Abstract
Key message
Research describes the practical application of the codA negative selection marker in Soybean. Conditions are given for codA selection at both the shooting and rooting stages of regeneration.
Abstract
Conditional negative selection is a powerful technique whereby the absence of a gene product allows survival in otherwise lethal conditions. In plants, the Escherichia coli gene codA has been employed as a negative selection marker. Our research demonstrates that codA can be used as a negative selection marker in soybean, Glycine max. Like most plants, soybean does not contain cytosine deaminase activity and we show here that wild-type seedlings are not affected by inclusion of 5-FC in growth media. In contrast, transgenic G. max plants expressing codA and grown in the presence of more than 200 μg/mL 5-FC exhibit reductions in hypocotyl and taproot lengths, and severe suppression of lateral root development. We also demonstrate a novel negative selection-rooting assay in which codA-expressing aerial tissues or shoot cuttings are inhibited for root formation in media containing 5-FC. Taken together these techniques allow screening during either the regeneration or rooting phase of tissue culture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Negative selection systems employing dominant marker genes that result in the death of transformed cells have been described in plants. Some are lethal under all conditions (non-conditional negative selection systems), while others require the presence of specific agents to manifest toxicity (conditional negative selection systems) (Babwah and Waddell 2000). The codA gene from Escherichia coli is a substrate-dependent conditional negative selectable marker. The codA gene encodes a cytosine deaminase that catalyses the deamination of cytosine into uracil (Andersen et al. 1989) and is able to convert the harmless compound 5-fluorocytosine (5-FC) into a highly cytotoxic metabolite 5-fluorouracil (5-FU). The suicide effect of cytosine deaminase in the presence of 5-FC has been exploited in negative selection procedures in both eukaryotic and prokaryotic organisms (Mullen et al. 1992; Stougaard 1993; Hartzog et al. 2005; Braks et al. 2006; Dubeau et al. 2009).
Homologues of codA have not been found in plant species including Arabidopsis, Pisum sativum (pea), Hordeum vulgare (barley), Beta vulgaris (sugar beet), and Glycine max (soybean), making it likely that this gene could be employed widely in plants as a negative selective marker (Stougaard 1993; Koprek et al. 1999). For Lotus japonicus, Nicotiana sylvestris (Stougaard 1993) and Arabidopsis thaliana (Perera et al. 1993; Kobayashi et al. 1995), transformed seedlings expressing codA were inhibited in growth in the presence of 5-FC. Expression of codA in Brassica napus resulted in a reduction of root and hypocotyl lengths in the presence of 5-FC, and a severe suppression of true leaf development (Babwah and Waddell 2000). To assess the feasibility of using the Cre site-specific recombinase for elimination of marker genes from the tobacco plastid genome, Corneille et al. (2001) transformed the plastid genome with a codA gene bordered by two directly oriented loxP sites. These investigators found that 5-FC resistance could be successfully used to identify cells with Cre-induced codA deletions. Dutt et al. (2008) showed in grapevines that a combination of positive (kanamycin resistance mediated by nptII) and negative (codA) selection could be used to produce transgenic plants free of marker genes. Additionally, codA has been used as a conditional negative selectable marker in tobacco, barley, potato, and strawberry (Koprek et al. 1999; Schlaman and Hooykaas 1997; Rommens et al. 2004; Schaart et al. 2004). No toxic effects of 5-FC on wild-type plants were observed.
As ongoing research, we are developing site-specific recombinase technology (Thomson and Ow 2006; Blechl et al. 2012; Wang et al. 2011) for precise genome modification of soybean to allow successive rounds of genomic integration into a predetermined site. This technology requires a positive and negative selection scheme to select plants that have successfully undergone targeted exchange via recombinase-mediated cassette exchange (Wang et al. 2011). Although cytosine deaminase activity is not found in G. max plants (Stougaard 1993), the utility of codA gene as an effective negative selectable marker has not been previously investigated. In this study, we introduce codA into soybean to assess its potential as a conditional negative selection marker. We report 5-FC concentrations and protocols that can be used to identify soybean that expresses the exogenous codA gene.
Materials and methods
G. max seedling growth on germination media containing 5-FU or 5-FC
Germination medium (GM) consisted of 3.2 g/L Gamborg B5 medium with vitamins (bioWORLD) (Gamborg et al. 1968), 20 g/L sucrose, 4 g/L Phytoblend (Caisson Labs) at a final pH of 5.8. The selection agents 5-FC (AK Scientific) and 5-FU (Sigma) were dissolved in deionized water at 65 °C at concentrations of 10 mg/mL. These solutions were filter sterilized and immediately added to GM at approximately 65 °C. The media were dispensed into Solo sundae cups with lids (TS5R and DLR100, Dart Container Corporation).
