Abstract
The aim of this study was to determine whether the functional integrin-α-M (ITGAM) rs1143679 polymorphism is associated with susceptibility to systemic lupus erythematosus (SLE), lupus nephritis (LN), and rheumatoid arthritis (RA). A series of meta-analyses were conducted to test for associations between the ITGAM rs1143679 polymorphism and SLE, LN, or RA. A total of 24 comparisons involving 7,738 patients and 8,309 controls for SLE, and 2,663 patients and 2,694 controls in RA were considered. Meta-analysis showed a significant association between the ITGAM rs1143679 A allele and SLE in all subjects (OR 1.773, 95 % CI 1.656, 1.901, p < 1.0 × 10−9). After stratification by ethnicity, the A allele was found to be significantly associated with SLE in European, Latin American, and Asian. A significant association was also found between the ITGAM A allele and lupus nephritis in Europeans (OR 2.131, 95 % CI 1.565, 2.903, p = 1.6 × 10−7). However, no association was found between RA and the ITGAM rs1143679 polymorphism. Our meta-analyses confirm that the ITGAM rs1143679 polymorphism is associated with SLE susceptibility in different ethnic groups and demonstrate that the polymorphism is associated with LN in European.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic lupus erythematosus (SLE) is a prototypical autoimmune disease in which immune regulation is disrupted and which is characterized by multisystem involvement mediated by autoantibodies and immune complex deposits. Rheumatoid arthritis (RA) is a chronic inflammatory disease that predominantly involves synovial joints and affects up to 1 % of adults worldwide. Although the etiologies of RA and SLE are not fully understood, it is clear that genetic components play a role in the development of both diseases [1, 2].
The integrin-α-M (ITGAM, also known as CD11B) gene located at chromosome 16p11.2 encodes the CD11b chain of a leukocyte-specific integrin (Mac-1, alpha M beta 2, CR3). CR3/Mac-1 modulates leukocyte adhesion and migration and plays a key role in the phagocytosis of complement-coated particles such as apoptotic cells [3]. R77H (histidine) variant protein, which corresponds to the ITGAM rs1143679 G/A polymorphism, contains histidine instead of arginine at position 77 in the extracellular domain of the CD11b integrin. This amino acid substitution significantly impairs the ability of the integrin to mediate cell adhesion to ligands and severely reduces phagocytosis of complement-coated particles [4, 5].
The functional ITGAM rs1143679 polymorphism has been reported to be associated with SLE, lupus nephritis (LN), and RA in different ethnic groups, although these findings are not consistent across all studies [6–15]. Gene allele frequencies often differ substantially between populations, and ethnic group-specific association studies are therefore required to confirm genetic associations in different populations. In order to overcome the limitations of individual studies, resolve inconsistencies, and reduce the likelihood of false-positive or false-negative associations due to random error, we performed meta-analysis [16–18]. In the present study, we used meta-analysis to investigate whether the functional ITGAM rs1143679 polymorphism contributes to disease risk of SLE, LN, or RA in different populations.
Materials and methods
Identification of eligible studies and data extraction
We performed a search for studies that examined associations between the ITGAM rs1143679 polymorphism and SLE and RA. The literature was searched using the MEDLINE and EMBASE citation databases to identify articles published from January 2000 to December 2013 in which the ITGAM polymorphism was analyzed in SLE or RA patients. Search strategy was as follows: (“antigens, cd11b”[MeSH Terms] OR (“antigens”[All Fields] AND “cd11b”[All Fields]) OR “cd11b antigens”[All Fields] OR “itgam”[All Fields]) AND (“polymorphism, genetic”[MeSH Terms] OR (“polymorphism”[All Fields] AND “genetic”[All Fields]) OR “genetic polymorphism”[All Fields] OR “polymorphism”[All Fields]) AND ((“lupus erythematosus, systemic”[MeSH Terms] OR (“lupus”[All Fields] AND “erythematosus”[All Fields] AND “systemic”[All Fields]) OR “systemic lupus erythematosus”[All Fields] OR (“systemic”[All Fields] AND “lupus”[All Fields] AND “erythematosus”[All Fields])) OR (“arthritis, rheumatoid”[MeSH Terms] OR (“arthritis”[All Fields] AND “rheumatoid”[All Fields]) OR “rheumatoid arthritis”[All Fields] OR (“rheumatoid”[All Fields] AND “arthritis”[All Fields]))). References in identified studies were also investigated to identify additional studies not indexed by MEDLINE and EMBASE. No language or country restrictions were applied. Studies were included if (1) they were case–control studies of the association between the ITGAM rs1143679 polymorphism and SLE or RA, (2) the data were original (to ensure independence among studies), and (3) they provided enough data to calculate odds ratios (OR). We excluded the following: (1) studies that contained overlapping data, (2) studies in which the number of the allele could not be ascertained, and (3) studies in which family members were studied, as such analyses are based on linkage considerations. Data were extracted from original studies by two independent reviewers. Any discrepancy between the reviewers was resolved by consensus or a third reviewer. The following information was extracted for each study: author, year of publication, ethnicity of the study population, demographics, the number of cases and controls, and genotype or allele frequency information for the ITGAM rs1143679 polymorphism. Allele frequencies were calculated from the corresponding genotype distributions.
