Abstract
Key message
Endophytic microbes Bacillus sp. LZR216 isolated from Arabidopsis root promoted Arabidopsis seedlings growth. It may be achieved by promoting the lateral root growth and inhibiting the primary root elongation.
Abstract
Plant roots are colonized by an immense number of microbes, including epiphytic and endophytic microbes. It was found that they have the ability to promote plant growth and protect roots from biotic and abiotic stresses. But little is known about the mechanism of the endophytic microbes-regulated root development. We isolated and identified a Bacillus sp., named as LZR216, of endophytic bacteria from Arabidopsis root. By employing a sterile experimental system, we found that LZR216 promoted the Arabidopsis seedlings growth, which may be achieved by promoting the lateral root growth and inhibiting the primary root elongation. By testing the cell type-specific developmental markers, we demonstrated that Bacillus sp. LZR216 increases the DR5::GUS and DR5::GFP expression but decreases the CYCB1;1::GUS expression in Arabidopsis root tips. Further studies indicated that LZR216 is able to inhibit the meristematic length and decrease the cell division capability but has little effect on the quiescent center function of the root meristem. Subsequently, it was also shown that LZR216 has no significant effects on the primary root length of the pin2 and aux1-7 mutants. Furthermore, LZR216 down-regulates the levels of PIN1-GFP, PIN2-GFP, PIN3-GFP, and AUX1-YFP. In addition, the wild-type Arabidopsis seedlings in the present of 1 or 5 µM NPA (an auxin transport inhibitor) were insensitive to LZR216-inhibited primary root elongation. Collectively, LZR216 regulates the development of root system architecture depending on polar auxin transport. This study shows a new insight on the ability of beneficial endophytic bacteria in regulating postembryonic root development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Like animals, plants have their own microbiome that protects them from various adverse environmental conditions (Berendsen et al. 2012). The plant rhizosphere is a noticeable ecological environment as myriad microorganisms colonize in and out of the roots of growing plants. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere (Berg and Smalla 2012). Plant growth-promoting rhizobacteria (PGPR) are naturally occurring soil microorganisms that colonize in/on roots and stimulate shoot and root growth. Such bacteria have been able to enhance crop yields (Kloepper et al. 1980, 1989). Within the rhizobacteria, some microbes can promote plant growth and provide a better environment for plant growth through indirect or direct methods (Couillerot et al. 2009; Richardson et al. 2009). Some PGPR can help plants withstand environment stresses including heavy metal contaminations or other pollutants (Jing et al. 2007; Tak et al. 2013). Some PGPR can promote plants in nutrition uptake via associative nitrogen fixation and phosphate solubilization (Richardson et al. 2009). They can suppress a broad spectrum of bacterial, fungal, and nematode diseases. Similarly, beneficial rhizobacteria from rice rhizosphere confer high protection against biotic and abiotic stresses inducing systemic resistance in rice seedlings (Lucas et al. 2014). Plant growth-promoting bacteria confer resistance to salt stress in tomato plants (Mayak et al. 2004). Therefore, utilizing PGPR is a new and promising approach for improving plant growth and development.
Some microbial species can alter root system architecture by producing phytohormones including auxin, ethylene, and cytokinins (Bowen and Rovira 1999). The effect of hormones from bacterial suspensions on root morphogenesis includes enhancing root hairs and lateral roots (Persello-Cartieaux et al. 2003). Understanding the function of PGPR on root development is important. One strategy is to inoculate roots with PGPR in vitro and monitor the resultant effects on plants. This method showed that many PGPR reduce the growth rate of the primary root (Dobbelaere et al. 1999) and increase the number of lateral roots (Combes-Meynet et al. 2011). Root surface areas were increased by inoculation with PGPR. So plants uptake minerals and water more favorably, and the fresh weight of the whole plant can be increased (Walker et al. 2012). Studies have shown that the molecular mechanisms of those observations are dissected at the genetic and molecular levels (López-Bucio et al. 2007). Root growth depends on two basal developmental processes: cell division in the root apical meristem and elongation of cells that leave the root meristem. The root system architecture is defined by the primary root formation, lateral root (LR), and root hair (RH) formation. Auxin is known to alter the root system architecture, including primary root elongation (Alonso et al. 2003) and root hair formation and elongation (Pitts et al. 1998; Rahman et al. 2002, 2007).