For sterilization, seeds dispensed to 100 × 15 mm Petri dishes were incubated for approximately 16 h in the presence of chlorine gas (produced by mixing 100 mL of 4 % sodium hypochlorite with 4 mL 12 N hydrochloric acid in a beaker), in a bell jar setup in a fumehood. Sterile seeds of wild-type G. max cv. Bert were germinated by placing on GM containing 5-FC (200, 400, 600, 800, or 1000 μg/mL) or 5-FU (50, 100, 200, or 300 μg/mL), and incubated in a growth chamber with 16/8 light–dark photoperiod at 24 °C. After 9 days, hypocotyl and root lengths were measured (n = 4 per treatment), and the Welch t test with Bonferroni correction was employed to compare each treatment (supplementation with 5-FU or 5-FC) to control (no supplementation). Representative seedlings were photographed.
Construction of the positive/negative selection vector for soybean transformation
The pCAMBIA390 vector (http://www.cambia.org/daisy/bios/585.html) was modified with recombinase recognition sites Bxb1 attP (Yau et al. 2011) for integration and CinH res (Moon et al. 2011) for excision, flanking the multiple cloning site (MCS), and termed pCTAG. The codA coding sequence was amplified from the E. coli K-12 genome by PCR with primers F 5′-ATGTCGAATAACGCTTTACA-3′ and R 5′-TCAACGTTTGTAATCGATGG-3′. The amplified product was fused in frame downstream from the F2A peptide-skipping domain (from aphthovirus, FMDV; Ryan and Drew 1994), which in turn was fused in frame downstream from an hptII coding sequence that lacked a stop codon. This hptII2AcodA coding sequence was inserted between the double enhanced CaMV 35S promoter and transcription terminator. The positive–negative gene cassette was cloned into the MCS of pCTAG, resulting in the vector pCTAG-35HC. An expression cassette consisting of a −60 bp minimal CaMV 35S promoter fused to the GUSPlus coding sequence (http://www.cambia.org/daisy/bioforge_gusplus/3850.html) and Nos 3′ transcription terminator was constructed and cloned into pCTAG-35HC. The resulting plasmid, pCTAG-GHC, was introduced into Agrobacterium tumefaciens strain AGL1 (Lazo et al. 1991) by electroporation.
Plant transformation and regeneration and growth of transgenic plants
Agrobacterium-mediated transformation of soybean cultivar Bert, using the hptII gene as a positive selectable marker, followed the procedure of Zeng et al. (2004) with a few modifications. Seeds were germinated on GM. Cotyledonary nodes were wounded with a scalpel and incubated in Agrobacterium infection medium (0.32 g/L Gamborg B5 medium with vitamins (PhytoTechnology Laboratories), 1 mg/L Gamborg vitamins (PhytoTechnology Laboratories), 30 g/L sucrose, 3.9 g/L 2-(N-morpholino) ethanesulfonic acid (MES), 0.25 mg/L gibberellic acid (GA3; Gold Biotechnology), 1.67 mg/L 6-benzylaminopurine (BAP; bioWORLD), 40 mg/L acetosyringone (bioWORLD), pH 5.4) in a 50-mL sterile tube, sonicated for 2–4 s, then incubated at room temperature for 30 min with gentle shaking (rotary shaker at 60 rpm). Sonication treatment increased the number of shoots recovered after hygromycin selection (data not shown). After infection, explants were kept in co-cultivation medium (0.32 g/L Gamborg B5 medium with vitamins, 1 mg/L Gamborg vitamins, 30 g/L sucrose, 3.9 g/L MES, 0.25 mg/L GA3, 1.67 mg/L BAP, 40 mg/L acetosyringone, 400 mg/L cysteine (PhytoTechnology Laboratories), 154.2 mg/L dithiothreitol (DTT), 4 g/L Phytoblend, pH 5.4) for 5 days before transfer to shoot initiation medium. Shoot initiation (3.2 g/L Gamborg B5 medium with vitamins, 30 g/L sucrose, 0.59 g/L MES, 1.67 mg/L BAP, pH 5.7), elongation (4.4 g/L MS with vitamins (Caisson Labs), 30 g/L sucrose, 0.59 g/L MES, 50 mg/L asparagine (Sigma), 100 mg/L l-pyroglutamic acid (MP Biomedicals), 0.1 mg/L indole-3-acetic acid (IAA; Gold Biotechnology), 0.5 mg/L GA3, 1 mg/L zeatin (Gold Biotechnology), pH 5.7 and rooting (4.4 g/L MS with vitamins, 20 g/L sucrose, 0.59 g/L MES, 50 mg/L asparagine, 100 mg/L l-pyroglutamic acid, 1 mg/L indole-3-butyric acid (IBA; Sigma), pH 5.6) media were solidified with 4 g/L Phytoblend and included 200 µg/mL ticarcillin (Gold Biotechnology). A concentration of 10 µg/mL hygromycin B (Gold Biotechnology) was used for selection during shoot initiation and shoot elongation. Rooted primary transformants (T0) were transferred to potting medium Sunshine mix #1 (Crop Production Services) in 4″ deep tech square pots (McConkey Grower Products) and placed in a growth chamber at 24 °C with a 16/8 light–dark photoperiod. 2 or 3 weeks later, plants were moved to the greenhouse and grown with supplemental halide lighting that provided a 16/8 light–dark photoperiod at 24 °C. 4 or 5 months later, T1 seeds were harvested. The T2 generation was derived by selfing.
