Abstract
Key message
A switchgrass vascular tissue-specific promoter (PvPfn2) and its 5′-end serial deletions drive high levels of vascular bundle transgene expression in transgenic rice.
Abstract
Constitutive promoters are widely used for crop genetic engineering, which can result in multiple off-target effects, including suboptimal growth and epigenetic gene silencing. These problems can be potentially avoided using tissue-specific promoters for targeted transgene expression. One particularly urgent need for targeted cell wall modification in bioenergy crops, such as switchgrass (Panicum virgatum L.), is the development of vasculature-active promoters to express cell wall-affective genes only in the specific tissues, i.e., xylem and phloem. From a switchgrass expression atlas we identified promoter sequence upstream of a vasculature-specific switchgrass profilin gene (PvPfn2), especially in roots, nodes and inflorescences. When the putative full-length (1715 bp) and 5′-end serial deletions of the PvPfn2 promoter (shortest was 413 bp) were used to drive the GUS reporter expression in stably transformed rice (Oryza sativa L.), strong vasculature-specificity was observed in various tissues including leaves, leaf sheaths, stems, and flowers. The promoters were active in both phloem and xylem. It is interesting to note that the promoter was active in many more tissues in the heterologous rice system than in switchgrass. Surprisingly, all four 5′-end promoter deletions, including the shortest fragment, had the same expression patterns as the full-length promoter and with no attenuation in GUS expression in rice. These results indicated that the PvPfn2 promoter variants are new tools to direct transgene expression specifically to vascular tissues in monocots. Of special interest is the very compact version of the promoter, which could be of use for vasculature-specific genetic engineering in monocots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cell walls of lignocellulosic feedstock crops such as switchgrass (Panicum virgatum L.) have been altered for increased efficiency for yield of biofuel. An important practical constraint when changing cell walls is to avoid off-target effects. The absolute required bioenergy crop traits are high biomass, efficient use of water and nutrients, resilience to abiotic and biotic stresses, adaptability to marginal lands, and compatibility with modern agricultural systems: all traits that make switchgrass an attractive bioenergy crop in the first place (Keshwani and Cheng 2009). A few biotechnological studies have focused on increasing switchgrass biomass, resulting in modest improvements (Do et al. 2016; Poovaiah et al. 2015; Wuddineh et al. 2015a). Using overexpression or RNA interference (RNAi), most reverse genetics experiments on switchgrass have targeted cell wall components with the goal of reducing cell wall recalcitrance and increasing biofuel production (Baxter et al. 2015; Shen et al. 2012; Wuddineh et al. 2015b, 2016).
However, there has been little variety of promoters used for controlling transgene expression in switchgrass. The most commonly used promoters in switchgrass transformation are constitutive promoters. These include cauliflower mosaic virus (CaMV) 35S promoter (Mazarei et al. 2008), maize (Zea mays) ubiquitin 1 gene (Ubi-1) promoter (Fu et al. 2012), rice (Oryza sativa L.) polyubiquitin gene (rubi2 and rubi3) promoter (Li and Qu 2011), switchgrass polyubiquitin gene (PvUbi1 and PvUbi2) promoters (Mann et al. 2011), and rice actin 1 (OsAct1) promoter (Ogawa et al. 2014). These promoters originated from either plant viruses or plant housekeeping genes, and drive constant gene expression in all tissues, at all times, and under all conditions. Nonetheless, continuous high-level of overexpression of some transgenes in all tissues may not be necessary and could cause undesirable side effects. For example, it could cause homology-dependent gene silencing that results in silencing of either the transgene or its endogenous homolog or both (Xi et al. 2009). It could also generate unintended phenotypes including altered transgenic plant growth and development (Jeon et al. 2000), compromised seed germination and yield (Robert et al. 2013), and abnormal morphology (Kosugi and Ohashi 2003). Moreover, overexpression of stress- or defense-related genes could result in unexpected activation of defense genes in the absence of pathogens (Xiao et al. 2001) or even fitness costs (Zavala et al. 2004).
