Abstract
Modern antibiotics have been developed with the aim of destroying cellular function; however, the risk of antibiotic-resistance is increasing continuously. As a result, antimicrobial peptide (AMP) is considered a novel strategy to substitute traditional drugs. This study focused on revealing the antibacterial mechanism(s) of periplanetasn-4, an AMP identified from Cockroach. To elucidate whether periplanetasin-4 generates reactive oxygen species (ROS), a crucial stress factor for cell death, intracellular ROS was measured in Escherichia coli. The degree of membrane and DNA damage was determined using the properties that ROS causes oxidative stress to cell components. Unlike normal cell death, membrane depolarization was observed but DNA fragmentation did not occur. In addition, accumulation of nitric oxide (NO), a free radical with high toxicity, was measured and the byproduct of NO also induced severe intracellular damage. Periplanetasin-4-induced NO also impacted on cytosol calcium levels and triggered lipid peroxidation and DNA oxidation. These features were weakened when NO synthesis was interrupted, and this data suggested that perplanetasin-4-induced NO participates in E. coli cell damage. Moreover, this AMP-induced NO stimulates expression of SOS repair proteins and activation of RecA, a bacterial caspase-like protein. Features of nitrosative damage did not occur especially without dinF gene which is associated with oxidative stress. Therefore, it was indicated that when there is a NO signal, dinF promotes cell death. In conclusion, the combined investigations demonstrated that the antibacterial mechanism(s) of periplanetasin-4 was a NO-induced cell death, and dinF gene is closely related to cell death pathway.
Graphic Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most organisms on Earth are known to produce and secrete antimicrobial peptides (AMPs) to prevent invasion from outside microorganisms. This compound primarily prevents intrusions from various forms of pathogenic bacteria before an immune response occurs in a higher organism [1, 2]. In particular, AMP has the advantage of being effective against bacteria that are resistant to traditional antibiotics and rarely cause the emergence of new resistant bacteria [3, 4]. Therefore, the research and development of AMP is becoming significant as an alternative to the risk of antibiotic-resistant bacteria. So far, the mechanism of AMP is mostly disturbing bacterial membrane [5, 6]. It is reported that peptide, which contains a large amount of cationic amino acids, combines with the negatively charged membrane to form wormhole, thereby interrupting cells as the membrane permeability changes [7, 8]. In addition to working by penetrating the membrane of target cells, it is also known to inhibit the bacteria proliferation by various mechanisms such as inhibition of protein synthesis or nucleic acid [9]. The periplanetasin-4 derived from American cockroach is an AMP with 13 amino acids. In previous study, this substance was reported to interfere with the communication between mitochondria and vacuole of Candida albicans [10]. Except for this mechanism, however, the antibacterial mode of action is poorly understood.
Nitric oxide (NO) is a typical signaling molecule involved in cellular function or metabolism. NO production affects bioavailability, and both excess and lack of NO are known to influence health and disease [11]. In general, endogenous NO is produced together in the pathway of L-arginine degradation to L-citrulline, and enzyme called NOS (Nitrox oxide synthase) participates in this process. In mammalian cells, 3 isoforms of NOS present (neuronal NOS, endothelial NOS, inducible NOS) that are regulated by intracellular calcium/calmodulin [11]. These enzymes were though to exist only in eukaryotes, but recently, a NOS-like enzyme similar to mammalian NOS (mNOS) was discovered in prokaryotes [12]. In particularly, enzyme present in bacteria is named bacterial NOS (bNOS), and it is reported that bNOS have many properties in common with mNOS [13]. Although bNOS has no N-terminal hook region unlike mNOS, it is the same in that it produces endogenous NO, which plays various roles in bacteria. NO and its byproducts are widely used as antimicrobial agent due to their broad-spectrum antibacterial activity [14]. In addition, their oxidative and nitrosative stress induces cell death, negatively affecting the cell membrane or DNA of microorganism. To counter such DNA damage, E. coli has a DNA repair system called SOS response. It is known that when hallmarks of DNA damage appear due to NO and byproducts of NO, SOS signals is initiated in E. coli [15, 16]. Using the aforementioned properties of NO, many bacterial infection treatments are being developed.
Using the features of periplanetasin-4 and NO described earlier, several experiments were conducted in this study to determine whether periplanetasin-4 induce cell death in Escherichia coli. Additionally, we also confirmed how NO influences the bacterial cell death and correlates with proteins and genes involved in DNA damage.
Materials and Methods
Preparation of Compound and Bacterial Strains
Whole transcription sequencing of E. coli-infected Periplaneta americana, and the identification of potential AMPs were performed by Kim et al. [17]. Among the identified AMPs, periplanetasin-4 (LRHKVYGYCVLGP-NH2) was selected for further analysis. The periplanetasin-4 was synthesized by Anygen Co., Ltd. (Gwangju, Republic of Korea) and dissolved in distilled water for experiments. For inhibition of NO production, cells were cotreated with 1 mM Nω-Nitro-L-arginine methyl ester (L-NAME; Sigma-Aldrich). This NOS inhibitor was concurrently added and was used as a negative control [18, 19]. Bacteria E. coli wildtype BW25113 was obtained from Coli Genetic Stock Center and ΔdinF which is mutated by E. coli K-12 strain BW25113 (constructed by P1 transduction) was obtained from KEIO collection [20].