Arabidopsis has been chosen as a model system to study plant–microbe interactions because of its short life cycle and the availability of many mutants impaired in hormone response pathways, including auxin and ethylene (Persello-Cartieaux et al. 2003; Ryu et al. 2005). Several PGPR can produce auxin and influence the root system architecture (Barazani and Friedman 1999; Patten and Glick 2002). Inoculation with Phyllobacterium brassicacearum strain STM196 stimulated the expression of several auxin biosynthesis pathway enzymes in Arabidopsis shoots with a potential redistribution of auxin into roots (Contesto et al. 2010). Auxin transport inhibitor treatment blocked auxin accumulation at sites of synthesis in shoots of Arabidopsis, inoculated with Bacillus subtilis, suppressed the levels of auxin in roots and reduced the growth-promoting effects (Zhang et al. 2007). Application of the auxin transport inhibitor, 1-naphthylphthalamic acid (NPA), restricted auxin accumulation to sites of synthesis (Keller et al. 2004). Inoculation of Arabidopsis auxin transport mutants aux1-7 and pin2 with Trichoderma virens reduced growth compared with wild-type plants (Contreras-Cornejo et al. 2009). The auxin-responsive promoter DR5::GUS lines as the object of study provide insights indirectly into changes of auxin levels, signaling, or responses during plant–microbe interaction. Besides the above important information, very little is known about the cellular, developmental, and physiological responses of plants to endophytic bacteria. We identified a new endophytic bacteria strain of Bacillus subtilis LZR216 from Arabidopsis thaliana, which is able to promote the growth of Arabidopsis plants. Our results showed that Bacillus subtilis. LZR216 can dramatically change the root system architecture, including increased growth of lateral roots and inhibited primary root elongation involving polar auxin transport.
Materials and methods
Isolation and identification of endophytic microbes
Arabidopsis (Col-0) seeds were surface-sterilized with 70 % ethanol for 30 s and subsequent 2 % bleach for 15 min, washed three times with autoclaved water, and then kept at 4 °C for 2 days for stratification. After that, the seeds were transferred to 1/2 MS solid medium for growth. 5-day-old seedlings were transferred to new tissue culture bottles containing 1/2 MS medium for further growth. The endophytic bacteria were isolated from roots of 4-week-old Arabidopsis seedlings. Each sample was triturated with autoclaved mortar and pestle and with autoclaved water, transferred to 5 mL of King’s B (KB) liquid medium, and then cultured at 28 °C with a constant shaking at 150 rpm. After 48 h, several concentrations of bacterial suspensions were plated on KB agar media and then incubated at 28 °C for another 48 h until colonies formed. To confirm the formation of endophytic bacteria, PCR reactions were performed with the universal 16S primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′). The PCR products were sequenced and the resultant 16s rDNA sequences were compared to GenBank databases using BLAST.
Plant materials and methods
Wild-type Col-0, mutants tir1-1, pin2, aux1-7, and axr1-12, and several transgenic plants DR5::GUS, CycB1;1::GUS, PIN1-GFP, PIN2-GFP, PIN3-GFP, and AUX1-YFP were used in this study. Seeds were surface-sterilized and stratified at 4 °C for 2 days on 1/2 MS agar-solidified medium. To test the effect of LZR216 on plant growth, 5-day-old seedlings were transferred and positioned vertically in a growth chamber under a long photoperiod (16 h light/8 h dark) and 65–70 % relative humidity at 22 °C. For inhibition of polar auxin transport, indicated concentrations of NPA were added in the media.
Bacillus sp. LZR216 was cultured on KB liquid medium with constant shaking at 150 rpm for 24 h at 28 °C. After 24 h of growth, cells were collected in autoclaved water, washed twice, and resuspended in autoclaved water after centrifugation at 7500 rpm for 10 min. The bacterial titer was adjusted to an optical density at 600 nm of 0.5 (5 × 108 colony-forming units mL−1). For treatment, 20 µL of bacterial suspension (autoclaved water as a control) was spotted at 1/2 MS petri plates. In order to test the effect of LZR216 on promoting Arabidopsis growth, seedlings were grown on horizontal plates for 12 days, and then the shoot fresh weight and total leaves surface area were measured. To determine the effect of LZR216 on the root system architecture, seedlings were grown on vertical plates for 7 days, and then the primary root length and lateral root number per plant were measured.
