Abstract
Infundibulicybe trachyspora is described as a new species from northeastern China. The species is characterized by clitocyboid to omphalioid habit, carneous, greyish-yellow to brownish pileus, brown to dark reddish-brown, longitudinally fibrillose-striate stipe, non-amyloid, non-smooth spores, the absence of cystidia and the presence of clamp connections. A comprehensive description of the species is provided together with photo-illustrations and comparisons with phenotypically similar and phylogenetically related species. The nuclear ribosomal internal transcribed spacer (ITS) region and the nuclear, large subunit rDNA (nrLSU) region of the new species was sequenced and analyzed. The phylogenetic analysis supported the novelty of the species and its placement within the genus. Furthermore, a discussion on the proposal to establish a new section is made, and a key is provided for the Infundibulicybe species reported from China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Infundibulicybe Harmaja was initially proposed for Clitocybe sect. Infundibuliformes (Fr.) in the restricted delimitation used by Harmaja [1, 2]. The species of this genus are mainly characterized as slightly depressed to infundibuliform pileus, decurrent lamellae, smooth, inamyloid spores that are mostly lacrymoid in shape, abundant clamp connections and the lack of hymenial cystidia, and in particular, mycelia that cannot reduce nitrate and cyanophobic spore walls [2, 3]. Matheny et al. [4] recognized the genus in the Agaricales and as a sister group of the Tricholomatoid clade based on multilocus analysis of a six-gene region supermatrix was re-certified by Binder et al. [5]. Subsequently, Dentinger et al. proposed the suborder Tricholomatineae Aime, Dentinger & Gaya for four families, including Infundibulicybe species as “Incertae sedis” within it [6]. He et al. [7] also treated the genus as “Incertae sedis” suggesting it remained with an uncertain familial placement.
Recent molecular analysis has increased the accuracy of fungal identification and the accumulated sequence data also provide an advantage of promptly discovering new or rare species [8,9,10,11]. Presently, 26 species have been recognized in Infundibulicybe [2, 3, 12, 13]. In China, only 8 of those species have been identified (with some species under Clitocybe): Infundibulicybe alkaliviolascens (Bellù) Bellù, I. altaica (Singer) Harmaja (≡Clitocybe altaica Singer), I. geotropa (Bull.) Harmaja (=Clitocybe geotropa (Bull.) Quél.), I. gibba (Pers.) Harmaja [=Clitocybe gibba (Pers.) P. Kumm.], I. hongyinpan L. Fan & H. Liu, I. rufa Q. Zhao, K.D. Hyde, J.K. Liu & Y.J. Hao, I. subsalmonea (Lamoure) N. Schwab (≡Clitocybe subsalmonea Lamoure), I. trulliformis (Fr.) Gminder {=Clitocybe trulliformis (Fr.) P. Karst. [as 'trullaeformis']} [12,13,14,15,16,17,18]. The present study describes a new species, Infundibulicybe trachyspora, from northeastern China based on morphology and molecular phylogenetic analyses and documents Infundibulicybe species reported in China.
Materials and Methods
Sampling and Morphological Analysis
The basidiocarps were collected in Sep 2020 in the forests dominated by Larix olgensis of northeastern China (Jilin Province). Specimens were preserved and deposited in the Herbarium Mycology of Jilin Agricultural Science and Technology University (HMJU). Conventional macro-morphological characters and detailed anatomy of the specimens were recorded from fresh collections and photographic illustrations. Color names are given in parentheses according to the color chart by Kornerup and Wanscher [19]. For microscopic studies, sections of dried basidiocarps were rehydrated in 3% KOH, subsequently stained in Congo red solution, Melzer’s reagent and Cotton blue, and then observed under an Olympus BX 53 (Tokyo, Japan) light microscope (Nikon, Tokyo, Japan) at either 40, 100, 400, 600, and 1000 magnifications. To evaluate the range of basidiospore size, 20 basidiospores each from one specimen of every collection were measured. The notation [n/m/p] indicates that measurements were made on “n” randomly selected spores from “m” basidiocarps of “p” collections. Q = the quotient of length and width in any one basidiospore; Qm = average of Q. The procedure for scanning electron microscopy followed that of Xu et al. [20], and an FEI Quanta 200FE-SEM (JEOL Ltd., Japan) was used at an accelerating voltage of 5–10 kV.