Antimicrobial Susceptibility Testing
The antimicrobial effects of periplanetasin-4 were determined using the Clinical and Laboratory Standard Institute (CLSI) guidelines. The following bacterial strains were used in this study: Enterococcus faecium (ATCC 19,434), Enterococcus faecalis (ATCC 29,212), Escherichia coli (ATCC 25,922), Pseudomonas aeruginosa (ATCC 27,853), and Staphylococcus aureus (ATCC 25,923). These were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Staphylococcus epidermidis (KCTC 1917) and Salmonella typhimurium (KCTC 1926) were obtained from the Korean Collection for Type Cultures (KCTC, Jeongeup-si, Jeollabuk-do, Korea). Bacterial strains were cultured in LB broth (BD) at 37 ℃ with aeration. Growing bacterial cells (2 × 106 cells/mL) were allocated into 96-microwell plates (0.1 mL/well). After 24 h of incubation at 37 °C, cell proliferation was determined by optical density at 600 nm using a microtiter ELISA reader. (BioTek Instruments, Winooski, VT, USA).
Evaluation of Intracellular ROS Levels
An ROS indicator, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Molecular Probes), was applied to assess intracellular ROS accumulation. Upon cleavage of the acetate groups by intracellular esterases and oxidation, the non-fluorescent H2DCFDA is converted to the highly fluorescent 2′,7′-dichlorofluorescein. After 2 and 4 h incubation with 5 μM periplanetasin-4, cells were washed with phosphate-buffered saline (PBS; pH 7.4, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) and stained with 10 μM H2DCFDA for 1 h at 37 °C. Then the cells were washed with PBS and the fluorescent cells were detected with a spectrofluorophotometer (Shidmadzu RF-5301PC; Shimadzu, Kyoto, Japan) at wavelengths of 485 nm (excitation) and 535 nm (emission).
Analysis of Bacterial Membrane Potential Alteration
bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)] (Molecular Probes) exhibits increased fluorescence by binding membranes or intracellular proteins of depolarized cells. To evaluate the changes in bacterial membrane potential, DiBAC4(3) was used and dissolved in dimethyl sulfoxide (DMSO). E. coli cells (2 × 106 cells/mL) were treated with 5 μM periplanetasin-4 and incubated for 2 h and 4 h at 37° C with shaking at 120 rpm. After incubation, the cells were centrifuged at 12,000 rpm for 5 min and the supernatants were removed. The cell pellets were resuspended twice in PBS and the final volume was adjusted to 1 mL with PBS. These bacterial solutions were stained with 5 μg/mL DiBAC4(3). The fluorescence intensity was measured utilizing a FACSVers flow cytometer (Becton Dickinson, NJ, USA).
Measurement of DNA Fragmentation
To assess DNA fragmentation, a terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay was performed using an In Situ Cell Death Detection Kit, Fluorescein (Roche Applied Science, Basel, Switzerland). E. coli cells were treated with 5 μM periplanetasin-4 and then incubated for 4 h at 37 °C with shaking at 120 rpm. The cells were then fixed with 2% paraformaldehyde for 1 h. The fixed cells were harvested, suspended in PBS, and incubated with 100 μL permeabilization solution (0.1% sodium citrate and 0.1% Triton-X 100) on ice for 2 min. The cells were washed twice with PBS, mixed with label solution and enzyme solution, and incubated for 1 h at 37 °C. The samples were collected and suspended in PBS, and fluorescence intensity was measured utilizing the FACSVerse flow cytometer.
Assessment of Hydroxyl Radical, Hydrogen Peroxide and NO Level
Hydroxyl radical accumulation were assessed using 3′-(p-hydroxyphenyl) fluorescein (HPF, Molecular Probes). E. coli cells were treated with 5 μM periplanetasin-4 for 4 h at 37 °C and then the cells were washed with PBS and stained with 10 μM HPF, which was dissolved in Dimethylformamide (DMF) (JUNSEI Chemical Co., Tokyo, Japan).
Hydrogen peroxide level was measured using Amplex Red Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The E. coli cells were treated with 5 μM periplanetasin-4 described above and incubated for 2 h. The cell suspension was extracted using lysis buffer (10 mM Tris–HCl, and 1 mM EDTA [pH 8.0], 100 mM NaCl, and 0.1% Triton-X 100). The absorbance of Amplex Red as measured using an ELISA microplate Reader at 570 nm.