GUS histochemical staining
Histochemical assay of the GUS activity was performed as described by Ulmasov et al. (1997) with minor modifications. Five-day-old seedlings were treated with LZR216 for another 7 days, and then incubated in GUS staining buffer containing 1 mM X-Gluc, 100 mM sodium phosphate (pH 7.5), 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 10 mM EDTA, and 0.1 % Triton X-100. After incubation at 37 °C for 12 h, the stained roots were cleared in a mixture of chloral hydrate:glycerol:water (8:1:2, g/v/v) and observed with Nomarski optics.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted with Trizol (TaKaRa) from shoots and roots, and then treated with RNase-free DNase (Promega, Madison, WI, USA). First-strand cDNA was synthesized with the PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa, Mountain View, CA, USA). Quantitative real-time PCR (qRT-PCR) was performed using the SYBR PrimeScript RT-PCR Kit (Perfect Real Time; TaKaRa). PCR was performed using a CFX 96 Real-Time system (Bio-Rad, Hercules, CA, USA) with the following standard cycling conditions: 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. The cycle threshold 2(−ΔΔC(T))-based method was used for relative quantitation of gene expression. Expression levels of genes were normalized to ACTIN2 levels. The primer sequences used in the study are shown in supplementary table S1.
Fluorescence microscopy
Confocal images were captured with a confocal laser scanning microscope (Olympus, FV1000) equipped with a 20× (water immersion) objective lens, a HeNe laser (543 nm), and an argon laser (488, 514 nm). Emission filters for GFP, YFP, and FM4-64 were set at 505–530, 490–510, and 560 nm, respectively. All images within a single experiment were captured with the same gain and exposure settings. For FM4-64 staining, seedlings were incubated for 10 min with 4 μM FM4-64 (Invitrogen) from a 10 mM stock solution in water.
Statistical analysis
Each experiment was repeated at least three times. Values were expressed as mean ± SE. Statistical analyses were done with one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test for independent samples or with Student’s t test when two means were compared (SPSS version 17.0). In all cases, the confidence coefficient was set at P < 0.05.
Results
Isolation and identification of endophytic bacterium Bacillus sp. LZR216
Endophytic bacterium was isolated from roots of 4-week-old Arabidopsis tissue culture seedlings, one of which was named as LZR216. For comparison, other six Bacillus in Bacillaceae, including Virgibacillus sp., Bacillus sp., Halobacillus sp., Alicyclobacillus sp., and Aneurinibacillus thermoaerophilus were selected from the GenBank for sequence alignment. Based on the phylogenetic tree constructed with MEGA6, this endophytic bacterium LZR216 belong to Bacillus sp. (Fig. 1).
LZR216 phylogenetic tree of the partial 16S rDNA sequences. The sequence data for several closely related type cultures were recovered from GenBank and included in the tree. The accession numbers for the sequences are as follows: Paenibacillus apiarius strain AECSP01, AB748948; Paenibacillus xylanilyticus strain AECSP02, AB748941; Virgibacillus halodenitrificans strain GUFBSama06-18, KF495554; Virgibacillus sp. GUFBSama06-11, KF495547; Virgibacillus sp. enrichment culture clone DSFB408, KJ850505; Virgibacillus sp. enrichment culture clone ZSTA205, KJ850502; Virgibacillus sp. B6B(2010), HQ433456; Bacillus sp. AECSB16, AB748970; Bacillus sp. AECSB15, AB748969; Bacillus sp. AECSB02, AB748939; Bacillus atrophaeus strain AECSB18, AB748959; Bacillus sp. AECSB10, AB748955; Halobacillus sp. S4B(2010), HQ433444; Halobacillus naozhouensis isolate 0612R1-8.2, HG515393; Halobacillus naozhouensis isolate 0612P19-10, HG515391; Halobacillus sp. JC256, HG974547; Halobacillus sp. JC250, HG974541; Uncultured Alicyclobacillus sp. clone METASED16, KJ907769; Alicyclobacillus sp. PS1, KF193534; Alicyclobacillus sp. PSS3, KF193533; Alicyclobacillus sp. CPP55, KF193532; Alicyclobacillus cellulosilyticus, AB622813; Aneurinibacillus thermoaerophilus strain Billoris-VO22, KJ913706; Aneurinibacillus thermoaerophilus strain Billoris-VO18, KJ913705; Aneurinibacillus thermoaerophilus strain Billoris-VO16, KJ913703; Aneurinibacillus thermoaerophilus strain VO26, KJ842104; Aneurinibacillus thermoaerophilus strain VO25, KJ842103
Bacillus sp. LZR216 affects Arabidopsis seedling growth
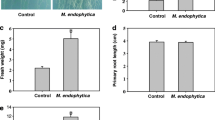
Because Bacillus sp. LZR216 is an endophytic bacterium, we proposed that LZR216 may promote Arabidopsis growth. To verify this hypothesis, Arabidopsis seeds were germinated and vertically grown for 5 days on petri dishes containing agar-solidified 1/2 MS medium. After that, the seedlings were transferred to a new 1/2 MS medium spotted with 20 µL distilled sterilized water (as the control) or 20 µL 5 × 108 CFU mL−1 LZR216 (OD600 = 0.5) suspension. Twelve days later, a significant increase in shoot biomass and total leaf surface area was observed in LZR216-treated seedlings compared to the control (Fig. 2), indicating that Bacillus sp. LZR216 can promote Arabidopsis shoot growth.