Evaluation of statistical analyses
The Chi-square test was used to determine whether the observed genotype frequencies in controls conformed to Hardy–Weinberg (H–W) expectations. We explored the association between the ITGAM rs1143679 polymorphism and risk of disease (SLE or RA) using the following allelic contrast models: A (the variant allele) versus G (the common allele), the AA genotype versus the AG + GG genotype, and the AA + AG genotype versus the GG genotype. The latter two correspond to the recessive and dominant effects of the A allele, respectively. We also performed meta-analyses using the following additive models: the AA genotype versus the GG genotype and the AG genotype versus the GG genotype. Point estimates of risk, ORs, and 95 % confidence intervals (CIs) were obtained for each study. Cochran’s Q statistic was used to assess within- and between-study variation and heterogeneity [19]. The heterogeneity test was used to assess the probability of the null hypothesis that all studies were evaluating the same effect. The random-effects model was used for meta-analysis when a significant Q statistic (p < 0.10) indicated heterogeneity across studies, while the fixed-effect model was used when heterogeneity was not indicated. The fixed-effect model assumes that genetic factors have similar effects on disease susceptibility across all studies and that observed variations between studies are caused by chance alone [20]. The random-effects model assumes that studies show substantial diversity, and assesses both within-study sampling errors and between-study variances [21]. The random-effects model is used in the presence of significant between-study heterogeneity. We quantified the effect of heterogeneity by using the recently developed I 2 measure, where \( I^{2} = 100\,\% \times (Q - df)/Q \) [22]. The I 2 measure ranges between 0 and 100 %, and it represents the proportion of inter-study variability attributable to heterogeneity rather than chance. I 2 values of 25, 50, and 75 % were defined as low, moderate, and high estimates, respectively. Statistical manipulations were performed using the comprehensive meta-analysis computer program (Biosta, Englewood, NJ, USA).
Evaluation of heterogeneity and publication bias
While funnel plots are often used to detect publication bias, this technique requires a range of studies of varying sizes and involves subjective judgment. Accordingly, we evaluated publication bias by using Egger’s linear regression test [23], which measures funnel plot asymmetry using a natural logarithm scale of ORs.
Results
Studies included in the meta-analysis
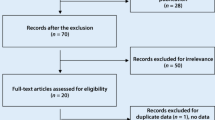
We identified 58 articles by electronic and manual searching and selected 20 of these for a full-text review based on title and abstract details. After the full-text review, 10 of these 20 articles were excluded. Six of these studies did not contain genotype or allele data for the ITGAM polymorphism [24–29], and four contained duplicate data [29–32]. We contacted corresponding authors of the original articles to ask for the raw data. However, one Korean study revealed no polymorphic data [24], and we could not get available genotype or allele data from other authors. Of the remaining studies, one study had data for nine different groups [8], one study contained data for five groups [10], one study contained data for three groups [11], and one study contained data for two groups [9]. Groups from different studies were treated independently. We excluded a set of European American subjects from one study [10] to avoid possible data overlap with another set of subjects [11]. We analyzed 24 separate comparisons in total, including 21 comparisons for SLE and three comparisons for RA (Fig. 1). The analyses included 7,738 patients and 8,309 controls for SLE, and 2,663 patients and 2,694 controls for RA [6–15]. The SLE analyses included 13 Europeans, four Latin Americans, two Asian populations, one African American, and one Gullah population; the RA analyses included two European populations and one Latin American population (Table 1). We performed a subgroup meta-analysis if at least two studies were available. Accordingly, ethnicity-specific meta-analyses were also conducted for European, Latin American, and Asian populations. Selected characteristics of these studies are summarized in Table 1.