For detecting of intracellular NO generation, non-fluorescent probe, 4-Amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA, Molecular Probes) was used. E. coli cells were treated with 5 μM periplanetasin-4 for 4 h at 37 °C and then the cells were washed with PBS and stained with 10 μM DAF-FM. The fluorescence intensity was measured utilizing the FACSVers flow cytometer.
Quantification of Superoxide Dismutase (SOD) Activity
E. coli cells were treated with 5 μM periplanetasin-4 and then incubated for 2 h at 37 °C with shaking at 120 rpm. The cell suspension which treated with periplanetasin-4 for 2 h was lysed using beads with lysis buffer (10 mM Tris–HCl, and 1 mM EDTA [pH 8.0], 100 mM NaCl, and 0.1% Triton‐X 100). After centrifugation, the supernatants were observed for SOD using the SOD Assay Kit (Sigma‐Aldrich) according to the manufacturer's instructions. After incubation for 30 min at 37 °C, absorbance was measured using microtiter ELISA Reader.
Assessment of Intracellular Calcium Level
Fura-2AM (Molecular Probes) were used to measure the calcium levels in the cytosol. To observe the intracellular calcium uptake, E. coli cells were treated with 5 μM periplanetasin-4 for 2 h. The cells were then washed twice with Krebs buffer (132 mM NaCl, 10 mM HEPES, 4 mM KCl, 10 mM NaHCO3, 6 mM glucose, 1.4 mM MgCl2 and 1 mM CaCl2; pH 7.2) and treated with 1% bovine serum albumin and 0.01% pluronic F-127 (Molecular Probes). The cells were stained with 5 µM Fura-2AM according to manufacturer's instructions and incubated for 40 min, after which the cells were washed twice with calcium-free Krebs buffer. Fluorescence intensities of calcium levels were monitored with a spectrofluorophotometer using the RFPC software for calculations at wavelengths of 340 nm (excitation) and 510 nm (emission) for Fura-2AM.
Measurement of Lipid Peroxidation and DNA Oxidation
Lipid peroxidation was quantified based on malondialdehyde (MDA) levels. After treatment with 5 μM periplanetasin-4 for 2 h, the cell suspension was centrifuged at 12,000 rpm for 5 min. Then, the pellet was sonicated twice on ice in lysis buffer (2% Triton-X 100, 1% SDS, 100 mM NaCl, 10 mM Tris–HCl, and 1 mM EDTA pH 8.0). The mixture was centrifuged, and the supernatant was added to an equal volume of 0.5% (w/v) thiobarbituric acid (TBA) solution in 5% TCA. The mixture was heated at 95 °C for 30 min and then cooled on ice. The absorbance of the reaction mixture was measured at 532 and 600 nm.
DNA oxidative damage was evaluated by measuring the levels of hydroxydeoxyguanosine (8-OHdG). 8-OHdG is one of the major forms of ROS-induced DNA oxidative lesions, has been widely used as a ubiquitous marker of oxidative stress. After 2 h incubation with 5 μM periplanetasin-4, DNA was extracted following as described in our previous research [21]. 8-OHdG levels were measured using the Oxiselect Oxidative DNA Damage ELISA Kit (Cell Biolabs Inc., San Diego, CA, USA), according to the manufacturer’s instructions. The absorbance of 8-OHdG was measured using an ELISA microplate Reader at 450 nm.
Expression of SOS Response Genes
Activation of RecA protein was detected using the CaspACE fluorescein isothiocyanate (FITC-VAD-FMK) in situ marker (Promega, Madison, WI, USA). FITC-VAD-FMK, FITC-conjugated peptide pan-caspase inhibitor, is transported into cells and binds to the active site of caspase as a substrate to investigate RecA expression. RecA has a classical binding site with caspase substrate. The cells were incubated with 5 μM periplaentasin-4 for 2 h at 37 °C, Next, the cells were washed twice and incubated with CaspACE FITC-VAD-FMK for 30 min. After centrifugation, the cells were resuspended in PBS and the fluorescence was analyzed using a FACSverse flow cytometer.
Quantitation of solubilized proteins was performed using Bradford’s assay (Bio-Rad, Hercules, CA, USA). Each 50 μg protein sample was separated by 12% (w/v) SDS‐PAGE, followed by electroblotting onto nitrocellulose membranes. The membrane was blocked in 3% skimmed milk for 1 h at room temperature. Immunodetection was performed using rabbit polyclonal LexA antibodies (1:3000; Thermo fisher) and GADPH loading control antibodies (1:5000; Thermo fisher) for 12 h at 4 °C. horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2000; Biovision, Milpitas, CA, USA) was used as a secondary antibody for 12 h at 4 °C. Pierce ECL Plus Western Blotting Substrate (Thermo Scientific, Waltham, MA, USA) was then added, and the membranes were exposed to an X-ray film [22]. The relative amount of LexA was numerically quantified with the ImageJ program (http://rsb.info.nih.gov/ij).