PCR analysis
Genomic DNA was extracted by grinding 1–2 cm2 young leaf fragments in 400 µL of buffer (200 mM Tris HCl pH 7.5, 250 mM NaCl, 25 mM EDTA, 0.5 % SDS). After centrifugation, the isopropanol precipitated pellet was washed with 70 % ethanol and resuspended in 50 µL of sterile deionized water. 2 µL of genomic DNA in a 25-µL volume was used per PCR reaction. The primers codAORF70F60 (a: 5′-catctgcaggacggaaaaat-3′) and codAORF1137R60 (b: 5′-gataatcaggttggcgctgt-3′) were used for the detection of codA sequences in soybean genomic DNA, using a 62 °C annealing temperature, 1 min extension time and 35 cycles for amplification. As a control, template DNA from non-transgenic ‘Bert’ plants was included in each PCR test.
Southern blot analysis
For Southern blot analysis, genomic DNA was extracted from leaf tissue of transgenic and wild-type plants (Dellaporta et al. 1983). 5 µg of DNA was digested with EcoRI for 6 h at 37 °C and separated by electrophoresis in a 0.8 % (w/v) agarose gel. The DNA was then transferred to a Hybond-N membrane (Amersham Biosciences Corp., Piscataway, NJ, USA) and hybridized with 32P-labeled codA sequence [amplified with Taq polymerase (Promega) using codAORF70F60 (a: 5′-catctgcaggacggaaaaat-3′) and codAORF1137R60 (b: 5′-gataatcaggttggcgctgt-3′)], as described previously by Stougaard (1993). Southern blot washes for the codA probe used 2× SSC for 2 min in shaking incubator at room temperature two times. Blots were placed on X-ray film (Phenix) with intensifying screens (Kodak) and placed in the −80 °C freezer for 2 days prior to development.
Histochemical assay for β-glucuronidase activity
The histochemical assay for GUSPlus gene expression was performed according to Jefferson (1987). Soybean tissues of mature plants that had set seed (120 days) were covered in X-gluc (5-bromo-4-chloro-3-indolyl-glucuronide) solution, vacuum infiltrated for 5 min, then kept at 37 °C overnight. After staining, the green tissues were treated with several changes of 70 % ethanol to remove chlorophyll.
codA negative selection—seedling assay
Sterilized T1, T2 (GHC21, 39), and/or T3 (GHC22) seeds from codA transgenic lines were transferred to GM with 200 μg/mL 5-FC. After 9 days, plants were scored as sensitive/resistant to 5-FC and tissue samples were taken for DNA extraction and PCR analysis for codA. Representative plants were photographed.
Reversibility to negative selection of transgenic plants and recovery from 5-FC cytotoxic effects—recovery assay
To test for reversibility to negative selection and to determine whether codA-transformed lines could recover from the cytotoxic effects of 5-FC, nine-day-old T2 or T3 seedlings were removed from 5-FC selection medium and transferred directly to Sunshine #1 potting media. The seedlings were maintained in a growth chamber with 16/8 light–dark photoperiod at 24 °C. Three days later, the seedlings were transferred to the greenhouse and grown to reproductive maturity for seed recovery and maintenance of codA-transformed lines.
codA negative selection—rooting assay
To test whether rooting on 5-FC could be used as a negative selection in soybeans, sterilized seeds were placed on GM in a growth chamber with a 16/8 light–dark photoperiod at 24 °C. After 9–10 days, aerial tissue of germinated seed was collected for this assay, defined as the first true leaf and apical meristem (referred to as the ‘shoot’), which was cut-off at the stem above the cotyledons and transferred to rooting medium (4.4 g/L MS medium and vitamins, 20 g/L sucrose, 0.59 g/L MES, 4 g/L Phytoblend, pH 5.6), supplemented with 300 μg/mL 5-FC. The cuttings were returned to the growth chamber for rooting and assessed 2 weeks later.