Meta-analysis of the association between the ITGAM rs1143679 polymorphism and SLE
Meta-analysis was performed on all SLE patients and on SLE patients in each ethnic group. A significant association was observed between the A allele and SLE in all subjects (OR 1.773, 95 % CI 1.656, 1.901, p < 1.0 × 10−9; Table 2; Fig. 2). After stratification by ethnicity, the A allele was found to be significantly associated with SLE in European, Latin American, and Asian (Table 2; Fig. 3). An association was observed between the AA + AG genotype and SLE in all subjects (OR 1.856, 95 % CI 1.682, 2.048, p < 1.0 × 10−9; Table 2). After stratification by ethnicity, associations between the AA + AG genotype and SLE were observed in Europeans and in Latin Americans (Table 2). Furthermore, the ITGAM rs1143679 polymorphism was found to be associated with SLE in Europeans under the recessive and additive models, and under homozygote contrast (Table 2).
Meta-analysis of the association between the ITGAM rs1143679 polymorphism and lupus nephritis
Three studies were included in the meta-analysis of the association between the ITGAM rs1143679 polymorphism and LN [6, 7, 12] (Table 3). Meta-analysis showed an association between the A allele of the ITGAM polymorphism and LN in Europeans (OR 2.131, 95 % CI 1.565, 2.903, p = 1.6 × 10−7; Table 3; Fig. 4). However, one Latin American study showed no evidence of association between the A allele and LN in Latin Americans (OR 1.440, 95 % CI 0.961, 2.157, p = 0.077; Table 3; Fig. 3).
Meta-analysis of the association between the ITGAM rs1143679 polymorphism and RA
Three studies were included in the meta-analysis of the association between the ITGAM rs1143679 polymorphism and RA [13–15] (Table 3). Meta-analysis showed no evidence of association between the A allele and RA, either in the overall study population or in Europeans (OR 1.078, 95 % CI 0.967, 1.191, p = 0.177; OR 1.064, 95 % CI 0.950, 1.191, p = 0.287; Table 3; Fig. 4).
Heterogeneity and publication bias
Within the control groups, the genotype distributions of the ITGAM rs1143679 polymorphism were consistent with H–W equilibrium in all studies. Between-study heterogeneity was observed in meta-analyses of AA vs. AG + GG and AA vs. GG (Table 3). However, the heterogeneity was resolved when the analysis was stratified by ethnicity. It was difficult to interpret the funnel plot, which is used to detect publication bias, because the number of studies included in the analysis was relatively small. However, Egger’s regression test showed no evidence of publication bias in the meta-analyses of association between the ITGAM rs1143679 polymorphism and susceptibility to SLE, LN, or RA (Egger’s regression test p > 0.1; Fig. 5).
Discussion
In the present study, we subjected previously published data to meta-analysis to evaluate the genetic association between the ITGAM rs1143679 polymorphism and susceptibility to SLE and RA in different ethnic populations. The prevalence of the A allele of this polymorphism was calculated for different populations, and an ethnic-specific meta-analysis was performed. The prevalence of the A allele was found to vary among ethnic groups, from 0.7 % in Asians to 12.4 % in Europeans. The mean frequency in all controls was 8.8 %. Interestingly, the A allele was not observed in Korean or Japanese populations, consistent with the idea that associations between diseases and the ITGAM rs1143679 polymorphism are dependent on the genotype frequencies in different ethnic groups [10, 24]. Our ethnic-specific meta-analysis shows that an association exists in European, Latin American, and Asian populations. It has been suggested that homozygosity for the A allele is dose-dependently associated with the risk of diseases. Consistent with this, we observed the presence of the dose effect of the A allele in the populations analyzed. Unfortunately, ethnic-specific meta-analysis was not possible in African American and Hispanic populations due to limited data. Our meta-analysis found no association between the ITGAM rs1143679 polymorphism and RA in the overall study population or in Europeans, suggesting that ITGAM may be not a general autoimmune gene.
One possible explanation for the association between SLE risk and the ITGAM rs1143679 A allele (R77H variant) is that the decreased binding of CR3/Mac-1 to iC3b in this variant gives rise to defects in leukocyte trafficking and reduced phagocytosis. The R77H (histidine) variant of the protein is known to be less efficient at mediating cell adhesion and clearing complement-covered particles [4, 5]. Failure to adequately clear apoptotic cells or immune complexes may then lead to activation of dendritic cells, inflammatory cytokine production, as well as uptake and presentation of nuclear antigens to T cells [4, 33].