Statistical Analysis
All experiments were performed in triplicate and the data were represented as the means ± SD. Statistical significance was determined via Student’s t-test. P < 0.05 was considered to indicate statistical significance.
Results
Antibacterial Effect of Periplanetasin-4
To assess whether periplanetasin-4 has antibacterial effects on pathogenic bacteria, the MIC values were performed based on the CLSI method. As shown in Table.1, it was confirmed that periplanetasin-4 exhibited broad-spectrum antibacterial effects with MIC values in ranging from 2.5 to 5 μM in eight bacterial strains. Periplanetasin-4 has potent antibacterial activity towards gram-positive bacterial as well as gram-negative bacterial. Among others, in our research, E. coli was also used as a bacterial model to examine the antibacterial mode of action of periplanetasin-4.
The Antibacterial Effect of Periplanetasin-4 has No Relevance to DNA Fragmentation by ROS
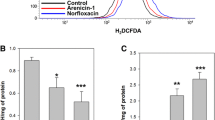
Various bactericidal agents influence accumulation or overproduction of ROS, which causes damage to intracellular components, DNA, and proteins. To determine whether periplanetasin-4 increases intracellular ROS levels during periplanetasin-4-induced cell death, a ROS fluorescent probe, H2DCFDA was used. Thus, this data indicated that periplaneasin-4 elevated intracellular ROS levels in accordance with time passed (Fig. 1A). Also, ROS-mediated membrane damage occurs quite frequently [23, 24]. DiBAC4(3), a sensitive slow-response probe for measuring cellular membrane potential that and can enter depolarized cells, was applied to estimate whether periplanetasin-4-induced ROS causes bacterial membrane depolarization. We observed that the ability of periplanetasin-4 to disrupt bacterial membrane potentials is a gradually increasing of membrane depolarization in time-dependent manner, which was consistent with the onset of cell death (Fig. 1B). Furthermore, we used TUNEL assay to assess DNA damage after periplanetasin-4 treatment. Incomplete DNA cleavage may result in chromosomal aberrations and genomic instability [25]. There was no significant difference in the percentages of TUNEL-positive cells between untreated cells and periplanetasin-4 treated cells (Fig. 1C). These results indicated that there is a specific point in time that leads to the accumulation of ROS without causing DNA fragmentation and membrane depolarization.
The antibacterial effect of periplanetasin-4. Intracellular ROS accumulation and membrane potential was measured. (A) Spectrofluorophotometric analysis of ROS accumulation was conducted using H2DCFDA. (B) Flow cytometric analysis of DiBAC4(3)-stained C. albicans following 2 and 4 h incubated with periplanetasin-4. (C) periplanetasin-4-induced DNA breaks as detected by TUNEL. The data are displayed as the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01 versus control
Periplanetasin-4-Induced Superoxide is Salient Oxidative Stress Substance
There are various factors that cause oxidative stress in cells, but superoxide, hydroxyl radical, and hydrogen peroxide are the most representative ROS. Hydroxyl radicals are typically produced by reducing oxygen or by Fenton reaction. Also, superoxide is a byproduct of mitochondrial respiration, which is highly toxic, so aerobic organisms have superoxide-scavenging enzyme called SOD. Moreover, hydrogen peroxide is a molecule that is involved in both the Fenton reaction and catalysis of superoxide. As previously stated, H2DCFDA assay showed that periplanetasin-4 produces intracellular ROS. Then, further experiments were conducted to determine which ROS affect bacterial cells. In our experiments, the level in the hydroxyl radicals via HPF staining was not changed (Fig. 2A). The internal hydrogen peroxide level did not show significant difference as well (Fig. 2B). Furthermore, SOD activity did not exhibit changes significantly (Fig. 2C). These results demonstrated that superoxide is the main causative ROS by not producing hydroxyl radicals and hydrogen peroxide. Furthermore, no difference of SOD activity suggested that the ROS defense system is not functioning properly in the cell, and that superoxide is present in the cell without changing to harmless hydrogen peroxide or oxygen.
The source of ROS by periplanetasin-4. (A) Flow cytometric analysis of hydroxyl radical detected by the HPF staining, (B) hydrogen peroxide production was measured by an Amplex Red assay (C) Analysis of SOD activity using an ELISA reader at 450 nm. The data are displayed as the mean ± SD from three independent experiments
Periplanetasin-4-Induced NO Triggers Oxidative Damage
To examine whether NO could be generated in E. coli cells when exposed to periplanetasin-4, NO was quantified using a reagent probe DAF-FM [26]. The increase of DAF-FM fluorescence indicates that intracellular NO level was produced in the cells treated with periplanetasin-4. This result confirmed that high levels of NO are produced in the cytosol within 2 h (Fig. 3A). In addition, to investigate the effect factor of bacterial NO, L-NAME was used as an inhibitor of NOS [27]. As shown in Fig. 3B, periplanetasin-4-induced cytosolic calcium ion accumulation and periplanetasin-4 treatment with L-NAME decreased the calcium ion levels in E. coli. This observation means that bNOS is dependent on calcium levels and NO signaling stimulated by periplanetasin-4 also influence the cytosol calcium level. NO immediately reacts with superoxide anion and produces peroxynitrite. To assess lipid peroxidation as a measurement of peroxynitrite activity by periplanetasin-4, MDA is routinely used by its reaction with thiobarbituric acid reactive substances (TBARS) [28]. As shown in Fig. 3C, the MDA levels in the periplanetasin-4-treated cells were increased compared to that in untreated cells for all tested cells. Furthermore, DNA oxidation was dependent upon NO level because DNA oxidation was decreased in the presence of L-NAME (Fig. 3D). These data suggested that the peroxidation of lipid and DNA oxidation by periplanetasin-4 are due to a combination of NO and superoxide anion.