Results
Sensitivity of wild-type G. max seedlings to 5-FU and 5-FC
In order to determine whether 5-FU had any observable effects on the growth and development of G. max seedlings, wild-type seeds were germinated on media containing various concentrations of the base analog. By 9 days of growth on medium containing 50 µg/mL 5-FU, seedlings showed significant reductions in hypocotyl and root growth (Welch t test, P < 0.005; Fig. 1a, b). At higher concentrations of 5-FU, the reductions in hypocotyl and root lengths were more severe. No lateral roots formed in the presence of 100, 200, and 300 µg/mL 5-FU (Fig. 1a, b). These results revealed that 50 µg/mL 5-FU inhibits hypocotyl and root growth and overall development of wild-type seedlings.
Phenotypes of wild-type G. max on 5-FU- and 5-FC-containing media. Nine-day-old seedlings from wild-type G. max germinated on GM containing various concentrations of 5-FU (a) and 5-FC (c). Hypocotyl length (black bars) and root length (white bars) of wild-type G. max seedlings germinated on media containing different concentrations of 5-FU (b) and 5-FC (d). Variation is expressed as standard deviation for measurements of four plants. Asterisk indicates significant difference from no treatment (Welch t test with Bonferroni correction, P < 0.05)
To determine the optimal concentration of 5-FC for negative selection, wild-type G. max seeds were germinated in media supplemented with 5-FC at various concentrations. After 9 days, wild-type seeds germinated and grew normally even at 5-FC concentrations of 200–400 µg/mL (Fig. 1c, d). As compared to control medium without 5-FC supplementation, hypocotyl lengths were shorter on medium supplemented with 600 µg/mL 5-FC (Welch t test, P < 0.01). Root lengths became significantly shorter than control only at 800 µg/mL 5-FC (Welch t test, P < 0.05). Thus, hypocotyl growth is more sensitive to 5-FC treatment than root growth (Fig. 1d). Lateral root formation was distinctly suppressed in the presence of 600 and 800 µg/mL 5-FC. In GM supplemented with 1000 µg/mL 5-FC, the lateral roots were almost completely suppressed, and the taproot was severely shortened. Because root and hypocotyl lengths were not significantly different on GM containing 200 µg/mL 5-FC as compared to medium without 5-FC (Fig. 1c), we chose 200–300 µg/mL 5-FC to assess codA negative selection in subsequent experiments.
G. max transformation
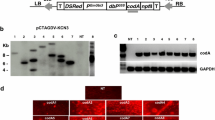
To obtain soybean containing an active codA gene, the vector pCTAG-GHC, diagrammed in Fig. 2a, was constructed for Agrobacterium-mediated transformation and selection analysis (“Materials and methods”). The positive/negative selection components, consisting of the coding region for the hptII gene (for positive hygromycin resistance selection) fused in frame with the F2A peptide-skipping domain (Ryan and Drew 1994) and the E. coli codA coding sequence (Genbank Accession No. S56903.1), were inserted between the double-enhanced promoter (dbP 35S) and transcription terminator sequences derived from the 35S gene of cauliflower mosaic virus (CaMV). The T-DNA portion of the vector also contains the −60 minimal CaMV 35S gene promoter (Min P 35S) driving the GUSPlus coding sequence with the nopaline synthase (Nos) transcription terminator (Fig. 2a). The minimal promoter-GusPlus cassette was included to detect whether the T-DNA inserts into a genomic region of high transcriptional activity. Ten hygromycin-resistant plants were recovered from 300 transformed wild-type cotyledons. Of these lines, all were positive for the codA gene as assayed by PCR (data not shown). However, only three lines, specifically GHC21, GHC22, and GHC39, were confirmed by Southern blot analyses (Fig. 2b). Thus, GHC21, GHC22, and GHC39 were chosen for further analyses.