Effects of Bacillus sp. LZR216 inoculation on the growth of Arabidopsis seedlings. a Representative images of 17-day-old seedlings after 12 days of growth under control and LZR216-inoculated conditions. b Effects of Bacillus sp. LZR216 inoculation on shoot fresh weight. c Effects of Bacillus sp. LZR216 inoculation on total leaf surface area. Data are mean ± SE of three independent experiments (n = 10). Asterisks indicate statistically significant differences compared with the control (Student’s t test; P < 0.05). The experiments were repeated three times with similar results
It has been shown that many PGPB strains could cause alterations in the root system architecture of host plants by promoting the formation of secondary roots and thus improving the root exploratory capacity (Zamioudis et al. 2013). The effect of LZR216 on root system architecture of Arabidopsis was tested. As shown in Fig. 3a, b, the primary root length of seedlings exposed to LZR216 was reduced by approximately 37 % after inoculation with LZR216 for 1 week compared to the control, suggesting a suppressive effect of LZR216 on the primary root elongation. Furthermore, in the time course experiment, we found that the inhibition of LZR216 on the primary root elongation was significantly exhibited after 6-day treatment (Fig. 3e). By the contrary, LZR216 observably enhanced both the number and the density of lateral roots, with more than six- and ninefold increase, respectively, after 7-day treatment (Fig. 3c, d). With the growth time elongation, the lateral root number as well as the density increased rapidly in LZR216-treated seedlings, but only slightly in control plants (Fig. 3f, g).
Effects of LZR216 on root system architecture in Arabidopsis seedlings. a Representative images of 12-day-old seedlings after 7 days of growth under control and LZR216-inoculated conditions. b Effects of LZR216 on primary root length. c Effects of LZR216 on lateral root numbers. d Effects of LZR216 on lateral root density. e Time course of primary root length in response to LZR216. f Time course of lateral root numbers in response to LZR216. g Time course of lateral root density in response to LZR216. In e, f, and g, at time point 0, LZR216 bacteria were spotted on the plates. Data are mean ± SE of three independent experiments (n = 30). Asterisks indicate statistically significant differences compared with control treated (Student’s t test; P < 0.05). The experiments were repeated three times with similar results
LZR216 negatively affects cell division in Arabidopsis roots
To clarify the mechanism by which LZR216 restrains the primary root growth, the effect of LZR216 on quiescent center (QC) was first analyzed. It was found that LZR216 has little effect on the expression of QC-localized marker WOX5-GFP in the meristem (Fig. 4a). Next, the effect of LZR216 on the cortical cell length was measured in the elongation zone (EZ) of primary roots. The results showed that LZR216 can markedly induce the elongation of cortical cells (Fig. 4b, c), suggesting a positive role of such endophytic microbe on cortical cell elongation. Meanwhile, meristematic zone length was also measured. Interestingly, the meristematic zone in LZR216-treated roots is significantly shorter than that in the control (Fig. 4d, e), suggesting that LZR216 inhibits the cell division. Consistently, in transgenic plant CYCB1;1::GUS, one cell cycle marker, LZR216 obviously represses the GUS activity compared to the control (Fig. 4f). Taken together, these data indicate that LZR216 inhibition of primary root may mainly result from its negative effect on cell division.
Effects of LZR216 on WOX5-GFP, CYCB1;1::GUS and meristem size of Arabidopsis roots. a Confocal images showing the expression patterns of the QC-localized marker WOX5-GFP under control and LZR216-inoculated conditions. b Cortical cell elongation in the elongation zone (EZ) of roots grown in the presence or absence of LZR216. c Quantitative graphs of b. d Meristem size of control- and LZR216-treated roots. e Quantitative graphs of d. f Expression of cell division marker CYCB1;1::GUS in the primary root meristem of control plants and LZR216-inoculated plants. Data are mean ± SE of three independent experiments (n = 30). Asterisks indicate statistically significant differences compared with the control (Student’s t test; P < 0.05). The experiments were repeated three times with similar results
Auxin is involved in LZR216 regulation of root system architecture
Phytohormone auxin plays an important role in the regulation of root system architecture, including the increase of lateral root and root hair development (Casimiro et al. 2001; Reed et al. 1998). The above results indicate that LZR216 inhibits the primary root elongation but enhances the lateral root formation. So it is reasonable to investigate whether auxin plays any roles in the LZR216 regulation of root growth. Firstly, using transgenic plants, expressing the synthetic auxin reporters DR5::GUS and DR5::GFP, we found that a strong auxin response was detected toward the root tips in the root meristem of primary roots (Fig. 5a), suggesting an induction role of LZR216 on auxin response. To test whether LZR216 affects auxin biosynthesis at the transcriptional level, the transcripts of NIT1, TAA1, and YUCCA1, key genes in auxin biosynthesis, were tested using qRT-PCR. The results indicated that LZR216 treatment notably induces the transcriptional levels of NIT1, TAA1, and YUCCA1 compared to the control (Fig. 5b). Hence, these data indicated that the LZR216 enhancement of auxin responses may be achieved by, at least partially, enhancing the auxin biosynthesis at the transcriptional level.