NOS inhibition diminishes activity of periplanetasin-4. (A) NO detection with DAF-FM staining (B) Calcium level was measured using Fura-2AM. E. coli cells were treated with periplanetasin-4 with or without L-NAME. (C) Lipid peroxidation was measured by TBARS assay, increase in MDA levels indicates peroxidation of lipids. (D) DNA oxidation was assessed by quantitating 8-OHdG, decrease of 450 nm absorbance indicates oxidation of DNA. The data represents the average, standard deviation, and p values from three independent experiments (*P < 0.05, **P < 0.01 compared to untreated samples; ##P < 0.01 compared to L-NAME cotreated sample)
Periplanetasin-4 Stimulates DNA Repair System
When E. coli undergoes DNA damage, the DNA repair pathway, SOS response begins. The key regulating proteins involved in this response are RecA and LexA [29]. In some papers, especially RecA is considered as a caspase-like protein in E. coli that acts like a caspase in eukaryotic cells [22]. In addition, LexA is SOS response repressor, and when there is DNA damage, SOS response begins as LexA is self-cleavage. To verify RecA activation in periplanetasin-4-induced cell death, affinity for the caspase substrate was determined using FITC-conjugated peptide pan-caspase inhibitor, FITC-VAD-FMK. As shown in Fig. 4A, periplanetasin-4 activated RecA co-protease activity, and the activation was decreased when periplanetasin-4-treated cells were pretreated with L-NAME. This result demonstrated that periplanetasin-4 activates RecA expression, and the expression is related with NO. Furthermore, we monitored the LexA protein expression using western blotting. According to our result, LexA expression was diminished in periplanetasin-4-treated cells. In periplanetasin-4-treated cells pretreated with L-NAME, LexA protein band appeared thicker than cells treated with periplanetasin-4 alone (Fig. 4B). Our findings listed above indicated that periplanetasin-4 stimulates SOS response proteins, and this response was suppressed when NO synthesis is inhibited.
Periplanetasin-4 causes DNA repair gene activation. E. coli cells were treated with periplanetasin-4 with or without L-NAME. (A) Activation of RecA was assessed using FITC-VAD-FMK. (B) Analysis of LexA expression levels by western blotting (a) Untreated (b) Periplanetasin-4 (c) Periplanetasin-4 + L-NAME
dinF is Essential to Stimulate the Cell Death Pathway Involved by Periplanetasin-4
SOS response and oxidative damage is based on the relationship with dinF [30]. We observed the periplanetasin-4 activity using the cells in absence of dinF. The cells deleted dinF gene were exhibited the resistance to periplanetasin-4. DNA oxidation, SOS response activation, and calcium ion elevation did not show significant changes (Fig. 5). This data suggested that dinF operates as a key regulator of cell death by periplanetasin-4. Moderate DNA damage stimulates expression of din genes and severe DNA damage stimulates expression of SOS repair genes [31]. In this case, dinF gene accelerates the cell death pathway and requires NO signaling. Lack of this gene might not cause NO accumulation leading to intracellular calcium elevation, RecA activation with LexA autocleavage. Therefore, in the cells in absence of dinF, DNA or protein could not be oxidized, so they have tolerance to periplanetasin-4.