T-DNA section of vector pCTAG-GHC (not to scale) and predicted single copy T-DNA insertion structure. a PCR primer pairs used for detection of codA in genomic DNA of transformed lines are shown as a and b with product size indicated above the dotted line. The att sites are shown as gray arrowheads and the hybridization probe used in Southern analyses as a gray rectangle. The size of the T-DNA from the unique EcoRI site to the right border is indicated above the dashed line. b Autoradiogram of blot of genomic DNA from non-transformed cultivar Bert (WT) and three transformed lines (numbered GHC21, 39 and 22). Total DNA from T0 (GHC21 and GHC39) or T3 (GHC22) digested with the EcoRI and hybridized to the 32P-labeled codA probe. The sizes of the hybridizing fragments indicated to the right. E, EcoRI
Genetic and phenotypic analysis of codA transgenic lines
GHC21 and GHC39 T0 plants were normal in appearance and produced many seeds, but the GHC22 T0 plant was shorter than wild-type plants, and produced few seeds (data not shown). T1 seeds from each line were germinated in soil to observe the growth and development of progeny derived by self-fertilization of the regenerated plants. Germination frequencies for both GHC21 and GHC39 transgenic lines were similar to those of wild-type seeds. T1 seedlings of both transgenic lines grew well and were phenotypically indistinguishable from wild-type seedlings. Only a single GHC22 T1 seed was viable. The resultant GHC22 T1 plant was healthy and produced viable T2 seeds, which were used in further analyses. GHC22 T2 progeny were phenotypically similar to wild-type plants and had normal seed set.
The presence of transgenes in the T0 plants GHC21, GHC22, and GHC39 was verified by PCR amplification with primer set a and b to detect codA (Fig. 2a; “Materials and methods”) and by sequencing the resultant amplicons (data not shown). The number of T-DNA insertions in the transgenic plants was confirmed by hybridization of EcoRI-digested genomic DNA with a 32P-labeled codA probe (gray bar in Fig. 2a). EcoRI cuts once within the target T-DNA (Fig. 2a), releasing a fragment at least 4.17 kb in length that extends beyond the right border into soybean genomic DNA adjacent to the insertion sites. The EcoRI digests of DNA from GHC21 and GHC39 T0 plants yielded single codA bands at 8.0 kb and 4.8 kb, respectively (Fig. 2b). DNA from the GHC22 T0 plant contained multiple codA bands (Supplemental Figure 1), in agreement with codA segregation in the T2 generation (Table 1). However, Southern blot analysis on the T3 generation, derived from a single GHC22 T2 plant, showed a single 4.3 kb EcoRI fragment for codA (Fig. 2b). Segregation of codA in the T3 generation (Table 1), derived from the same GHC22 T2 plant, confirmed that this GHC22 line has a single T-DNA insert.
For GHC21, GHC22, and GHC39, DNA was extracted from the progeny of each (i.e., T1 seedlings for GHC21/GHC39 and T2 seedlings for GHC22) and assayed by PCR for codA (Table 1). For GHC21 progeny (T1 seedlings), segregation of codA conformed to a 3:1 ratio (χ 2 test, P > 0.05) expected for a single locus, whereas transgene segregation in progeny of GHC22 (T2 seedlings) and GHC39 (T1 seedlings) varied significantly from a 3:1 ratio (χ 2 test, P < 0.05; Table 1). For each line, a codA-positive individual was picked at random for analysis of codA segregation in the subsequent generation (i.e., T2 for GHC21/GHC39 and T3 for GHC22). When codA inheritance was re-assessed by PCR in the T2 generation of GHC21 and GHC39 and in the T3 generation of GHC22, all segregated in a 3:1 ratio (χ 2 test, P > 0.05; Table 1). No additional lines were sought after recovery of three independent lines, each carrying a single, hemizygous transgene locus.
Histochemical staining for β-glucuronidase activity was also done in the T2 generation of GHC21 and GHC39 and in the T3 generation of GHC22, derived from the same parents used in the codA segregation analysis (Fig. 3). Staining for GUSPlus revealed expression patterns that ranged from non-detectable for GHC21, to medium for GHC22, to very high for GHC39 (Fig. 3).