DNA Extraction, PCR and Sequencing
Genomic DNA was isolated from dried specimens using an M5 Fungal Genomic DNA Kit (Mei5 Biotechnology Co., Ltd, Beijing, China) according to the manufacturer’s instructions. Polymerase chain reaction (PCR) and DNA sequencing were performed with the primers ITS1 and ITS4 [21] for the ITS region; LROR and LR7 [22] for the nrLSU region. The cycling parameters were as follows, ITS: 5 min at 94 ℃ for one cycle; 30 s at 94 ℃, 30 s at 53 ℃, 30 s at 72 ℃ for 33 cycles, 5 min at 72 ℃ for one cycle; nrLSU: 4 min at 94 ℃ for one cycle; 45 s at 94 ℃, 40 s at 46 ℃, 40 s at 72 ℃ for 30 cycles, 4 min at 72 ℃ for one cycle. The PCR amplification products were examined on a 1% agarose gel and checked by a JY 600 electrophoresis apparatus (Beijing JUNYI Electrophoresis Co., Ltd, Beijing, China). Sequencing was completed by BGI Co., Ltd, Beijing, China.
Phylogenetic Analyses
The newly generated sequences in this study were compared with the representative sequences in the GenBank database using the BLASTn algorithm. Since the accessible gene markers of Infundibulicybe species are limited in GenBank databases, this study mainly focuses on the ITS and nrLSU regions for phylogenetic analyses to obtain a representative topological structure. Based on the BLASTn results and outcomes of recent phylogenetic studies focused on Infundibulicybe [3, 12, 13, 18, 23,24,25], sequences were retrieved from GenBank databases, then were aligned automatically with MAFFT 7.0 [26] using ‘–auto’ strategy and normal alignment mode. Aligned sequences were visually inspected and manually adjusted using MEGA 7.0. ModelFinder [27] determined the best-fitted substitution model using the Akaike information criterion (AIC). GTR + F + I + G4 was chosen as the best model for both ITS and ITS-LSU analyses. Phylogenetic analyses using Bayesian Inference (BI) analyses and Maximum Likelihood (ML) analyses were subsequently conducted on MrBayes 3.2.2 [28] and IQ-TREE [29], respectively. Four incrementally heated simultaneous Monte Carlo Markov Chains (MCMC) were run over 10 M generations for the BI analyses. Trees were sampled every 1000 generations resulting in a broad sampling of 10,001 trees. The first 2500 trees were discarded as burn-in (25%). For the ML analyses, statistical supports were obtained using nonparametric bootstrapping with 1000 replicates. The resulting consensus trees were visualized using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). The significance threshold was set at ≥ 0.90 for Bayesian posterior probability (PP) and ≥ 70% for ML bootstrap proportions (BP). All the sequences used in this study are listed in Table 1.
Results
Phylogenetic Analyses
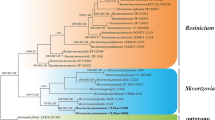
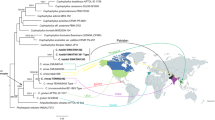
The ITS data matrix comprised 49 sequences (including 41 from GenBank). This dataset was 860 bp long and contained 404 (47.0%) variable sites. The combined ITS-LSU dataset matrix comprised 33 sequences (including 26 from GenBank). Both BI and ML approaches resulted in the same tree topology, as such only the ML trees are shown with Bayesian PP values (left) and MLBP values (right) for each node (Figs. 1, 2). Phylogenetic analysis based on ITS and ITS-nrLSU sequences showed similar results, the new species grouped in the genus Infundibulicybe and represented a distinct monophyletic lineage.
50% majority rule Bayesian phylogenetic analysis of Infundibulicybe based on ITS sequences, with Singerocybe species as outgroup taxa. Nodes were annotated if supported by ≥ 0.90 Bayesian PP (left) or ≥ 70% ML BP (right) values. For each sequenced taxon, the Genbank number is given. The new species are in red (Color figure online)
50% majority rule Bayesian phylogenetic analysis of Infundibulicybe based on ITS-nrLSU sequences, with Mycena species as outgroup taxa. Nodes were annotated if supported by ≥ 0.90 Bayesian PP (left) or ≥ 70% ML BP (right) values. For each sequenced taxon, the collection number/Specimen voucher number is given. The new species are in red (Color figure online)
Taxonomy
Infundibulicybe trachyspora J. Z. Xu, J. C. Qin & Yu Li, sp. nov.
Mycobank No. MB 842,075.