Effects of LZR216 on auxin level in Arabidopsis roots. a Representative images of DR5::GUS (bar 30 μm) and DR5::GFP (bar 50 μm) with or without LZR216 treatment. b Quantitative real-time PCR analysis of the relative transcript abundance of NIT1, TAA1 and YUCAA1 genes in WT seedlings. 5-day-old seedlings were treated with 20 µL 5 × 108 CFU mL−1 suspension in water (20 µL sterilized water as control) for 7 days. All gene transcripts were normalized to ACTIN2. Data are mean ± SE of three independent experiments. Asterisks indicate statistically significant differences compared with control treated (Student’s t test; P < 0.05). The experiment was repeated three with similar results
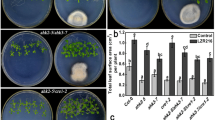
To determine how LZR216 affects auxin responses in the regulation of Arabidopsis root system, effect of LZR216 on the growth of four auxin signaling mutants was tested. Through measuring the primary root length, lateral root number per plant, lateral root density, shoot fresh weight, and total leaf surface area per seedling, we found that the responses of these mutants to LZR216 were more or less impaired compared to wild type. For example, after LZR216 treatment, the lateral root number per plant was increased with about threefold in wild type, but only with 1.7-, 1.6-, 1.1-, and 2.5-fold in mutants tir1-1, pin2, aux1-7, and axr1-12, respectively (Fig. 6). These data suggest that auxin signaling and polar transport are necessary for the LZR216 effect on plant growth.
Influence of auxin signaling and transport on LZR216-mediated primary root length and LR formation in Arabidopsis roots. a Primary root length under control and LZR216-inoculated conditions. b Lateral root length under control and LZR216-inoculated conditions. c Lateral root density under control and LZR216-inoculated conditions. d Shoot fresh weight under control and LZR216-inoculated conditions. e Total leaf surface area under control and LZR216-inoculated conditions. Data are mean ± SE of three independent experiments (n = 30), bars with different letters are significantly different at the level of P < 0.05. The experiments were repeated three times with similar results
LZR216 regulation of root system architecture is likely attributed by affecting polar auxin transport
Given the necessity of auxin polar transport for LZR216-mediated plant growth, the expression levels of AUX1 and PIN2 together with other two efflux carriers PIN1 and PIN3 were tested using transgenic plants AUX1-YFP, PIN2-GFP, PIN1-GFP, and PIN3-GFP, respectively, in the presence or absence of LZR216 treatment. LZR216 significantly reduces the expressions of these four proteins compared to the control (Fig. 7), suggesting LZR216-induced alteration of the root system architecture involves polar auxin transport. To further confirm this notion, we used the specific inhibitor of polar auxin transport, NPA. As shown in Fig. 8a, b, the LZR216-induced increase of total leaf surface area per seedling was significantly impaired in the presence of 1 or 10 µM NPA, suggesting that LZR216-mediated growth promotion is interrupted if the auxin transport is blocked. Similarly, in the absence of NPA, LZR216 represses the growth of primary roots; but in the presence of NPA, with concentrations from 0.5 to 5 µM, the primary root length of LZR216-treated seedlings becomes comparable with the control plants (Fig. 8c). Furthermore, the ability of LZR216-stimulated LR formation is severely attenuated by 0.5 or 1 µM NPA. The LR formation in response to LZR216 is almost completely abolished after 5 µM NPA treatment (Fig. 8d, e). Together, we concluded that polar auxin transport is necessary for LZR216 in the regulation of plant growth.