GUSPlus gene expression in GHC transgenic lines. Results of the histochemical assay for β-glucuronidase activity in leaves, stems, roots, and seeds of wild-type plants and transgenic soybean plants carrying the MinP 35S::GUSPlus::nos expression cassette. Tissues are from T2 plants of GHC21 and GHC39 and from T3 plants of GHC22
5-FC selection assays
To determine whether codA could be used as a negative selectable marker to identify progeny with and without a transgene, T2 or T3 seeds from transgenic lines GHC21 and GHC39 or GHC22, respectively, were germinated on medium supplemented with 200 μg/mL 5-FC. Seedlings that inherited the codA gene displayed reduced taproot and hypocotyl lengths, and severe suppression of lateral root development, whereas codA null (segregated siblings) and wild-type G. max were unaffected and developed normally (Fig. 4a–d). Minimally, 18 seedlings for each line were scored for a codA-sensitive/resistant phenotype, and PCR genotyping showed complete agreement with phenotype analysis (Table 1).
Phenotypes and PCR analysis of transgenic codA seedlings on 5-FC-containing media. Top panels (a–d) seedlings 9 days after germination on selection GM containing 200 μg/mL 5-FC. Bottom panels (a–d) PCR analysis of same plants with primers a and b (Fig. 2a). a T2 seedlings of GHC21. b T3 seedlings of GHC22. c T2 seedlings of GHC39. d Wild-type G. max control. Top panels (e–h) shoot cuttings (subset) after 2 weeks at 24 °C in rooting media containing 200 µg/mL 5-FC (GHC39) or 300 µg/mL 5-FC (GHC21, GHC22, Bert). Bottom panels (e–h) PCR analysis and 5-FC rooting phenotype of cuttings (n = 8 per genotype) tested for rooting in the presence of 5-FC (+, root formation). M DNA size markers, P positive plasmid DNA, N genomic DNA from wild-type ‘Bert.’ The 0.98 kb is the expected PCR size for codA
We also developed a novel codA negative selection-rooting assay. As an alternative assay, we examined codA-transformed aerial tissue (“Materials and methods”) for rooting in the presence of 200–300 µg/mL 5-FC. Wild-type and codA-transformed aerial tissue cuttings (defined as ‘shoots’ hereafter) (n = 8 for each line) were transferred to rooting medium containing 5-FC. After 2 weeks of growth, shoots were scored for the presence/absence of roots. Wild-type ‘Bert’ displayed normal rooting in 200–300 µg/mL 5-FC rooting medium (Fig. 4h). On medium supplemented with 200 µg/mL 5-FC, codA-positive cuttings (as verified by PCR) from GHC39 T2 seedlings had no roots, whereas codA null cuttings formed extensive root networks, similar to wild type (Fig. 4g). Shoot cuttings from codA null segregants rooted equally well in the presence and absence of selection. Inhibition of root production for codA-containing cuttings from lines GHC21 (T2) and GHC22 (T3) was achieved in the presence of 300 µg/mL 5-FC. In contrast, their codA null siblings formed as many roots under selection as wild-type cuttings (Fig. 4e–h). These results show that codA negative selection is effective in G. max during the rooting process.
Conditional negative selection of codA seedlings grown on 5-FC substrate
To examine if the cytotoxic effects of codA selection were reversible, segregating seedlings (GHC21/GHC39 T2 generation and GHC22 T3 generation) were grown for 9 days on medium supplemented with 200 μg/mL 5-FC, then transferred to soil and grown to maturity. One week after transfer, 5-FC-sensitive individuals (presence of codA confirmed by PCR) were shorter than wild-type plants and 5-FC-resistant codA null segregants (Fig. 5a). However, after 2 weeks in soil, the 5-FC-sensitive plants had fully recovered and were indistinguishable from the 5-FC-resistant plants (Fig. 5b). Wild-type ‘Bert’ and codA null seedlings exhibited no negative effects when germinated on 200 μg/mL 5-FC and no lag in growth after transfer to soil, compared to transplants that had never been exposed to 5-FC (Fig. 5a, b). All codA-positive GHC21 (15 individuals) and GHC22 (16 individuals) plants recovered from the effects of exposure to 5-FC, and 13 of 14 GHC39 codA-positive individuals recovered. At maturity, seed set was examined and all plants were found to be similar to wild-type plants grown under the same conditions. In conclusion, codA is effective as a conditional negative selectable marker to the 5-FC substrate.