Unlike constitutive promoters, tissue-specific promoters drive transgene expression specifically in target tissues, and sometimes at high levels (Benfey and Chua 1989). They can also be used in multigene transformation experiments to maximize the transgenic effects by reducing the likelihood of transcriptional silencing (Peremarti et al. 2010). Thus, identification and characterization of switchgrass tissue-specific promoters are needed for switchgrass transgenic modification. So far, vascular bundle-specific promoters CCR from Eucalyptus globulus, RSs1 from rice, and rolC from Agrobacterium rhizogenes had been used for engineering plants to down-regulate lignin content in switchgrass (Torre et al. 2014) and to improve resistance to pests in rice, tobacco (Nicotiana tabacum L.), and chickpea (Cicer arietinum L.) (Saha et al. 2007), respectively. The Arabidopsis profilin 2 (AtPfn2) promoter has also been used to drive GUS expression specifically in the vascular bundles of most tissues/organs of plants, such as roots, leaves, sepals, petals, stamen filaments, and stalks of developing seeds (Ramachandran et al. 2000). Profilin is a small actin-, polyproline- and inositol phospholipid-binding protein (Sohn et al. 1995; Tanaka and Shibata 1985) and ubiquitously expressed in various eukaryotic cells (Christensen et al. 1996; Huang et al. 1996; Kovar et al. 2000; Ramachandran et al. 2000; Staiger et al. 1993). It is involved in cell elongation, cell shape maintenance, polarized root hair growth, and flowering time (Christensen et al. 1996).
In this study, we identified a vascular bundle-specific profilin promoter from the switchgrass genome and expression atlas database. The prolifin expression profiles were further characterized in switchgrass tissues using RT-PCR and real-time RT-PCR. Stably transgenic rice was selected as a model heterologous system to assess the performance of the 1715 bp switchgrass promoter and four 5′-end deletion constructs using a GUS reporter gene. Our data led us to conclude that the promoters, especially the 413-bp truncated promoter offer valuable xylem- and phloem-specific expression in various monocot organs, which should facilitate more precise genetic engineering of cereal crops. The biological function of the long span (> 1200 bp) upstream sequence from the compact 413-bp promoter remains enigmatic.
Materials and methods
Sequence analysis and in silico expression profiles
The deduced amino acid sequences of the switchgrass PvPfn2 genes were obtained from the switchgrass genomic DNA database (V1.0) on the Phytozome website (http://www.phytozome.net) using BlastP; the Arabidopsis thaliana vascular bundle-specific profilin (AtPfn2; accession number: U43326; Ramachandran et al. 2000) was the query sequence. The amino acid sequences of these switchgrass genes were aligned together with AtPfn2 using ClustalX 2.0 (Fig. 1).
Comparison of the deduced amino acid sequences of the switchgrass PvPfn2 and AtPfn2. The homologous sequences of AtPfn2 in the switchgrass genome were obtained by using the amino acid sequence of AtPfn2 as the query sequence to Blast search against the Phytozome switchgrass genomic DNA sequence database (http://www.phytozome.net/search.php). The numbers indicate the positions of the amino acid residues
The genomic DNA sequences, cDNA (including 5′ and 3′ UTR) sequences, and promoter sequences (the sequences upstream the translational start sites) of these switchgrass homologs were downloaded from Phytozome. In order to search for their unique transcript IDs, the switchgrass cDNA sequences were used to query the Noble Foundation Switchgrass Gene Atlas (Zhang et al. 2013) PviUT V1.2 (http://switchgrassgenomics.noble.org). The returned switchgrass gene expression profiles were then downloaded from the atlas. In addition, the promoter sequences were used as the query sequences to search the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (Lescot et al. 2002) and PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) (Higo et al. 1999) databases for known cis-regulatory elements (or motifs) within the promoter regions.
Plant materials and growth conditions
Switchgrass cv. ‘Alamo’ was used for tissue-specific RNA and genomic DNA extraction, while japonica rice (Oryza sativa L.) cv. ‘Taipei 309’ (‘TP309’) was used for stable rice transformation. Potted plants were grown in a 40% sand and 60% 3B media mix (Sun Gro Horticulture, Agawam, MA, USA). Transgenic rice plants were grown in a growth chamber at 27 °C under fluorescent white light and 12 h photoperiods for 2 weeks. Then, 1-month-old plants were grown in a greenhouse at 25–29 °C under 12-h photoperiods in 50% average relative humidity. Plants were fertilized once per week with Peters® Professional All Purpose Plant Food (Spectrum Group, St. Louis, MO, USA).
RT-PCR and real-time RT-PCR
Total RNA was extracted from root, leaf blade, internode, node, and inflorescences of switchgrass (cv. ‘Alamo’) at R1 growth stage (Moore et al. 1991) using Tri-Reagent (Molecular Research Center, Cincinnati, OH, USA). RNA quality was analyzed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). Two micrograms of RNA from each tissue was treated with amplification-grade DNase I (Invitrogen™, Carlsbad, CA, USA) in order to remove the genomic DNA and then used for reverse transcription with High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s protocol.