Antibacterial activity of periplanetasin-4 in the absence of dinF. E. coli ΔdinF cells were treated with periplanetasin-4 with or without L-NAME. (A) NO detection with DAF-FM staining (B) Spectrofluorophotometric analysis of ROS accumulation was conducted using H2DCFDA. (C) Lipid peroxidation was measured by TBARS assay, increase in MDA levels indicates peroxidation of lipids. (D) DNA oxidation was assessed by quantitating 8-OHdG, decrease of 450 nm absorbance indicates oxidation of DNA. (E) Activation of RecA was assessed using FITC-VAD-FMK. (F) Analysis of LexA expression levels by western blotting (a) Untreated (b) Periplanetasin-4 (c) Periplanetasin-4 + L-NAME. The data represents the average, standard deviation, and P values from three independent experiments
Discussion
Cockroaches have potential as a solution in overcoming microbial contamination, even with pathogens that are multi-drug resistant. Since they reside in unsanitary environments, cockroaches had to develop their defense systems for their survival [32]. Indeed, the extracts of various organs of cockroaches body have exhibited potent antibacterial activities against methicillin-resistant S. aureus and neuropathogenic E. coli [33]. We had reported previously the antimicrobial peptides isolated from American cockroach [17]. They express a broad antimicrobial activity with low cytotoxicity. Periplanetasin-2 alleviated intestinal inflammatory reaction in the toxin A- injected mouse enteritis model and showed activation of apoptotic response in fungi and bacteria [34,35,36]. Periplanetasin-4 also inhibited the cell toxicities caused by toxin A in human colonocytes [35]. Here we synthesized periplanetasin-4 and used it to explore how bacterial cell death with low cytotoxicity is induced. First of all, the bactericidal activity was evinced mainly using an ROS indicator. Periplanetasin-4 rapidly stimulates ROS production and then the membrane depolarization is followed. Disruption of the electrochemical gradient due to the improper trafficking of membrane proteins leads to the changes in membrane potential associated with hydroxyl radical formation [37]. Internal hydrogen peroxide concentration is responsible for the hydrogen peroxide-mediated death of bacterial cells [38]. The gap that ROS generating without detection of membrane depolarization was noted. At same time, DNA fragmentation is insignificantly enhanced. These phenomena can be explained that ROS induced by periplanetasin-4 stimulates other intracellular damage pathways as well as membraned targeted mechanism.
Oxygen is essential for life, however it can harm cell own ability to function normally or participate in its devastation due to the creation of ROS [38]. Furthermore, ROS interrupt cellular macromolecules such as lipids, proteins and nucleic acids, and can inactivate metabolism by inhibiting mononuclear iron enzymes and [4Fe–4S] dehydratases [39]. Among several ROS, we explored the critical ROS to perform intracellular damage by periplaentasin-4. Hydroxyl radical and hydrogen peroxide did not exhibit the difference after periplanetasin-4 treated. Additionally, SOD activity also did not change after periplanetasin-4 treated. Previously, ROS accumulation is accordant closely with superoxide anion overproduction. Peroxynitrite is created when NO and superoxide rapidly interact [40]. NO is a membrane permeant molecule and plays a multifunctional role in physiological regulation such as neurotransmission, vasodilation, wound healing, and immune response. They induce nitrosative stress which brings out a great number of toxic effects on bacteria [41,42,43]. Peroxynitrite is a powerful oxidant which induces lipid peroxidation, DNA cleavage, and direct modification of membrane proteins. These reactions result in cellular responses ranging from cell signaling modulations to oxidative damage, perpetrating an apoptosis or necrosis [40, 43]. The potent antibacterial activity by periplanetasin-4 have a possibility to involve NO production. Consequently, periplaentasin-4 treated cells could be observed calcium-dependent NO signal stimulation. NO take in iron–sulfur cluster-dependent enzymes and restrain main enzymes that include iron in their catalytic centers. NO participate in numerous cellular response, including copper metabolism, apoptosis-like cell death, and high-temperature stress tolerance [44]. Proteins homologous to mNOS have been identified and studied in prokaryotic sources [12]. The bNOS are in many ways similar to their mNOS counterpart. NOS enzyme in mammals exists in three isoforms encoded by distinct genes. Neuronal (nNOS or Type 1) and endothelial (eNOS or Type 3) are both constitutive and calcium dependent [45]. Furthermore, lipid peroxidation and DNA oxidation were induced by periplanetasin-4 and then, the result was diminished using NOS inhibitor. Indeed, intracellular damage pathway is affected by NO level.
DNA damage response activates pro-death signaling [25]. SOS response is a DNA damage repair system in which RecA filaments united in ssDNA induce self-cleavage of the LexA protein and the cell cycle is arrested. Cleavage of LexA, is related to ATP-DnaA complex, promotes expression of the SOS genes, causing a cell division pause for the time required to repair the damages [46]. LexA and RecA, which are noteworthy because LexA is an inhibitor of the SOS response. Among them, RecA, SOS response regulator and an essential bacterial DNA recombinase, can bind to classic caspase substrates [47]. The caspase ability of RecA was increased after periplanetasin-4 treatment. This result indicated that periplanetasin-4 induces NO-dependent SOS response activation.
DnaA-dependent transcription of SOS genes demand to frustrate over-expression of the SOS genes to preserve genome integrity [48]. Each of the SOS genes is in close proximity to its promoter/operator site where the LexA repressor protein is sticked [30, 49]. Because it makes a transcriptional unit with lexA, it would be supposed that transcription of dinF is firmly regulated by LexA. Additionally, dinF protects cells from hydrogen peroxide, which induces cell death in E. coli. Periplanetasin-4 treated cells did not significantly exhibit the intracellular damage pathway when dinF protein is deleted.