Phenotypes of wild-type (Bert) and transgenic codA (GHC21, GHC39, and GHC22) G. max seedlings after transfer to soil from 5-FC selection medium. a Plants 1 week after transfer from 200 µg/mL 5-FC selection medium into potting media. b Plants 2 weeks after transfer to soil. c PCR analysis of resistant (R) and sensitive (S) phenotypes on 5-FC selection. N wild-type ‘Bert’ plants geminated on medium without 5-FC, N200 wild-type ‘Bert’ plants germinated on 200 μg/mL 5-FC selection media, R plants resistant to 5-FC, S plants sensitive to 5-FC
Discussion
In this study, we have introduced the codA gene into an important legume crop, G. max, and defined two conditions in which the negative selection agent 5-FC can be used successfully for transgene detection. All codA-expressing transgenic plants were sensitive to germination in the presence of 5-FC, the precursor of the cytotoxin 5-FU. Although codA transgenic seedlings grew well in the absence of 5-FC substrate, they displayed shortened taproot and hypocotyl lengths, and severe suppression in lateral root development on medium supplemented with 200 μg/mL 5-FC (Fig. 4a–d). Molecular analyses of segregating populations confirmed a 5-FC-sensitive/resistant phenotype correlated with the presence/absence of the codA transgene (Fig. 4a–d). The differences between the sensitive and resistant phenotypes, as assessed by 9 days growth of hypocotyls and roots, are distinct and easily observed. These results indicate that 5-FC sensitivity in G. max is sufficient to distinguish transgenic lines from wild-type progeny. The sensitive phenotype of transgenic G. max during germination is similar to that reported for codA-transformed Lotus, tobacco, and Brassica (Stougaard 1993; Babwah and Waddell 2000).
Although codA-expressing seedlings are sensitive to 200 μg/mL 5-FC at 9 days, over 90 % of seedlings fully recover when transferred to soil suggesting that the cytotoxic nature of this selection is reversible. Stougaard (1993) reported that only 40 % of lotus plants recovered from the effects of 5-FC negative selection under their conditions. Babwah and Waddell (2000) reported that 67 % (T1) and 100 % (T2) B. napus plants recover from the effects of 5-FC negative selection. The high frequency with which our codA-transformed G. max recovered from the effects of 5-FC may be due to the short period of time (9 days) needed to identify sensitive individuals. In any case, the complete recovery of sensitive plants after transfer to potting media means that codA can be used to identify transgenic plants without killing them.
Additionally, a novel negative selection-rooting assay is presented. We demonstrate rooting of G. max shoot cuttings on 5-FC medium occurs only in the absence of a codA transgene (Fig. 4). We observed that 300 μg/mL 5-FC completely abolished rooting in GHC21, GHC22, and GHC39 transgenic lines that contain codA, while not affecting the root development of wild type or codA null segregants. Potentially this assay has use in tissue culture selection and regeneration.
In this study, wild-type G. max plants were shown to develop normally in the presence of 5-FC at concentrations below 600 μg/mL. In contrast, even low levels (50 μg/mL) of 5-FU, a cytotoxic compound that is the product of deamination of 5-FC, inhibit growth of wild-type ‘Bert’ plants (Fig. 1). The tolerance of wild-type G. max to the presence of 5-FC suggests that G. max lacks enzymatic activity capable of converting 5-FC to 5-FU. These results are in agreement with findings from in vitro assays, which indicated that cytosine deaminase activity is absent from G. max (Stougaard 1993).
The vector used in this research included the GUSPlus reporter gene under control of the minimal (−60 bp) version of the CaMV 35S promoter. GUSPlus expression was seen clearly in all the major organs of both GHC22 and GHC39 plants, but was not detected by histochemical staining of organs from the GHC21 line (Fig. 3). Ongoing experiments will determine the sites of transgene insertion in GHC22 and GHC39 and verify if the GUSPlus minimal promoter cassette was activated due to integration near a native soybean promoter or enhancer in these lines.
Our research results indicate that codA gene is stably inherited and acts as a strong and effective marker for conditional negative selection with the 5-FC substrate. The ability to use negative selection will be very useful for developing precise genomic engineering strategies for soybean.
Author contribution statement
JT designed the approach, constructed the plasmids, and interpreted the research results. JT supervised MS, prepared, and submitted the manuscript. MS performed codA transformation, data interpretation. JMM performed data collection, analysis, and edited the manuscript. AB analyzed the genetic data and co-wrote the manuscript. SKH analyzed data and edited the manuscript.