Type China, Jilin Province, Jilin City, Zuojia Town, (44°3′55′′ N, 126°6′8′′ E, elevation 349 m), on the ground under mixed forests dominated by Larix olgensis Henry, 1 Sep 2020, J.Z. Xu (holotype, HMJU 744, Genbank accession numbers: ITS, MW736885; LSU, MW880692; RPB1, OL677427; RPB2, OL677428).
Etymology The epithet derives from the non-smooth spores.
Diagnosis Distinguished by the clitocyboid to omphalioid habit, a carneous, greyish-yellow to brownish pileus, a brown to dark reddish-brown, longitudinally fibrillose-striate stipe, non-amyloid, non-smooth spores and the presence of clamp connections.
Description Basidiocarp clitocyboid to subomphalioid. Pileus 4.5–8 cm diam., at first plane to slightly depressed in the center, finally subinfundibuliform to umbilicate, sagging at the margin, carneous (3B3) then greyish-yellow to brownish (4B3), and dark brown (6D5) at the center. Surface dry, smooth, glabrous, some with a ring stripe, slightly appearing hygrophanous when water-soaked. Margin always involute, irregular, eroded. Lamellae decurrent, up to 0.3 cm broad, dark cream to yellowish-white (1A2, 1A3), moderately crowded, with lamellulae of 1 or 3 lengths, edges concolorous, entire. Stipe 3.5–7 × 0.5–0.75 cm, cylindrical, concolorous with the center of pileus or slightly lighter, brown to dark reddish-brown (6D5, 7E8), solid, some slightly twisty, surface with fibrous longitudinal stripes.
Basidiospores [60/6/3] 6.5–8.1(8.5) × (3.6)3.8–4.4(4.7) μm, Q = (1.69)1.71–1.87(1.95), Qm = 1.78, cyanophilous, lacrymoid to shortly ellipsoid, apiculate, non-amyloid, surface non-smooth under a light microscope, hyaline. Basidia (20.7)21.5–29.3(33.2) × (4.4)4.7–5.9 μm, subclavate to clavate, hyaline, infertility numerous, 4-spored, sterigmata up to 3.5 μm long. Hymenial cystidia absent. Hymenophoral trama regular to subregular, consisting of parallel, cylindrical to clavate, thin-walled hyphae, hyphae 3.1–9.6 μm wide. Pileipellis as a cutis composed of repent, cylindrical, subparallel, septate, thin-walled hyphae, hyphae 3.9–11.0 μm wide. Stipitipellis similar to the pileipellis. Clamp connections are present.
Habitat Scattered on the ground under mixed forests dominated by Larix olgensis.
Distribution Currently, only known from northeastern China (Jilin Province).
Additional specimen examined China, Jilin Province, Jilin City, Zuojia Town, on the ground under mixed forests dominated by Larix olgensis, 1 Sep 2020, J.Z. Xu, HMJU 850, GenBank accession numbers: ITS, MW880701; LSU, MW880707; same location, 1 Sep 2020, J.Z. Xu, HMJU 851, Genbank accession numbers: ITS, MW880702; LSU, MW880708; same location, 1 Sep 2020, J.Z. Xu, HMJU 758, Genbank accession numbers: ITS, MW913424.
Notes Infundibulicybe trachyspora, characterized by the clitocyboid to subomphalioid habit, a carneous, greyish-yellow to brownish pileus, moderately crowded, narrow lamellae, a brown to dark reddish-brown, longitudinally fibrillose-striated stipe, cyanophilous, non- amyloid and smooth spores and the presence of clamp connections. Morphologically, I. trachyspora shares similarities with Infundibulicybe gigas (Harmaja) Harmaja in crowed and narrow lamellae and lacrymoid to short ellipsoid spores. But I. gigas showed bigger size in pileus and stipe as compared in I. trachyspora (I. gigas, pileus diameter 9–28 cm, stipe 4–11 × 1.2–3.8 cm; I. trachyspora, pileus diameter 9–28 cm, stipe 3.5–7 × 0.5–0.75 cm) [2, 30]. Infundibulicybe lapponica resembles I. trachyspora having a medium-sized pileus, but it differs in the minutely scaly to the areolate surface of the pileus, which is smooth in I. trachyspora [1]. The new species recently described from China, I. rufa, resembles I. trachyspora in the smooth pileus and the longitudinally striated stipe. However, I. rufa produces a reddish pileus and stipe, I. trachyspora has a carneous, greyish-yellow to brownish pileus and a brown to dark reddish-brown stipe, I. rufa shows ixocutis hyphae in pileipellis, but I. trachyspora shows cutis hyphae. In addition, I. rufa differs from I. trachyspora by the incurved, wavy to undulate pileus margin (appeared to the species I. kotanensis M. Ishaq, M. Fiaz & A.N. Khalid, also described from Pakistan) [12, 25]. Infundibulicybe hongyinpan is also a new species from China growing on the ground under mixed forests dominated by Larix olgensis in autumn, just like I. trachyspora. But the pileus color of I. hongyinpan is reddish-brown, of I. trachyspora is calcareous, greyish-yellow to brownish. There is a more remarkable feature that I. trachyspora produces non-smooth spores absent in all Infundibulicybe species.