The results of our meta-analysis of the association of the ITGAM rs1143679 polymorphism with SLE risk are in good agreement with a similar meta-analysis performed by Fan et al. [34]. Compared with the previous study, our analyses included three additional studies on the ITGAM rs1143679 polymorphism, 586 more SLE patients, and 779 more controls [6, 7, 12]. One comparison study was excluded due to possible duplicate data [10]. We performed additional meta-analyses to examine potential association of the ITGAM rs1143679 polymorphism with disease risk for LN and for RA. We performed further ethnic-specific meta-analyses. Finally, meta-regression and sensitivity analyses were further performed to detect heterogeneity and potential publication bias.
Since common genetic variants are shared between RA and SLE, it is needed to explain why ITGAM rs1143679 polymorphism is associated only with SLE. First, recent study revealed that none of the nine recently identified SLE risk factors showed association with RA, suggesting that common genetic factors affecting the pathogenesis of these two disorders are limited [15]. Second, there is also a possibility that different polymorphisms in the same gene are involved in different diseases [35]. Third, large sample sizes were included in SLE studies, resulting in the identification of variants with even small effects, while small number of RA studies may be underpowered to detect the association.
The present study has some limitations that should be noted. Firstly, heterogeneity, confounding factors, and publication bias might have distorted the meta-analysis. Secondly, since our ethnic-specific meta-analyses included data only from European, Latin American, and Asian patients, our findings may be applicable only to these ethnic groups. Thirdly, as most of the available studies were performed in populations of European descent, further studies would be needed to understand disease risk associations in other ethnic populations. Finally, the number of available studies was too small to examine the association between the ITGAM rs1143679 polymorphism and RA, especially in different ethnic groups. Only three suitable ITGAM polymorphism studies could be identified for RA; two of these were conducted in European populations and one in a Latin American population.
In conclusion, our meta-analyses confirm that the ITGAM rs1143679 polymorphism is associated with susceptibility to SLE and LN in several ethnic groups, especially in European, and we found no evidence for association between the ITGAM rs1143679 polymorphism and susceptibility to RA. These findings suggest a need for further studies to examine associations between ITGAM polymorphisms and SLE, LN, and RA susceptibility in different ethnic groups. Larger scale studies in different ethnic populations will also help to elucidate the roles of specific polymorphisms of the ITGAM gene in the pathogeneses of SLE, LN, and RA.
References
Lee YH, Nath SK (2005) Systemic lupus erythematosus susceptibility loci defined by genome scan meta-analysis. Hum Genet 118(3–4):434–443
Choi SJ, Rho YH, Ji JD, Song GG, Lee YH (2006) Genome scan meta-analysis of rheumatoid arthritis. Rheumatology (Oxford) 45(2):166–170
Tan SM (2012) The leucocyte beta2 (CD18) integrins: the structure, functional regulation and signalling properties. Biosci Rep 32(3):241–269
MacPherson M, Lek HS, Prescott A, Fagerholm SC (2011) A systemic lupus erythematosus-associated R77H substitution in the CD11b chain of the Mac-1 integrin compromises leukocyte adhesion and phagocytosis. J Biol Chem 286(19):17303–17310
Rhodes B, Furnrohr BG, Roberts AL, Tzircotis G, Schett G, Spector TD et al (2012) The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Ann Rheum Dis 71(12):2028–2034
Toller-Kawahisa JE, Vigato-Ferreira IC, Pancoto JA, Mendes-Junior CT, Martinez EZ, Palomino GM et al (2014) The variant of CD11b, rs1143679 within ITGAM, is associated with systemic lupus erythematosus and clinical manifestations in Brazilian patients. Hum Immunol 75(2):119–123
Warchol T, Lianeri M, Lacki JK, Olesinska M, Jagodzinski PP (2011) ITGAM Arg77His is associated with disease susceptibility, arthritis, and renal symptoms in systemic lupus erythematosus patients from a sample of the Polish population. DNA Cell Biol 30(1):33–38
Suarez-Gestal M, Calaza M, Endreffy E, Pullmann R, Ordi-Ros J, Sebastiani GD et al (2009) Replication of recently identified systemic lupus erythematosus genetic associations: a case-control study. Arthritis Res Ther 11(3):R69
Yang W, Zhao M, Hirankarn N, Lau CS, Mok CC, Chan TM et al (2009) ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum Mol Genet 18(11):2063–2070
Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM et al (2009) Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE). Hum Mol Genet 18(6):1171–1180
Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS et al (2008) A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet 40(2):152–154
Jarvinen TM, Hellquist A, Koskenmies S, Einarsdottir E, Panelius J, Hasan T et al (2010) Polymorphisms of the ITGAM gene confer higher risk of discoid cutaneous than of systemic lupus erythematosus. PLoS One 5(12):e14212
Anaya JM, Kim-Howard X, Prahalad S, Chernavsky A, Canas C, Rojas-Villarraga A et al (2012) Evaluation of genetic association between an ITGAM non-synonymous SNP (rs1143679) and multiple autoimmune diseases. Autoimmun Rev 11(4):276–280
Phipps-Green AJ, Topless RK, Merriman ME, Dalbeth N, Gow PJ, Harrison AA et al (2009) No evidence for association of the systemic lupus erythematosus-associated ITGAM variant, R77H, with rheumatoid arthritis in the Caucasian population. Rheumatology (Oxford) 48(12):1614–1615
Suarez-Gestal M, Calaza M, Dieguez-Gonzalez R, Perez-Pampin E, Pablos JL, Navarro F et al (2009) Rheumatoid arthritis does not share most of the newly identified systemic lupus erythematosus genetic factors. Arthritis Rheum 60(9):2558–2564
Lee YH, Harley JB, Nath SK (2006) Meta-analysis of TNF-alpha promoter -308 A/G polymorphism and SLE susceptibility. Eur J Hum Genet 14(3):364–371
Nath SK, Harley JB, Lee YH (2005) Polymorphisms of complement receptor 1 and interleukin-10 genes and systemic lupus erythematosus: a meta-analysis. Hum Genet 118(2):225–234
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG (2006) Association of TNF-alpha -308 G/A polymorphism with responsiveness to TNF-alpha-blockers in rheumatoid arthritis: a meta-analysis. Rheumatol Int 27(2):157–161
Davey Smith G, Egger M (1997) Meta-analyses of randomised controlled trials. Lancet 350(9085):1182
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ 315(7121):1533–1537
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
Kim K, Brown EE, Choi CB, Alarcon-Riquelme ME, Biolupus, Kelly JA et al (2012) Variation in the ICAM1-ICAM4-ICAM5 locus is associated with systemic lupus erythematosus susceptibility in multiple ancestries. Ann Rheum Dis 71(11):1809–1814
Ramos PS, Criswell LA, Moser KL, Comeau ME, Williams AH, Pajewski NM et al (2011) A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet 7(12):e1002406
Chung SA, Taylor KE, Graham RR, Nititham J, Lee AT, Ortmann WA et al (2011) Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet 7(3):e1001323
Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP et al (2008) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 40(2):204–210
Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S et al (2008) Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med 358(9):900–909
Sanchez E, Webb RD, Rasmussen A, Kelly JA, Riba L, Kaufman KM et al (2010) Genetically determined Amerindian ancestry correlates with increased frequency of risk alleles for systemic lupus erythematosus. Arthritis Rheum 62(12):3722–3729
Hughes T, Adler A, Kelly JA, Kaufman KM, Williams AH, Langefeld CD et al (2012) Evidence for gene-gene epistatic interactions among susceptibility loci for systemic lupus erythematosus. Arthritis Rheum 64(2):485–492
Sanchez E, Nadig A, Richardson BC, Freedman BI, Kaufman KM, Kelly JA et al (2011) Phenotypic associations of genetic susceptibility loci in systemic lupus erythematosus. Ann Rheum Dis 70(10):1752–1757
Sanchez E, Comeau ME, Freedman BI, Kelly JA, Kaufman KM, Langefeld CD et al (2011) Identification of novel genetic susceptibility loci in African American lupus patients in a candidate gene association study. Arthritis Rheum 63(11):3493–3501
Fagerholm SC, MacPherson M, James MJ, Sevier-Guy C, Lau CS (2013) The CD11b-integrin (ITGAM) and systemic lupus erythematosus. Lupus 22(7):657–663
Fan Y, Li LH, Pan HF, Tao JH, Sun ZQ, Ye DQ (2011) Association of ITGAM polymorphism with systemic lupus erythematosus: a meta-analysis. J Eur Acad Dermatol Venereol 25(3):271–275
Orozco G, Abelson AK, Gonzalez-Gay MA, Balsa A, Pascual-Salcedo D, Garcia A et al (2009) Study of functional variants of the BANK1 gene in rheumatoid arthritis. Arthritis Rheum 60(2):372–379
Acknowledgments
This study was supported in part by a grant of the Korea Healthcare technology R&D Project, Ministry for Health and Welfare, Republic of Korea (HI13C2124).
Conflict of interest
The authors have no financial and non-financial conflicts of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, Y.H., Bae, SC. Association between the functional ITGAM rs1143679 G/A polymorphism and systemic lupus erythematosus/lupus nephritis or rheumatoid arthritis: an update meta-analysis. Rheumatol Int 35, 815–823 (2015). https://doi.org/10.1007/s00296-014-3156-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-014-3156-2