The relative transcript abundance of the endogenous PvPfn2 gene among switchgrass tissues was estimated by reverse transcription polymerase chain reaction (RT-PCR) and real-time quantitative reverse transcription PCR (real-time RT-PCR) using sequence-specific primers (Table 1). The primer specificity was confirmed by agarose gel electrophoresis. A switchgrass actin gene (PvActC) was used as the internal control (Mann et al. 2011) for both RT-PCR and real-time RT-PCR. For RT-PCR, a 100-bp band was amplified by each primer pair at 95 °C for 5 min, followed by PCR cycles at 95 °C for 30 s, 55 °C for 20 s, and 72 °C for 20 s, then extended 2 min at 72 °C.
The real-time RT-PCR was conducted using the Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The standard curve method was used for relative transcript quantification normalized by PvActC transcript abundance as described (Mann et al. 2011). The quality cutoff of the real-time RT-PCR efficiency was set at > 95% with R2 > 0.99 for the standard curve.
Switchgrass genomic DNA extraction and vector construction
Genomic DNA was extracted from the leaf blade of switchgrass (cv. Alamo) at R1 growth stage using a CTAB method (Stewart and Vie 1993). The genomic DNA was used as templates for PCR amplification of the putative full-length promoter regions of PvPfn2 using sequence-specific primers (Table 1). After gel purification, the PCR product was then cloned into the pCR™8/GW/TOPO® (Invitrogen™, Carlsbad, CA, USA) vector following the manufacturer’s directions. After sequence confirmation, the promoter region was subcloned into the destination vector pMDC162 to drive uidA (GUS) expression using the Gateway® LR Clonase™ II enzyme (Invitrogen™, Carlsbad, CA, USA). The resultant promoter::GUS fusion constructs were confirmed by sequencing.
Based on the distribution patterns of the known motifs within the promoter region of PvPfn2, serial 5′-end deletions of the promoter was PCR amplified and cloned into the pCR™8/GW/TOPO® vector. After sequence confirmation, the serial deletions were subcloned into the binary vector pMDC162. The resultant truncated promoter::GUS fusion constructs were confirmed by sequencing. The full-length CaMV 35S promoter was cloned into pMDC162, which was used as the known-expression (positive) control.
Rice tissue culture and transformation
Each expression vector was electroporated into Agrobacterium tumefaciens strain GV3850 under the selection with 50 mg/L rifampicin and 50 mg/L kanamycin. Mature seeds of rice cv. ‘TP309’ were used for embryogenic callus induction and stable rice transformation as described (Hiei et al. 1994).
Histochemical GUS analysis
Histochemical GUS assays were conducted in T0 transgenic rice according to Jefferson et al. (1987) with modifications. The root, leaf blade, and sheath of each individual plant were sampled into 1.5 mL Eppendorf® microtubes 1 month after regeneration. All the plant materials were stained in 50 mM sodium phosphate at pH 7.0 containing 10 mM Na2-EDTA, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, 1 mg/mL X-Gluc (Amresco, Solon, OH, USA), 100 µg/mL chloramphenicol, 0.1% (V/V) Triton X-100 and 20% (V/V) methanol. The microtubes were placed at 37 °C for 24 h in dark. All the samples were cleared by replacing the GUS solution with 70% ethanol until optimal visualization and stored in 70% ethanol for imaging. The samples were sectioned to 10 µm thickness for light microscopy. The intensity of GUS activity was visualized with a stereomicroscope (Fisher Scientific™ Stereomaster™ Track Pole, Pittsburgh, PA, USA), and a digital camera (Infinity X-32, Lumenera Corporation, Ottawa, ON, USA).
Cross-sections of the histochemical GUS assays were conducted in the leaves of T1 transgenic rice as described in Wu et al. (2012) with modifications. The samples were cleared in 70, 50 and 30% ethanol, and then mounted in water before observation with a light microscope.
Results and discussion
Sequence analysis
The BlastP search for the AtPfn2 orthologues in the switchgrass genome returned 8 candidate sequences with high amino acid sequence similarity (Fig. 1). Pavirv00014318m, Pavirv00019052m, Pavirv00038285m, and Pavirv00038913m had the highest sequence identity to AtPfn2 (Supplementary Table S1). The deduced amino acid sequences of Pavirv00014318m and Pavirv00019052m were identical except that the latter contained an additional 45 amino acids at the 5′-end (Fig. 1). By comparing the cDNA sequences with the genomic DNA sequences, we found these four candidate genes shared the same exon/intron structures as AtPfn2 (Fig. 2a). Each candidate gene consisted of 131 amino acids in length, three exons and two introns at conserved positions (Fig. 2a). The existence of the four candidate sequences indicates the tetraploidy nature of the switchgrass genome (Triplett et al. 2012).
