Abstract
Filamentous fungi play an important role in the production of a range of useful extracellular hydrolytic enzymes for wide industrial applications. The Western Ghats region is known for its rich microbial biodiversity and could be a potential source of several useful fungi that could be exploited for the production of industrially important enzymes. From this soil, we aimed at the isolation of multienzyme producing fungi, optimization of the culture conditions using solid-state fermentation (SSF), partial purification of enzymes and characterization by zymography. Out of seven fungal strains, two isolates, namely Penicillium citrinum and Aspergillus clavatus, were found to produce amylase and cellulase enzymes maximally. The effect of different physicochemical parameters on the production of amylase and cellulase was investigated and the maximum production of multienzymes was achieved in wheat bran substrate. The newly formulated and optimized medium increased the multienzyme production in P. citrinum and A. clavatus as compared to medium with individually optimized parameters. Further, for the first time, different isoforms of amylase and cellulase have been identified from P. citrinum and A. clavatus by zymography. In summary, the present study showed that the filamentous fungi can utilize the industrial waste product such as wheat bran as the substrate for multienzymes production by SSF and could be a promising source of enzymes for biotechnological applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Several fungi have been recognized for decades as a vital source of natural biologicals for biomedical and industrial developments [1]. Filamentous fungi are skilled in secreting a huge amount of primary and secondary metabolites into the production medium which has been extensively exploited industrially for years. This practice of the utilization of these fungi has led us to generally consider several species of filamentous fungi as a safe and attractive host for the synthesis of novel products for human usage [2]. Cellulases are inducible enzymes hydrolyzing β-1,4-glycosidic bonds of intact cellulose and other related cellooligosaccharide derivatives which are classified as endoglucanase (EC 3.2.1.4), exoglucanase (EC 3.2.1.91) and β-glucosidase (EC 3.2.1.21). Endoglucanases hydrolyze the amorphous fraction of the cellulose chain reducing its degree of polymerization [3]. The β-glucosidases, which possesses wide industrial application, primarily hydrolyzes β-(1,4) chemical bonds between glucose units [4]. These enzymes synergistically hydrolyze cellulose into glucose and other soluble sugars. Among the lignocellulolytic enzymes, cellulases are found to be a potent source for a variety of industries such as chemicals, cosmetics, detergents, foods, and paper [5]. Amylases, the starch degrading hydrolases, are widely distributed in animals, plants, eukaryotes, and prokaryotes which play a major role in carbohydrate metabolism. These specifically cleave α-glycosidic linkage in starch and are categorized into α-amylase (EC 3.2.1.1) which randomly cleave the α-1,4 bonds between adjacent glucose units to produce maltose and maltodextrins, β-amylase (EC 3.2.1.2), and glucoamylase (EC 3.2.1.3). Fungal amylases are more reliable and preferred over other sources due to ease of production and cost-effectiveness. All these enzymes have wide applications in food, fermentation, paper, and textile industries [6,7,8].
The Western Ghats are known for the rich microbial diversity and hence considered as one of the biodiversity hotspots. Thus, the soil from this region could be a potential source of several useful fungi for industrial applications [9]. The combined effect and functional integration of multienzymes with distinct counterparts have established a major role in the industrial production of diverse biomolecules [10]. Several fungi have been exploited in the past for the isolation of useful industrially important enzymes such as amylase [8], β-glucosidases [4], and endoglucanases [3]. Keeping in view of all these, the present study was aimed at the isolation of filamentous fungi from soil samples of Western Ghat that are capable of producing extracellular enzymes, amylase, and cellulase.
Solid-state fermentation (SSF) is a better alternative over submerged fermentation for large-scale production of fungal metabolites as it resembles the natural environment for fungi to grow in the absence of free-flowing water. The fungi in SSF gives elevated production of metabolites by utilizing a cost-effective substrate with simpler equipment having lower operating expenses [11, 12]. Hence in this study, the selective isolation of fungi from this unique region was optimized for various physicochemical parameters to enhance the enzymatic activities of amylase and cellulase by using solid-state fermentation.
Materials and Methods
Soil Sample Collection
The soil samples were collected from seven different places of the Agumbe forest Western Ghats region (13.5027°N, 75.0903°E) in the state of Karnataka, India. The collection was performed by scraping a freshly exposed face from a depth of 3 inches below the surface with a sterilized scoop and transferred to sterile polythene bags. The collected samples were brought aseptically to the laboratory for further studies.