Effect of LZR216 on the protein level of PIN1/PIN2/PIN3/AUX1. a Five-day-old seedlings containing indicated markers were transferred to medium with or without LZR216 for 7 days and GFP expression was monitored. b Quantification of GFP/YFP fluorescence by image analysis of confocal sections as described in a. Images shown are representatives of at least three independent experiments. Bars 50 µm
Effects of the polar auxin transport inhibitor NPA on LZR216-promoted Arabidopsis growth and LZR216-altered root system architecture. a Photograph of 17-day-old Arabidopsis (Col-0) seedlings grown on the surface of 1/2 MS medium containing different concentrations of NPA. 5-day-old seedlings were treated with 20 µL autoclaved water or with 20 µL LZR216 bacterial suspension for 12 days and photographed. Bar 1 cm. b Total leaf surface area under control, and LZR216-induced conditions in different concentrations of NPA after 12 days of co-cultivation. c Primary root length under control and LZR216-induced conditions in different concentrations of NPA after 7 days of co-cultivation. d Lateral root formation under control and LZR216-induced conditions in different concentrations of NPA after 7 days of co-cultivation. e Lateral root density under control and LZR216-induced conditions in different concentrations of NPA after 7 days of co-cultivation. Data are mean ± SE of three independent experiments (n = 30). Within each set of experiments, bars with different letters are significantly different at the level of P < 0.05
Discussion
In natural ecosystems, the environment influences plant growth and development. Some beneficial microbes can enhance stress resistance against various adverse environment conditions (Berendsen et al. 2012). Rhizosphere microorganisms have significant effects on plant performance in agricultural and marginal soils by influencing growth and development of roots and improving nutrient availability in the rhizosphere (Tinker 1984; Rodríguez and Fraga 1999; Chaiharn et al. 2009). Most studies on mechanisms for plant growth promotion by PGPR are focused on bacterial traits without examining the detailed phenotypic, cellular, and physiological responses of host plants. This is because root architecture is difficult to observe, quantify, and/or interpret. Roots grow in soil, an opaque medium from which they cannot be extricated or readily observed without introducing artifacts and destroying the native root architecture. In recent years, significant progress has been made in our understanding of how the root system architecture alters in response to soil microbes. But studies on how endophytic microbes affect plant growth and development are still limited. In this work, we used Arabidopsis thaliana as a model system to characterize the phenotypic and physiological basis by which endophytic microbe Bacillus sp. LZR216 regulates plant growth and root system architecture.
PGPR have been extensively studied in respect of their capability to induce biomass production and systemic resistance in plants (Lugtenberg and Kamilova 2009; Pozo and Azcón-Aguilar 2007). Our results indicated that endophytic microbe Bacillus sp. LZR216 stimulates the growth of Arabidopsis (Fig. 2), suggesting that this bacterial strain likely acts as a novel plant growth-promoting bacterium. Strains of B. subtilis and Enterobacter cloacae promote plant growth by releasing several volatiles, of which acetoin shows the highest level of growth promotion (Ryu et al. 2003). Previously, it was shown that Pseudomonas fluorescens WCS417 enhances the shoot fresh weight and alters the root system architecture (Zamioudis et al. 2013). Similarly, our results showed that LZR216 inhibits the primary root elongation and promotes the lateral root formation (Fig. 3). There was a report that the expression of CYCB1;1::GUS is clearly reduced in plants inoculated with Bacillus megaterium (López-Bucio et al. 2007). Our microscopic survey showed that LZR216 decreases the expression of the CYCB1;1::GUS reporter in the meristematic zone of root tips and reduces the meristem zone length, and enhances the length of cells entering the elongation zone (Fig. 4). These results imply that LZR216 inhibits cell division and the meristem zone length.
Auxin is an essential plant hormone in the maintenance of stem cell identity and function. Both primary root elongation and lateral root formation require auxin transport. Transcripts of GH3 genes which are involved in IAA conjugation, PtaGH3-1, PtaGH3-2, PtaGH3-7, and PtaGH3-8, were induced in populus with Laccaria bicolor co-culture (Felten et al. 2009). Similarly, Phyllobacterium brassicacearum strain STM196 induces the expression of DR5::GUS in Arabidopsis primary as well as lateral root tips (Contesto et al. 2010). Our results showed that LZR216 enhances DR5::GUS and DR5::GFP levels in primary root tips (Fig. 5a), suggesting that auxin responses can be strengthened by the inoculation of this endophytic microbe. Furthermore, our results indicated that LZR216 treatment notably induces the transcriptional levels of NIT1, TAA1, and YUCCA1 (Fig. 5b), suggesting that LZR216 also regulates auxin biosynthesis at the transcriptional level. On the other hand, the effect of LZR216 was impaired in several mutants defective in auxin signaling or polar transport (Fig. 6). Ryu and co-workers (2005) also reported that the Arabidopsis growth promotion by several PGPR involves complex hormonal signaling. More recently, growth responses to Trichoderma virens or Phyllobacterium brassicacearum inoculation are reduced in axr1-3 compared to wild type (Contesto et al. 2010; Contreras-Cornejo et al. 2009). So affecting plant growth through auxin modulation may be a universal pathway for some endophytic microbes, including LZR216.