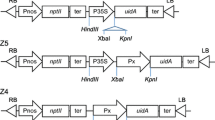
Deduced gene structure models for Pfn2 genes and the PvPfn2 promoter::GUS fusion constructs used in stable rice transformation experiments. a The gene structure of AtPfn2 and its switchgrass orthologs with the highest sequence identity. Boxes and lines represent exons and introns, respectively. Intron length (bp; base pairs) are indicated above each intron, whereas exon length (aa; amino acids) are indicated under each exon. The additional 45 amino acids on the 5′-end (N terminus) of Pavirv00019052m are not shown. b The full-length and serial deletions of the PvPfn2 promoter were cloned into the pMDC162 to drive GUS expression. Lines represent promoters with promoter length (bp) indicated above each promoter variant. NosT, Nos terminator
Expression profiles of PvPfn2 in switchgrass
The Pavirv00014318m and Pavirv00019052m sequences were most similar to the Noble Foundation Gene Atlas entry AP13ISTG56127 with sequence identities being 99 and 95%, respectively, while Pavirv00038285m and Pavirv00038913m had the highest (100 and 99%) sequence identity to AP13ISTG71490 and AP13CTG11882, respectively (Supplementary Table S2). In silico expression analysis showed an extremely low expression level of AP13ISTG71490 in all the tissues, while the expression of AP13CTG11882 was evident in all of the tissues with no pattern of tissue-specificity (Supplementary Fig. S1). However, we found that the expression level of AP13ISTG56127 was highest in internodes, followed by inflorescences, nodes, crowns, and roots. Its lowest expression was found in leaf blades, leaf sheaths and inflorescences of the glume and floret development stages (Supplementary Fig. S1).
RT-PCR and real-time RT-PCR primers for Pavirv00019052m all failed. However, the RT-PCR results revealed that Pavirv00014318m was highly expressed in switchgrass roots and nodes, weakly expressed in inflorescences and internodes, and undetectable in leaves (Fig. 3a). Its expression was comparable to the PvActC expression level in roots, but much weaker than PvActC in nodes. Similarly, the real-time RT-PCR analysis demonstrated that Pavirv00014318m expression was highest in roots, followed by nodes, inflorescences, internode, and leaf (Fig. 3b). Its expression in roots was about 62% of the PvActC expression level, while about 11–13% of PvActC expression in nodes and inflorescences and 1–2% of PvActC in leaves and internodes. As a result, Pavirv00014318m was selected as the potential switchgrass homolog of AtPfn2 and named as PvPfn2 hereafter, whose promoter and derivatives were used for functional analysis in this study.
Endogenous PvPfn2 mRNA expression levels in different tissues of wild-type switchgrass measured by RT-PCR and real-time RT-PCR. RNA was extracted from switchgrass cv. ‘Alamo’ at R1 growth stage. A switchgrass actin gene PvActC was used as the internal control for both RT-PCR and real-time RT-PCR, and the real-time RT-PCR data of PvPfn2 was normalized to the quantity of PvActC. The cDNA template was replaced with the same amount of ddH2O in the negative control. The relative quantification was performed using the standard curve method, and the bar represents the mean of three independent replicates with the standard errors of the noted mean
PvPfn2 consists of a 77-bp 5′ untranslated region (UTR), two introns (118 and 90 bp in length, respectively), three exons (123, 138 and 135 bp in length, respectively), and a 376-bp 3′ UTR, which was very similar to the gene structure of AtPfn2 (Sohn et al. 1995). A 1715-bp-long promoter region (including a 1638-bp promoter sequence and a 77-bp 5′ UTR) of PvPfn2 were obtained from the Phytozome database. The PlantCARE and PLACE databases were searched for putative known cis-regulatory elements (or motifs) within the promoter region of PvPfn2. Several cis-regulatory elements with putative or known functionality were identified (Supplementary Table S3). However, these motifs have been only deduced from in silico analysis and have yet to be functionally validated.
GUS expression driven by the full-length PvPfn2 promoter in stable transgenic rice
When the 1715-bp promoter region of PvPfn2 (i.e., PvPfn2p; Supplementary Fig. S2) fused with GUS, the transgenic rice callus had the same survival frequency and appearance on hygromycin selection as did the 35S::GUS transgenic callus. Likewise, there were no apparent phenotypic differences between transgenic T0 PvPfn2p::GUS and 35S::GUS rice lines, even though both types of lines set fewer seeds than the non-transformed rice plants.