Discussion
The Status of Infundibulicybe trachyspora
Based on morphology and phylogenetic analyses, I. trachyspora was maintained as a new taxon in the genus Infundibulicybe. In ITS and combined ITS-nrLSU phylogenetic analyses, Infundibulicybe group showed close relationships with Ampulloclitocybe clavipes (Pers.) Redhead, Lutzoni, Moncalvo & Vilgalys and Trichocybe puberula (Kuyper) Vizzini, which is consistent with previous studies [12, 23,24,25]. Ampulloclitocybe clavipes and I. trachyspora both have non-smooth spores and lack cystidia. However, A. clavipes produces bulbose-base stipe, I. trachyspora produces cylindrical stipe [31]. Trichocybe puberula and I. trachyspora share the same characteristics in the presence of clamp connections, but T. puberula differs from I. trachyspora by having cystidia and smooth spores [32].
So far, this genus has no subdivisions and there is no established opinion on the infrageneric classification of Infundibulicybe. Meanwhile, the non-smooth spores and the distance of I. trachyspora from the other Infundibulicybe species suggested that the non-smooth spored species may deserve its section. Additional specimens from different areas are needed to evaluate the status of I. trachyspora and reveal the infrageneric classification of Infundibulicybe.
Key to Chinese Species of Infundibulicybe
1a. The color of pileus is red-brown……………………………………………………………………0.2
1b. The color of pileus is not red-brown………………………………………………………………3.
2a. Pileus hygrophanous…………………………………………………………….. I. hongyinpan.
2b. Pileus not hygrophanous………………………………………………………………….. I. rufa.
3a. Pileus surface smooth………………………………………………………………. I. trachyspora.
3b. Pileus surface not smooth…………………………………………………………………………….0.4
4a. The base of stipe with a few rhizoids…………………………………….. I. trullaeformis.
4b. The base of stipe without rhizoids………………………………………………………………5.
5a. Cystidia present………………………………………………………………………………. I. altaica.
5b. Cystidia absent……………………………………………………………………………………………6.
6a. Stipe diameter > 2 cm …………………………………………………………………. I. geotropa.
6b. Stipe diameter < 2 cm…………………………………………………………………………………0.7
7a. Stipe fibrillose…………………………………………………………………… I. alkaliviolascens.
7b. Stipe not fibrillose………………………………………………………………….. I. subsalmonea.
References
Harmaja H (1969) The genus Clitocybe (Agaricales) in Fennoscandia. Karstenia 10:5–168. https://doi.org/10.29203/ka.1969.62
Harmaja H (2003) Notes on Clitocybe s. lato (Agaricales). Ann Bot Fenn 40:213–218
Vizzini A, Contu M, Musumeci E, Ercole E (2011) A new taxon in the Infundibulicybe gibba complex (Basidiomycota, Agaricales, Tricholomataceae) from Sardinia (Italy). Mycologia 103:203–208. https://doi.org/10.3852/10-137
Matheny PB, Curtis JM, Hofstetter V et al (2006) Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia 98:982–995. https://doi.org/10.3852/mycologia.98.6.982
Binder M, Larsson KH, Matheny PB, Hibbett DS (2010) Amylocorticiales ord. nov. and Jaapiales ord. nov.: Earlydiverging clades of Agaricomycetidae were dominated by corticioid forms. Mycologia 102:865–880. https://doi.org/10.3852/09-288
Dentinger BTM, Gaya E, O’Brien H, Suz LM, Lachlan R, Díaz-Valderrama JR, Koch RA, Aime MC (2016) Tales from the crypt: genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biol J Linn Soc 117:11–32. https://doi.org/10.1111/bij.12553
He MQ, Zhao RL, Hyde KD et al (2019) Notes, outline and divergence times of basidiomycota. Fungal Divers 99:105–367. https://doi.org/10.1007/s13225-019-00435-4
Bruns TD, White TJ, Taylor JW (1991) Fungal molecular systematics. Annu Rev Ecol Syst 22:525–564
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x