References
Andersen L, Kilstrup M, Neuhard J (1989) Pyrimidine, purine and nitrogen control of cytosine deaminase synthesis in Escherichia coli K12. Arch Microbiol 152:115–118
Babwah V, Waddell S (2000) Cytosine deaminase as a substrate-dependent negative selectable marker in Brassica napus. Theor Appl Genet 100:802–809
Blechl A, Lin J, Shao M, Thilmony R, Thomson J (2012) The Bxb1 recombinase mediates site-specific deletion in transgenic wheat. Plant Mol Biol Rep 30:1357–1366
Braks JAM, Franke-Fayard B, Kroeze H, Janse CJ, Waters AP (2006) Development and application of a positive–negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res 34:e39
Corneille S, Lutz K, Svab Z, Maliga P (2001) Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox site-specific recombination system. Plant J 27:171–178
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Dubeau MP, Ghinet MG, Jacques PE, Clermont N, Beaulieu C, Brzezinski R (2009) Cytosine deaminase as a negative selection marker for gene disruption and replacement in the genus Streptomyces and other Actinobacteria. Appl Environ Microbiol 75:1211–1214
Dutt M, Li ZT, Dhekney SA, Gray DJ (2008) A co-transformation system to produce transgenic grapevines free of marker genes. Plant Sci 175:423–430
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hartzog PE, Nicholson BP, McCusker JH (2005) Cytosine deaminase MX cassettes as positive/negative selectable markers in Saccharomyces cerevisiae. Yeast 22:789–798
Jefferson R (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Kobayashi T, Hisajima S, Stougaard J, Ichikawa H (1995) A conditional negative selection for Arabidopsis expressing a bacterial cytosine deaminase gene. Jpn J Genet 70:409–422
Koprek T, McElroy D, Louwerse J, Williams-Carrier R, Lemaux PG (1999) Negative selection systems for transgenic barley (Hordeum vulgare L.): comparison of bacterial codA and cytochrome P450 gene-mediated selection. Plant J 19:719–726
Lazo GR, Stein PA, Ludwig RA (1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Nat Biotechnol 9:963–967
Moon H, Abercrombie L, Eda S, Blanvillain R, Thomson J, Ow D, Stewart C (2011) Transgene excision in pollen using a codon optimized serine resolvase CinH-RS2 site-specific recombination system. Plant Mol Biol 75:621–631
Mullen CA, Kilstrup M, Blaese RM (1992) Transfer of the bacterial gene for cytosine deaminase to mammalian cells confers lethal sensitivity to 5-fluorocytosine-a negative selection system. Proc Natl Acad Sci USA 89:33–37
Perera RJ, Linard CG, Signer ER (1993) Cytosine deaminase as a negative selective marker for Arabidopsis. Plant Mol Biol 23:793–799
Rommens CM, Humara JM, Ye JS, Yan H, Richael C, Zhang L, Perry R, Swords K (2004) Crop improvement through modification of the plant’s own genome. Plant Physiol 135:421–431
Ryan MD, Drew J (1994) Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J 13:928–933
Schaart JG, Krens FA, Pelgrom KTB, Mendes O, Rouwendal GJA (2004) Effective production of marker-free transgenic strawberry plants using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotechnol J 2:233–240
Schlaman HRM, Hooykaas PJJ (1997) Effectiveness of the bacterial gene codA encoding cytosine deaminase as a negative selectable marker in Agrobacterium- mediated plant transformation. Plant J 11:1377–1385
Stougaard J (1993) Substrate dependent negative selection in plants using a bacterial cytosine deaminase gene. Plant J 3:755–761
Thomson JG, Ow DW (2006) Site-specific recombination systems for the genetic manipulation of eukaryotic genomes. Genesis 44:465–476
Wang Y, Yau Y-Y, Perkins-Balding D, Thomson JG (2011) Recombinase technology: applications and possibilities. Plant Cell Rep 30:267–285
Yau Y-Y, Wang Y, Thomson JG, Ow DW (2011) Method for Bxb1-mediated site-specific integration in planta. Methods Mol Biol Plant Chromosom Eng Methods Protoc 701:147–166
Zeng P, Vadnais DA, Zhang Z, Polacco JC (2004) Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep 22:478–482
Acknowledgments
We are grateful to Ron Chan, Bryan Hernandez, and Paul Duellman for technical assistance. This work was supported by the United Soybean Board project number 1420-532-5644, by the Minnesota Soybean Research and Promotion Council project number 7-14C, by the USDA Agricultural Research Service CRIS project 5325-21000-020, and by the Biotechnology Risk Assessment Program competitive grant 2010-33522-21773 from the USDA—National Institute of Food and Agriculture. Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Wang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shao, M., Michno, JM., Hotton, S.K. et al. A bacterial gene codA encoding cytosine deaminase is an effective conditional negative selectable marker in Glycine max . Plant Cell Rep 34, 1707–1716 (2015). https://doi.org/10.1007/s00299-015-1818-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1818-5