Histochemical GUS assays of young (1-month post-regeneration) T0 plants resulted in expected phenotypes of negative controls (Fig. 4a) as well as positive 35S::GUS controls, of which the latter exhibited constitutive GUS expression in both leaf blades and roots (Fig. 4b). Three representative lines (i.e., #5, 9 and 20) of stable transgenic rice containing PvPfn2p::GUS had GUS expression in leaf veins and vascular bundles (Fig. 4c–e). GUS expression was low or marginally detectable in roots. This observation was different from the RT-PCR and real-time RT-PCR analyses which detected higher PvPfn2 expression in roots and nodes than in leaves (Fig. 3). Possibly, the 1715-bp promoter region might only perform partial function of the real full-length promoter, and the sequences upstream the 1715-bp promoter region was likely associated with the enhanced expression in roots and nodes and the suppressed expression in leaves. Comparison of the cross-sections of the leaves of non-transgenic rice and T1 transgenic rice at the seed setting stage revealed that GUS expression was detected in both xylem and phloem in transgenic rice containing the full-length PvPfn2 promoter (Fig. 5). These expression patterns were similar to the AtPfn2p::GUS expression in transgenic Arabidopsis, where GUS was expressed in the vascular bundles of cotyledons and leaves (Christensen et al. 1996). However, the AtPfn2 promoter activities were also detected in the vascular bundles of roots and root hairs (Christensen et al. 1996), which was consistent with the PvPfn2 gene expression in switchgrass roots detected by RT-PCR and real-time RT-PCR analyses (Fig. 3).
Histochemical GUS analysis of leaf blade (A1–E1) and root (A2–E2) of T0 stable transgenic rice containing the full-length PvPfn2 promoter at the seedling stage. Non-transformed (N.T.) rice and stable transgenic rice containing 35S::GUS were used as the negative and positive control, respectively. The intensity of GUS expression was visualized with a stereomicroscope and a digital camera. A Non-transformed rice, B 35S promoter, C line 5 of the full-length PvPfn2 promoter, D line 19 of the PvPfn2 promoter, E line 20 of the PvPfn2 promoter. Each scale bar represents 0.5 mm for leaf blade or 0.1 mm for root
Cross-sections of histochemical GUS expression in the leaves of T1 stable transgenic rice containing the full-length or the shortest truncation of the PvPfn2 promoter at seed setting stage. Non-transformed (N.T.) rice was used as the negative control. Line 5 of the full-length PvPfn2 promoter and line 7 of the shortest truncation of PvPfn2 promoter (i.e., PvPfn2-4p) were used as representatives. The intensity of GUS staining was visualized with a stereomicroscope and a digital camera
Different tissues (stamens, lemma and palea, nodes, internodes, leaf blades, sheath, seeds, and roots) were also collected from the same three T0 lines at the seed setting stage for GUS expression analysis. Similarly, the non-transformed rice showed no GUS expression (Fig. 6a) while stable transgenic rice containing 35S::GUS exhibited an ectopic GUS expression in all the above tissues (Fig. 6b). However, GUS expression was mainly detected in the vascular bundles of stamen filament, lemma and palea, node, internode, leaf blade, sheath, seed husk, and stalk of developing seed of transgenic rice containing PvPfn2p::GUS (Fig. 6c–e). Compared to the GUS expression levels at 1 month after regeneration from callus, the PvPfn2p::GUS transgenic rice during seed setting was stronger than that of the younger tissues (Fig. 6c–e).
Histochemical GUS expression in different tissues of T0 stable transgenic rice containing the full-length PvPfn2 promoter at seed setting stage. Non-transformed rice and stable transformed rice containing CaMV 35S::GUS were used as the negative and positive control, respectively. The intensity of GUS staining was visualized with a stereomicroscope and a digital camera. A Non-transformed rice, B 35S promoter, C line 5 of the full-length PvPfn2 promoter, D line 19 of the full-length PvPfn2 promoter, E line 20 of the full-length PvPfn2 promoter. 1, 20 days after flowering (DAF) seeds; 2 and 3, 10 DAF seeds; 4, stamen; 5, lemma and palea; 6, cross section of node; 7, cross section of internode; 8, internode; 9, leaf blade; 10, sheath; 11, root elongation zone; 12, root cap. Each scale bar represents 1 mm for seeds (2; 3), stamen (4), lemma and palea (5), 0.5 mm for node (6), internode (7; 8), leaf blade (9) and sheath (10) or 0.1 mm for root (11; 12)
We know of several vascular bundle-specific promoters that have been functionally characterized in plants. The Arabidopsis trehalose-6-phosphate synthase (AtTPS1) promoter was associated with vascular bundles, but at a low level of expression (Bae et al. 2009). The Arabidopsis sucrose transporter (AtSUC2) promoter was used to drive GUS expression in the phloem of Arabidopsis (Truernit and Sauer 1995), while its protein was found to be synthesized in the plasma membrane of companion cells (Stadler and Sauer 1996). The activities of the rice RPP23 (Fukuda et al. 2004), the pumpkin PP1 (Clark et al. 1997) and the AtPP2-A1 (Zhang et al. 2011) promoters were detected in vascular tissues at different levels during different developmental stages. In comparison to these known promoters, the PvPfn2 promoter appears to be highly vascular bundle-specific with a strong and developmentally robust promoter strength and homogeneity, which would make it useful for cereal transformation for vasculature-specific traits.