Taylor JW, Jacobsona DJ, Krokena S et al (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. https://doi.org/10.1006/fgbi.2000.1228
Cho HJ, Lee H, Park MS et al (2020) Two New Species of Laccaria (Agaricales, Basidiomycota) from Korea. Mycobiology 48:288–295. https://doi.org/10.1080/12298093.2020.1786961
Zhao Q, Hao YJ, Liu JL, Hyde KD, Brooks S, Zhao YC (2016) Infundibulicybe rufa sp. nov. (Tricholomataceae), a reddish brown species from southwestern China. Phytotaxa 266:134–140. https://doi.org/10.11646/phytotaxa.266.2.7
Liu H, Guo S, Fan L (2019) Infundibulicybe hongyinpan sp. nov. a well-known edible fungus from Shanxi based on both nrDNA-ITS and morphological analysis [In Chinese]. J Shanxi Univ (Natural Science Edition) 42:275–280. https://doi.org/10.13451/j.cnki.shanxi.univ(nat.sci.).2018.03.16.007
Mao XL (1989) Part of the macro-fungus of Shennongjia [In Chinese]. Microbiology China 3:183–157
Zang M, Xia Y (1989) Notes on the fungi from Western Kunlun Mountain [In Chinese]. Acta Bot Yunnanica 11:397–406
Shao LP, Xiang CT (1997) Forest mushroom of China [In Chinese]. Northeast Forestry University Press, Heilongjiang Province
Li Y, Li TH, Yang ZL, Bau T, Dai YC (2015) Atlas of Chinese macrofungal resources [In Chinese]. Central China Farmers Publishing House, Henan Province
Wei TZ, Li BB, Wang WJ, Yao YJ (2015) Infundibulicybe alkaliviolascens, a new agaric record of China [In Chinese]. J Fungal Res 13:284–288. https://doi.org/10.13341/j.jfr.2014.2063
Kornerup A, Wanscher JHK (1978) The methuen handbook of colour. Eyre Methuen, London
Xu JZ, Yu XD, Lu MZ, Hu JJ, Moodley O, Zhang CL, Gong L, Li Y (2019) Phylogenetic analyses of some Melanoleuca species (Agaricales, Tricholomataceae) in northern China, with descriptions of two new species and the identification of seven species as a first record. Front Microbiol 10:2167. https://doi.org/10.3389/fmicb.2019.02167
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, Massachusetts
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Vizzini A, Ercole E (2012) Paralepistopsis gen. nov. and Paralepista (Basidiomycota, Agaricales). Mycotaxon 120:253–267. https://doi.org/10.5248/120.253
Qin J, Feng B, Yang ZL et al (2014) The taxonomic foundation, species circumscription and continental endemisms of Singerocybe: evidence from morphological and molecular data. Mycologia 106:1015–1026. https://doi.org/10.3852/13-338
Ishaq M, Khan MB, Ullah S, Fiaz M, Khalid AN (2019) Infundibulicybe kotanensis sp. nov. (Tricholomataceae), a new species from Buner, Pakistan. Phytotaxa 418:195–202. https://doi.org/10.11646/phytotaxa.418.2.4
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Ronquist F, Teslenko M, van der Mark P et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Harmaja H (1978) New species and combinations in the pale-spored Agaricales. Karstenia 18:29–30
Redhead SA, Lutzoni F, Moncalvo JM et al (2002) Phylogeny of agarics: Partial systematics solutions for core Omphalinoid genera in the Agaricales (Euagarics). Mycotaxon 83:19–57. https://doi.org/10.1017/S0953756202006147
Vizzini A, Musumeci E, Murat C (2010) Trichocybe, a new genus for Clitocybe puberula (Agaricomycetes, Agaricales). Fungal Divers 42:97–105. https://doi.org/10.1007/s13225-010-0030-8
Funding
The authors thank the Science and Technology Department of Jilin Province (20200402086NC) for their fnancial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, J., Zhao, W., Yu, X. et al. Infundibulicybe trachyspora, a New Species from Northeastern China Based on Morphology and Molecular Phylogeny. Curr Microbiol 79, 130 (2022). https://doi.org/10.1007/s00284-022-02808-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02808-6