In conclusion, periplanetasin-4 raises superoxide anion levels. Although the interaction with superoxide and its by-products does not cause serious intracellular damage, the peroxynitrite by superoxide and NO induce significant intracellular damage. The funtion of periplanetasin-4 in E. coli is the initiation of calcium-dependent NO signal. NO signal-induced damage is diminished when NO synthesis is stopped. It was found that intracellular substances damage selectively occurred, and DNA repair proteins were concerned. Furthermore, the expression levels of the typical SOS repair proteins (RecA and LexA) have changed significantly. In the absence of dinF, periplanetasin-4 did not cause deterioration of intracellular molecules, after which superoxide and NO accumulated. We conclude that periplanetasin-4 prompts calcium-dependent NO signals in bacteria, and those signals are closely related to the din gene.
Conclusion
Today AMP is in the spotlight as a broad-spectrum antibiotic and a novel therapeutic agent. A common mechanism of AMP is to incapacitate the negatively charged bacterial membrane. This activity of AMP not only inhibits the functions of DNA, RNA, and proteins, but also leads to the production of intracellular ROS. The periplanetasin-4 used in this paper exhibits membrane potential alteration and superoxide generation without DNA cleavage. In addition to superoxide, it generated NO known to have antimicrobial effects, and found that the levels of cytoplasmic calcium, MDA, and 8-OHdG are increased due to the periplanetasin-4-induced NO. Additionally, the expression of SOS genes and activity of caspase-like protein (RecA) were detected, indicating that SOS response was initiated by NO-induced DNA damage. In particular, NO-induced features were not shown in cells lacking dinF, which are known to protect oxidative stress, demonstrating that dinF is involved in NO signals as well as ROS.
References
Scott MG et al (2002) The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J Immunol 169(7):3883–3891
Sperandio B et al (2008) Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J Exp Med 205(5):1121–1132
Lam SJ et al (2016) Combating multidrug-resistant Gram-negative bacteria with structurally nanoengineered antimicrobial peptide polymers. Nat Microbiol 1(11):1–11
Lázár V et al (2018) Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat Microbiol 3(6):718–731
Nizet V (2006) Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr Issues Mol Biol 8(1):11
Yeaman MR, Yount NY (2003) Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 55(1):27–55
Tamba Y, Yamazaki M (2005) Single giant unilamellar vesicle method reveals effect of antimicrobial peptide magainin 2 on membrane permeability. Biochemistry 44(48):15823–15833
Mangoni ML et al (2004) Effects of the antimicrobial peptide temporin L on cell morphology, membrane permeability and viability of Escherichia coli. Biochemical Journal 380(3):859–865
Uyterhoeven ET et al (2008) Investigating the nucleic acid interactions and antimicrobial mechanism of buforin II. FEBS Lett 582(12):1715–1718
Lee H, Hwang JS, Lee DG (2019) Periplanetasin-4, a novel antimicrobial peptide from the cockroach, inhibits communications between mitochondria and vacuoles. Biochem J 476(8):1267–1284
Luiking YC, Engelen MP, Deutz NE (2010) Regulation of nitric oxide production in health and disease. Curr Opin Clin Nutr Metab Care 13(1):97
Crane BR, Sudhamsu J, Patel BA (2010) Bacterial nitric oxide synthases. Annu Rev Biochem 79:445–470
Sudhamsu J, Crane BR (2009) Bacterial nitric oxide synthases: what are they good for? Trends Microbiol 17(5):212–218
Jones ML et al (2010) Antimicrobial properties of nitric oxide and its application in antimicrobial formulations and medical devices. Appl Microbiol Biotechnol 88(2):401–407
Kim H, Lee DG (2020) Nitric oxide–inducing Genistein elicits apoptosis-like death via an intense SOS response in Escherichia coli. Appl Microbiol Biotechnol 104(24):10711–10724
Kim H, Lee DG (2021) Lupeol-induced nitric oxide elicits apoptosis-like death within Escherichia coli in a DNA fragmentation-independent manner. Biochemical Journal 478(4):855–869
Kim I-W et al (2016) De novo transcriptome analysis and detection of antimicrobial peptides of the American cockroach Periplaneta americana (Linnaeus). PLoS One 11(5):e0155304
Almeida B et al (2007) NO-mediated apoptosis in yeast. J Cell Sci 120(18):3279–3288
Nishimura A, Kawahara N, Takagi H (2013) The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem Biophys Res Commun 430(1):137–143
Baba T et al (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology. https://doi.org/10.1038/msb4100050
Lee B, Lee DG (2017) Reactive oxygen species depletion by silibinin stimulates apoptosis-like death in escherichia coli. J Microbiol Biotechnol 27(12):2129–2140
Lee H, Lee DG (2019) SOS genes contribute to Bac8c induced apoptosis-like death in Escherichia coli. Biochimie 157:195–203