GUS expression driven by the serial deletions of the PvPfn2 promoter in stable transgenic rice
Based on the distribution patterns of the known motifs within the promoter region of PvPfn2, 5′-end serial deletions were produced to assess the functionality of various portions of PvPfn2p, including the core promoter region. Promoter fragments of 1313, 958, 712 and 413 bp in length, respectively, were obtained (Fig. 2b). These four serial deletions were named PvPfn2-1p, -2p, -3p, and -4p, respectively (Fig. 2b) and their activities were assessed in both T0 and T1 transgenic rice. In each of the T0 transgenic rice plants assayed, each promoter deletion conveyed strong GUS expression in the veins of leaves (Fig. 7). In each of the T1 transgenic rice plants assayed, even the shortest truncation of the PvPfn2 promoter (i.e., PvPfn2-4p) directed comparable GUS expression in both xylem and phloem of leaves (Fig. 5), implying it contained motifs dominating expression in the vascular tissues of leaves. These results demonstrated that the four promoter deletions could drive GUS expression in a vascular bundle-specific manner in stable transgenic rice. The minimal promoter region tested (i.e., 413 bp in length) appears to be a desirably compact and useful sequence to control vascular bundle-specific transgene expression in monocots. It would be interesting to perform additional 5′-deleted regions to better understand motif functionality within a limited context of sequence.
Histochemical GUS analysis in the leaf blades of stable transgenic rice with GUS being driven by different serial deletions of the PvPfn2 promoter. All samples were taken from T0 plants 1 month after regeneration form callus. The intensity of GUS staining was visualized with a stereomicroscope and a digital camera. A Non-transformed rice, B CaMV 35S promoter, C1–C3 three lines of PvPfn2-1p, D1–D3 three lines of PvPfn2-2p, E1–E3 three lines of PvPfn2-3p, F1–F3 three lines of PvPfn2-4p
The present study showed that the PvPfn2 promoter and the serial deletion promoters restrict specific transcription in xylem and phloem in a number of rice organs. The promoter and biological function should be useful to develop synthetic promoters since vasculature-specific promoters could be utilized for genetic engineering agronomically important crops for trait improvement (Liu and Stewart 2016; Liu et al. 2013). For example, the specific reintroduction of xylan biosynthesis in the xylem vessels of the Arabidopsis xylan-deficient mutants by driving the expression of the same xylan biosynthesis genes (i.e., glycosyltransferases) with a vessel-specific promoter could result in the restoration of the undesirable mutant phenotype (dwarf, decreased stem strength) with desirable low xylan and lignin phenotype (Peterson et al. 2012). The use of vasculature-specific promoter has also been shown to be a more efficient strategy in genetic engineering of lignin profiles than the use of the constitutive 35S promoter (Meyer et al. 1998). Moreover, vasculature (especially phloem)-specific expression of insecticidal or virus resistance proteins/RNAs had been demonstrated to be highly effective in the generation of genetically superior lines with enhanced resistance against hemipteran insects which are phloem sap-sucking pests or some viruses which replicate exclusively in or are dependent on their interactions with phloem cells (Saha et al. 2007). Vasculature-specific promoters could also be used for transgenic expression of antibacterial genes for enhanced resistance against phloem-associated Gram-negative bacteria such as the notorious citrus greening disease (Huanglongbing disease or HLB).
Author contribution statement
WX participated in the design, performed the experiments and drafted the manuscript. WL, RY and MM participated in the design, data analysis and manuscript revisions. DH assisted with the cross section analysis. XZ and CNS conceived and coordinated the study, revised the manuscript, and obtained the funding. All authors read and approved the final version.