Auxin influx and efflux transporter genes in populus are transiently induced by Laccaria bicolor co-culture (Felten et al. 2009). In Arabidopsis, when co-cultured with Laccaria bicolor, aux1, and pin3 mutants exhibited similar induction of lateral roots compared to wild type, but less stimulation was observed in the pin2 and pin2,3,4,7 mutant lines (Felten et al. 2009). These aforementioned results provide evidence that auxin transport machinery is involved in plant–microbe interactions. This study on LZR216-mediated root development provides important information on the mechanism by which candidate molecules with auxin activity are perceived and transported within the roots. Our results indicated that LZR216 can still inhibit the primary root elongation in tir1-1 and axr1-12 mutants, with similar extent exhibited in wild type (Fig. 6a), which may be because TIR1 is just one of auxin receptors, whereas AXR1 is not important for LZR216 inhibition of the primary root elongation. LZR216 has little effect on the primary root elongation of pin2 and aux1-7 mutants (Fig. 6a), indicating that polar auxin transport might be the key factor for LZR216 in affecting the root development of Arabidopsis seedlings. LZR216 inhibition of PIN1, PIN2, PIN3, and AUX1 accumulation in root tips strengthens this viewpoint. Moreover, NPA eliminated the roles of LZR216 on promoting plant growth and affecting root system architecture (Fig. 8), further suggesting that the LZR216 function in plants is very likely through the interference of the polar auxin transport.
Microorganisms interact with plants either beneficially or as pathogens and may influence plant growth and development. Perception of microorganisms by plants is highly coordinated through cellular processes that determine the final outcome of the relationship, ranging from parasitism to mutualism (Bais et al. 2004; Pozo and Azcón-Aguilar 2007). Important biological processes mediated by root exudates, such as communication with symbiotic microorganisms and alterations in chemical and physical properties of the soil justify such an energy investment (Bais et al. 2004, 2006). In this work, we used an endophytic microbe to study its effects on plant growth and development. We can understand how plant growth and development is affected when endophytic microbes overgrow. It is expected that endophytic microbe-induced alterations in the root system architecture would greatly facilitate plant nutrition, thereby conferring significant ecological benefits to host plants.
Author contribution statement
The experiments were conceived and designed by Yurong Bi, Jianfeng Wang, Yongqiang Zhang, and Ying Li. The experiments were performed by Jianfeng Wang, Yongqiang Zhang, Ying Li, Yanfeng Hu, Wenbin Nan, and Hong Zhang. The data were analyzed by Jianfeng Wang, Feng Wang, Ping Li, Chengzhou Zhao, and Hongyong Shi. Yurong Bi, Jianfeng Wang, Yongqiang Zhang, and Xiaomin Wang wrote the paper.
Abbreviations
- CFU:
-
Colony-forming units
- GFP:
-
Green fluorescent protein
- GUS:
-
β-Glucuronidase
- KB:
-
King’s B medium
- NPA:
-
1-N-Naphthylphthalamic acid
- PAT:
-
Polar auxin transport
- PGPB:
-
Plant growth-promoting bacteria
- PGPR:
-
Plant growth-promoting rhizobacteria
- QC:
-
Quiescent center
- YFP:
-
Yellow fluorescent protein
References
Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100:2992–2997
Bais HP, Park S-W, Weir TL, Callaway RM, Vivanco JM (2004) How plants communicate using the underground information superhighway. Trends Plant Sci 9:26–32
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266
Barazani O, Friedman J (1999) Is IAA the major root growth factor secreted from plant-growth-mediating bacteria? J Chem Ecol 25:2397–2406
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Berg G, Smalla K (2012) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Bowen GD, Rovira AD (1999) The rhizosphere and its management to improve plant growth. Adv Agron 66:1–102
Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, Bennett M (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13:843–852
Chaiharn M, Chunhaleuchanon S, Lumyong S (2009) Screening siderophore producing bacteria as potential biological control agent for fungal rice pathogens in Thailand. World J Microbiol Biotechnol 25:1919–1928
Combes-Meynet E, Pothier JF, Moënne-Loccoz Y, Prigent-Combaret C (2011) The Pseudomonas secondary metabolite 2, 4-diacetylphloroglucinol is a signal inducing rhizoplane expression of Azospirillum genes involved in plant-growth promotion. Mol Plant Microbe Interact 24:271–284
Contesto C, Milesi S, Mantelin S, Zancarini A, Desbrosses G, Varoquaux F, Bellini C, Kowalczyk M, Touraine B (2010) The auxin-signaling pathway is required for the lateral root response of Arabidopsis to the rhizobacterium Phyllobacterium brassicacearum. Planta 232:1455–1470
Contreras-Cornejo HA, Macías-Rodríguez L, Cortés-Penagos C, López-Bucio J (2009) Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol 149:1579–1592
Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moënne-Loccoz Y (2009) Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol 48:505–512
Dobbelaere S, Croonenborghs A, Thys A, Broek AV, Vanderleyden J (1999) Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 212:153–162
Felten J, Kohler A, Morin E, Bhalerao RP, Palme K, Martin F, Ditengou FA, Legué V (2009) The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol 151:1991–2005
Jing YD, He ZL, Yang XE (2007) Role of soil rhizobacteria in phytoremediation of heavy metal contaminated soils. J Zhejiang Univ Sci B 8:192–207
Keller CP, Stahlberg R, Barkawi LS, Cohen JD (2004) Long-term inhibition by auxin of leaf blade expansion in bean and Arabidopsis. Plant Physiol 134:1217–1226
Kloepper JW, Leong J, Teintze M, Schroth MN (1980) Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286:885–886
Kloepper JW, Lifshitz R, Zablotowicz RM (1989) Free-living bacterial inocula for enhancing crop productivity. Trends Biotechnol 7:39–44
López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farías-Rodríguez R, Macías-Rodríguez LI, Valencia-Cantero E (2007) Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin-and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol Plant-Microbe Interact 20:207–217
Lucas JA, García-Cristobal J, Bonilla A, Ramos B, Gutierrez-Mañero J (2014) Beneficial rhizobacteria from rice rhizosphere confers high protection against biotic and abiotic stress inducing systemic resistance in rice seedlings. Plant Physiol Biochem 82:44–53
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Mayak S, Tirosh T, Glick BR (2004) Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol Biochem 42:565–572
Patten CL, Glick BR (2002) Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol 68:3795–3801
Persello-Cartieaux F, Nussaume L, Robaglia C (2003) Tales from the underground: molecular plant-rhizobacteria interactions. Plant Cell Environ 26:189–199
Pitts RJ, Cernac A, Estelle M (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16:553–560
Pozo MJ, Azcón-Aguilar C (2007) Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol 10:393–398
Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S (2002) Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol 130:1908–1917
Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI (2007) Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J 50:514–528
Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118:1369–1378
Richardson A, Barea J-M, McNeill A, Prigent-Combaret C (2009) Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321:305–339
Rodríguez H, Fraga R (1999) Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv 17:319–339
Ryu C-M, Farag MA, Hu C-H, Reddy MS, Wei H-X, Paré PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100:4927–4932
Ryu C-M, Hu C-H, Locy R, Kloepper J (2005) Study of mechanisms for plant growth promotion elicited by rhizobacteria in Arabidopsis thaliana. Plant Soil 268:285–292
Tak H, Ahmad F, Babalola O (2013) Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. Rev Environ Contam Toxicol 223:33–52
Tinker PB (1984) The role of microorganisms in mediating and facilitating the uptake of plant nutrients from soil. Plant Soil 76:77–91
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971
Walker V, Couillerot O, Von Felten A, Bellvert F, Jansa J, Maurhofer M, Bally R, Moënne-Loccoz Y, Comte G (2012) Variation of secondary metabolite levels in maize seedling roots induced by inoculation with Azospirillum, Pseudomonas and Glomus consortium under field conditions. Plant Soil 356:151–163
Zamioudis C, Mastranesti P, Dhonukshe P, Blilou I, Pieterse C (2013) Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol 162:304–318
Zhang H, Kim M-S, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu C-M, Allen R, Melo IS (2007) Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839–851
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31170225; 31201145), Foundation of Science and Technology Program of Gansu Province (1208RJZA224), Foundation of Science and Technology Program of Gansu Province (1107RJYA005), the National High Technology Research and Development Program (2007AA021401), National Program on Key Basic Research Project (2012CB026105), and Fundamental Research Funds for the Central Universities (lzujbky-2012-104).
Conflict of interest
The content in this paper is new. It is not being submitted to any other journal. All the authors listed have approved the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Chong.
J. Wang and Y. Zhang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, J., Zhang, Y., Li, Y. et al. Endophytic microbes Bacillus sp. LZR216-regulated root development is dependent on polar auxin transport in Arabidopsis seedlings. Plant Cell Rep 34, 1075–1087 (2015). https://doi.org/10.1007/s00299-015-1766-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1766-0