Allen SA et al (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3(1):1–10
Kamat J et al (2000) Reactive oxygen species mediated membrane damage induced by fullerene derivatives and its possible biological implications. Toxicology 155(1–3):55–61
Roos WP, Thomas AD, Kaina B (2016) DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 16(1):20
Vumma R et al (2016) Antibacterial effects of nitric oxide on uropathogenic Escherichia coli during bladder epithelial cell colonization—a comparison with nitrofurantoin. J Antibiot 69(3):183–186
Pereira L et al (2017) β-Adrenergic induced SR Ca2+ leak is mediated by an Epac-NOS pathway. J Mol Cell Cardiol 108:8–16
Xiong Z-Q et al (2013) The mechanism of antifungal action of a new polyene macrolide antibiotic antifungalmycin 702 from Streptomyces padanus JAU4234 on the rice sheath blight pathogen Rhizoctonia solani. PloS One 8(8):e73884
Michel B (2005) After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3(7):e255
Rodríguez-Beltrán J et al (2012) The Escherichia coli SOS gene dinF protects against oxidative stress and bile salts. PloS one 7(4):e34791
Peeters SH, de Jonge MI (2018) For the greater good: Programmed cell death in bacterial communities. Microbiol Res 207:161–169
Moges F et al (2016) Cockroaches as a source of high bacterial pathogens with multidrug resistant strains in Gondar town, Ethiopia. BioMed Res Int. https://doi.org/10.1155/2016/2825056
Ali SM et al (2017) Identification and characterization of antibacterial compound (s) of cockroaches (Periplaneta americana). Appl Microbiol Biotechnol 101(1):253–286
Hong J et al (2017) The American cockroach peptide periplanetasin-2 blocks Clostridium Difficile toxin A-induced cell damage and inflammation in the gut. J Microbiol Biotechnol 27(4):694–700
Yoon IN et al (2017) The American cockroach peptide periplanetasin-4 inhibits Clostridium difficile toxin A-induced cell toxicities and inflammatory responses in the mouse gut. J Pept Sci 23(11):833–839
Yun J, Hwang J-S, Lee DG (2017) The antifungal activity of the peptide, periplanetasin-2, derived from American cockroach Periplaneta americana. Biochemical Journal 474(17):3027–3043
Kohanski MA et al (2008) Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135(4):679–690
Uhl L, Dukan S (2016) Hydrogen peroxide induced cell death: the major defences relative roles and consequences in E. coli. PloS one 11(8):e0159706
Smirnova GV et al (2015) Extracellular superoxide provokes glutathione efflux from Escherichia coli cells. Res Microbiol 166(8):609–617
Islam BU et al (2015) Pathophysiological role of peroxynitrite induced DNA damage in human diseases: a special focus on poly (ADP-ribose) polymerase (PARP). Indian J Clin Biochem 30(4):368
Bicker G (2001) Sources and targets of nitric oxide signalling in insect nervous systems. Cell Tissue Res 303(2):137–146
Bryan NS, Bian K, Murad F (2009) Discovery of the nitric oxide signaling pathway and targets for drug development. Front Biosci 14(1):1–18
Privett BJ et al (2012) Examination of bacterial resistance to exogenous nitric oxide. Nitric Oxide 26(3):169–173
Sun C et al (2018) Nitric oxide acts downstream of hydrogen peroxide in regulating aluminum-induced antioxidant defense that enhances aluminum resistance in wheat seedlings. Environ Exp Bot 145:95–103
Forstermann U, Sessa WC (2012) Nitric oxide synthases: regulation and function. Eur Heart J 33(7):829–37
Maslowska KH, Makiela-Dzbenska K, Fijalkowska IJ (2019) The SOS system: a complex and tightly regulated response to DNA damage. Environ Mol Mutagen 60(4):368–384
Asplund-Samuelsson J (2015) The art of destruction: revealing the proteolytic capacity of bacterial caspase homologs. Mol Microbiol 98(1):1–6
Brambilla E et al (2018) DnaA and LexA proteins regulate transcription of the uvrB gene in Escherichia coli: the role of DnaA in the control of the SOS regulon. Front Microbiol 9:1212
Janion C (2008) Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J Biol Sci 4(6):338
Acknowledgements
This work was supported by a grant from the Next-Generation BioGreen 21 Program (Project No. PJ01325603), Rural Development Administration, Republic of Korea. This manuscript also has been grammatically edited by a native speaker, a professional editing firm, Enago.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or nonprofit sectors.
Author information
Authors and Affiliations
Contributions
H. Lee, J.S. Hwang, and D.G. Lee conceived the study and designed the experiment. H. Lee performed the experiments and collected the data. H. Lee and D.G. Lee analyzed the data. H. Lee wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, H., Hwang, J.S. & Lee, D.G. dinF Elicits Nitric Oxide Signaling Induced by Periplanetasin-4 from American Cockroach in Escherichia coli. Curr Microbiol 78, 3550–3561 (2021). https://doi.org/10.1007/s00284-021-02615-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02615-5