References
Bae H, Sicher R, Natarajan S, Bailey B (2009) In situ expression of trehalose synthesizing genes, TPS1 and TPPB, in Arabidopsis thaliana using the GUS reporter gene. Plant Cell Tiss Organ Cult 98:311–319
Baxter HL, Poovaiah CR, Yee KL, Mazarei M, Rodriguez M Jr, Thompson OA, Shen H, Turner GB, Decker SR, Sykes RW, Chen F, Davis MF, Mielenz JR, Davison BH, Dixon RA, Stewart CN Jr (2015) Field evaluation of transgenic switchgrass plants overexpressing PvMYB4 for reduced biomass recalcitrance. Bioenerg Res 8:910–921
Benfey P, Chua N-H (1989) Regulated genes in transgenic plants. Science 244:174–181
Christensen HEM, Ramachandran S, Tan C, Surana U, Dong C, Chua N-H (1996) Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profiling. Plant J 10:269–279
Clark AM, Jacobsen KR, Bostwick DE, Dannenhoffer JM, Skaggs MI, Thompson GA (1997) Molecular characterization of a phloem-specific gene encoding the filament protein, Phloem Protein 1 (PP1), from Cucurbita maxima. Plant J 12:49–61
Do PT, Tar JRD, Lee H, Folta MK, Zhang ZJ (2016) Expression of ZmGA20ox cDNA alters plant morphology and increases biomass production of switchgrass (Panicum virgatum L.). Plant Biotechnol J 14:1532–1540
Fu C, Sunkar R, Zhou C, Shen H, Zhang JY, Matts J, Wolf J, Mann DGJ, Stewart CN Jr, Tang Y, Wang Z-Y (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10:443–452
Fukuda A, Okada Y, Suzui N, Fujiwara T, Yoneyama T, HayashiH (2004) Cloning of the phloem-specific small heat-shock protein from leaves of rice plants. Soil Sci Plant Nutr 50:1255–1262
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Huang S, McDowell JM, Weise MJ, Meagher RB (1996) The Arabidopsis profilin gene family: evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol 111:115–126
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jeon J-S, Lee S, Jung K-H, Yang W-S, Yi G-H, Oh B-G, An G (2000) Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol Breed 6:581–592
Keshwani DR, Cheng JJ (2009) Switchgrass for bioethanol and other value-added application: a review. Bioresource Technol 100:1515–1523
Kosugi S, Ohashi Y (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132:2012–2022
Kovar DR, Drøbak BK, Staiger CJ (2000) Maize profilin isoforms are functionally distinct. Plant Cell 12:583–598
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Li R, Qu R (2011) High throughput Agrobacterium-mediated switchgrass transformation. Biomass Bioenerg 35:1046–1054
Liu W, Stewart CN Jr (2016) Plant synthetic promoters and transcription factors. Curr Opin Biotechnol 37:36–44
Liu W, Yuan JS, Stewart CN Jr (2013) Advanced genetic tools for plant biotechnology. Nat Rev Genet 14:781–793
Mann DGJ, King ZR, Liu W, Joyce BL, Percifield RJ, Hawkins JS, LaFayette PR, Artelt BJ, Burris JN, Mazarei M, Bennetzen JL, Parrott WA, Stewart CN Jr (2011) Switchgrass (Panicum virgatum L.) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnol 11:74
Mazarei M, Al-Ahmad H, Rudis MR, Stewart CN Jr (2008) Protoplast isolation and transient gene expression in switchgrass (Panicum virgatum L.). Biotechnol J 3:354–359
Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C (1998) Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA 95:6619–6623
Moore KJ, Moser LE, Vogel KP, Waller SS, Johnson BE, Pedersen JF (1991) Describing and quantifying growth stages of perennial forage grasses. Agron J 83:1073–1077
Ogawa Y, Shirakawa M, Koumoto Y, Honda M, Asami Y, Kondo Y, Hara-Nishimura Y (2014) A simple and reliable multi-gene transformation method for switchgrass. Plant Cell Rep 33:1161–1172
Peremarti A, Twyman RM, Gomez-Galera S, Naqvi S, Farre G, Sabalza M, Miralpeix B, Dashevskaya S, Yuan D, Ramessar K, Christou P, Zhu C, Bassie L, Capell T (2010) Promoter diversity in multigene transformation. Plant Mol Biol 73:363–378
Peterson PD, Lau J, Ebert B, Yang F, Verhertbruggen Y, Kim JS, Varanasi P, Suttangkakul A, Auer M, Loque D, Scheller HV (2012) Engineering of plants with improved properties as biofuels feedstocks by vessel-specific complementation of xylan biosynthesis mutants. Biotechnol Biofuels 5:84
Poovaiah CR, Mazarei M, Decker SR, Turner GB, Sykes RW, Davis MF, Stewart CN Jr (2015) Transgenic switchgrass (Panicum virgatum L.) biomass is increased by overexpression of switchgrass sucrose synthase (PvSUS1). Biotechnol J 10:552–563
Ramachandran S, Christensen HEM, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124:1637–1647
Robert CAM, Erb M, Hiltpold I, Hibbard BE, Gaillard MDP, Bilat J, Degenhardt J, Cambet-Petit-Jean X, Turlings TCJ, Zwahlen C (2013) Genetically engineered maize plants reveal distinct costs and benefits of constitutive volatile emissions in the field. Plant Biotechnol J 11:628–639
