with MR spectroscopy. Radiology 258:767–775
Yokoo T, Shiehmorteza M, Hamilton G et al (2011) Estimation of hepatic proton-density fat fraction by using MR imaging at 3.0 T. Radiology 258:749–759
Bydder M, Yokoo T, Hamilton G et al (2008) Relaxation effects in the quantification of fat using gradient echo imaging. Magn Reson Imaging 26:347–359
Kuhn JP, Evert M, Friedrich N et al (2011) Noninvasive quantification of hepatic fat content using three-echo Dixon magnetic resonance imaging with correction for T2* relaxation effects. Invest Radiol 46:783–789
Deugnier Y, Turlin B (2007) Pathology of hepatic iron overload. World J Gastroenterol 13:4755–4760
Wood JC, Enriquez C, Ghugre N et al (2005) MRI R2 and R2* mapping accurately estimates hepatic iron concentration in transfusion-dependent thalassemia and sickle cell disease patients. Blood 106:1460–1465
Yu H, Shimakawa A, Hines CD et al (2011) Combination of complex-based and magnitude-based multiecho water-fat separation for accurate quantification of fat-fraction. Magn Reson Med 66:199–206
Hines CD, Yu H, Shimakawa A, McKenzie CA, Brittain JH, Reeder SB (2009) T1 independent, T2* corrected MRI with accurate spectral modeling for quantification of fat: validation in a fat-water-SPIO phantom. J Magn Reson Imaging 30:1215–1222
Kuhn JP, Hernando D, Munoz del Rio A et al (2012) Effect of multipeak spectral modeling of fat for liver iron and fat quantification: correlation of biopsy with MR imaging results. Radiology 265:133–142
Zhong X, Nickel MD, Kannengiesser SA, Dale BM, Kiefer B, Bashir MR (2013) Liver fat quantification using a multi-step adaptive fitting approach with multi-echo GRE imaging. Magn Reson Med 72:1353–1365
McCarville MB, Hillenbrand CM, Loeffler RB et al (2010) Comparison of whole liver and small region-of-interest measurements of MRI liver R2* in children with iron overload. Pediatr Radiol 40:1360–1367
Positano V, Salani B, Pepe A et al (2009) Improved T2* assessment in liver iron overload by magnetic resonance imaging. Magn Reson Imaging 27:188–197
Acknowledgments
The scientific guarantor of this publication is Dr. Benjamin Henninger. The authors of this manuscript declare relationships with the following companies: S. Kannengiesser, X. Zhong and G. Reiter are employees of Siemens Healthcare. They had no control of all data for the duration of the study. All the other authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article. One of the authors has statistical experience (C. Kremser). Nevertheless no complex statistical methods were necessary for this paper. Institutional review board approval was obtained. Written informed consent was obtained from all subjects (patients) in this study. Methodology: prospective, diagnostic study, performed at one institution.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Henninger, B., Zoller, H., Rauch, S. et al. Automated two-point dixon screening for the evaluation of hepatic steatosis and siderosis: comparison with R2*-relaxometry and chemical shift-based sequences. Eur Radiol 25, 1356–1365 (2015). https://doi.org/10.1007/s00330-014-3528-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-014-3528-8