Saha P, Chakraborti D, Sarkar A, Dutta I, Basu D, Das S (2007) Characterization of vascular-specific RSs1 and rolC promoters for their utilization in engineering plants to develop resistance against hemipteran insect pests. Planta 226:429–442
Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DGJ, Wang H, Jackson L, Tang Y, Stewart CN Jr, Chen F, Dixon RA (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193:121–136
Sohn RH, Chen J, Koblan KS, Bray PF, Goldschmidt-Clermont PJ (1995) Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J Biol Chem 270:21114–21120
Stadler R, Sauer N (1996) The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Botanica Acta 109:299–306
Staiger CJ, Goodbody KC, Hussey PJ (1993) The profilin multigene family of maize: differential expression of three isoforms. Plant J 4:631–641
Stewart CN Jr, Vie LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750
Tanaka M, Shibata H (1985) Poly (L-proline)-binding proteins from chick embryos are a profilin and a profilactin. Eur J Biochem 151:291–297
Torre F, Rodríguez RG, Villar B, Álvarez-Otero R, Grima-Pettenati J, Gallego PP (2014) Genetic transformation of Eucalyptus globulus using the vascular-specific EgCCR as an alternative to the constitutive CaMV35S promoter. Plant Cell Tiss Organ Cult 117:77–84
Triplett JK, Wang Y, Zhong J, Kellogg EA (2012) Five nuclear loci resolve the polyploid history of switchgrass (Panicum virgatum L.) and relatives. PLoS One 76:e38702
Truernit E, Sauer N (1995) The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of p-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196:564–570
Wu B, Zhang B, Dai Y, Zhang L, Shang-Guan K, Peng Y, Zhou Y, Zhu Z (2012) Brittle culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol 159:1440–1452
Wuddineh WA, Mazarei M, Turner GB, Sykes RW, Decker SR, Davis MF, Stewart CN Jr (2015a) Identification and molecular characterization of the switchgrass AP2/ERF transcription factor superfamily, and overexpression of PvERF001 for improvement of biomass characteristics for biofuel. Front Bioeng Biotechnol 3:101
Wuddineh WA, Mazarei M, Zhang J, Poovaiah CR, Mann DGJ, Ziebell A, Sykes RW, Davis MF, Udvardi MK, Stewart CN Jr (2015b) Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol J 13:636–647
Wuddineh WA, Mazarei M, Zhang J-Y, Turner GB, Sykes RW, Decker SR, Davis MF, Udvardi MK, Stewart CN Jr (2016) Identification and overexpression of a knotted1-like transcription factor in switchgrass (Panicum virgatum L.) for lignocellulosic feedstock improvement. Front Plant Sci 7:520
Xi Y, Fu C, Ge Y, Nandakumar R, Hisano H, Bouton J, Wang Z-Y (2009) Agrobacterium-mediated transformation of switchgrass and inheritance of the transgenes. Bioenerg Res 2:275–283
Xiao F, Tang X, Zhou J (2001) Expression of 35S::Pto globally activates defense-related genes in tomato plants. Plant Physiol 126:1637–1645
Zavala JA, Patankar AG, Gase K, Baldwin IT (2004) Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuate. Proc Natl Acad Sci USA 101:1607–1612
Zhang C, Shi H, Chen L, Wang X, Lu B, Zhang S, Liang Y, Liu R, Qian J, Sun W, You Z, Dong H (2011) Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol 11:11
Zhang J-Y, Lee Y-C, Torres-Jerez I, Wang M, Yin Y, Chou W-C, He J, Shen H, Srivastava AC, Pennacchio C, Lindquist E, Grimwood J, Schmutz J, Xu Y, Sharma M, Sharma R, Bartley LE, Ronald PC, Saha MC, Dixon RA, Tang Y, Udvardi MK (2013) Development of an integrated transcript sequence database and a gene expression atlas for gene discover and analysis in switchgrass (Panicum virgatum L.). Plant J 74:160–173
Acknowledgements
This study was supported by funding from the BioEnergy Science Center (BESC) to CNS, the National High Technology Research and Development Program (863 Program) of China (no. 2012AA101801-02) to XZ, and China Scholarship Council to WX. We thank the technical support from the UTIA Genomics Hub and the DNA Sequencing Lab at UTK. BESC is a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All regulations were followed in the research.
Conflict of interest
Some of the authors are inventors of promoter patents, some of which have been licensed by companies. The promoter sequences and their function in plants is the topic of a US patent application, which is assigned to the University of Tennessee Research Foundation.
Additional information
Communicated by Baochun Li.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, W., Liu, W., Ye, R. et al. A profilin gene promoter from switchgrass (Panicum virgatum L.) directs strong and specific transgene expression to vascular bundles in rice. Plant Cell Rep 37, 587–597 (2018). https://doi.org/10.1007/s00299-018-2253-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2253-1