Isolation and Screening of Fungi
The isolation of fungi from the soil sample was carried out by the serial dilution method. The diluted samples were primarily screened by inoculating on potato dextrose agar (PDA) medium with a spread plate technique and incubated at 28 ± 2 °C for 5–7 days [13]. Each fungal isolate was secondarily screened for the amylase and cellulase activity on starch agar medium and carboxymethyl cellulose (CMC) agar medium, respectively, by point inoculation method. The zone of clearance on a selective medium by isolate was considered as screening criteria [14, 15].
Identification of Selective Isolates
The fungal isolates were microscopically identified with lactophenol cotton blue stain based on the morphological characteristics like surface appearance, colony color, and texture. Appropriate references were made utilizing taxonomic descriptions and mycological features [16]. The potent amylase and cellulase producing fungi were identified, authenticated, and certified by the Agharkar Research Institute, Pune. The isolates were preserved at 4 °C in PDA slants for further use.
Inoculum Preparation
For the preparation of inoculum, a loop full of 72 h old culture grown on PDA medium having 106 spores mL−1 was suspended in Tween 80 (0.1%) and used for inoculation.
Enzyme Production and Extraction
Production and extraction of amylase and cellulase from each organism were carried out by the SSF method. The medium containing 10 g of wheat bran substrate with 10 mL of salt solution containing (g/L) KNO3 (2.0), MgSO4‧7H2O (0.5), K2HPO4 (1.0), ZnSO4‧7H2O (0.437), FeSO4‧7H2O (1.116), and MnSO4‧7H2O (0.203) at pH 7.0 in 250 mL Erlenmeyer flask was autoclaved at 121 °C for 30 min. The flasks were inoculated with 1.0 mL of spore suspension and incubated at 28 °C for eight days. The cultures were extracted with 100 mL of chilled sterile water by shaking for 2 h in a rotary shaker. The obtained filtrate was centrifuged at 12,000×g at 4 °C for 10 min. The supernatant was filtered using Whatman filter paper and the crude enzyme extract was stored at 4 °C until further use [17].
Amylase Assay
The amylase activity was determined by mixing 0.1 mL of the enzyme solution with 0.5 mL of substrate (1% soluble starch in 0.1 M phosphate buffer, pH 7.0) and incubated at 40 °C for 30 min. Further, 1 mL of 3,5-dinitrosalicylic acid (DNS) reagent was added to the mixture and heated for 5 min in a boiling water bath which was then diluted by adding distilled water (10 mL). The absorbance was read at 540 nm using spectrophotometer against a blank containing buffer. A calibration curve was made with glucose to convert colorimeter readings to a unit of activity. One unit (U) of amylase activity defines as the amount of amylase enzyme liberating 1 µM of glucose/minute under described assay conditions [18].
Endoglucanase Assay
The endoglucanase activity was performed by adding 0.5 mL of 50 mM sodium citrate buffer (pH 4.8), 100 µL of enzyme extract and 500 µL of 2% CMC into an assay tube. The samples were incubated at 50 °C for 30 min and 1.0 mL of DNS was added. The tubes were kept in a boiling water bath for 5 min and 10 mL of distilled water was added and subsequently, the absorbance was measured at 540 nm. One unit (U) of enzyme activity described as the amount of enzyme needed to liberate 1 µM of glucose from CMC/minute under the described assay conditions [19].
Total Cellulase Activity: Filter Paper Assay
The total cellulase activity (FPase) was estimated using Whatman No. 1 filter paper (1 cm × 6 cm) strip as a substrate. A rolled strip (50 mg) of the filter paper was dipped into 0.5 mL of sodium citrate buffer (50 mM, pH 6.0) and incubated with 0.1 mL of enzyme extract at 50 °C for 1 h. To terminate the reaction, 1.0 mL of DNS was added and boiled for 5 min. The developed color was read at 540 nm. One unit (U) of enzyme activity was defined as the amount of FPase required to liberate 1 µM of glucose from the substrate/minute under described assay conditions [20].
β-Glucosidase Assay
The β-glucosidase activity was carried out with p-nitrophenyl-β-d-glucopyranoside (pNPG) as the substrate in a microtiter plate as per the standard protocol [21]. The enzyme extract (25 µL) was mixed with 50 µL of sodium acetate buffer (50 mM, pH 5.0) and the reaction was initiated by adding 25 µL of pNPG (10 mM) followed by incubation at 50 °C for 30 min. The reaction was ceased by adding 100 µL of glycine–NaOH buffer (0.4 M, pH 10.8) and the developed yellow color was read at 405 nm using ELISA reader (MULTISKAN EX; Thermo Scientific). One unit (U) of β-glucosidase activity was expressed as the amount of enzyme required to release 1 µM of p-nitrophenol per minute under assay conditions.
One-Factor-At-a-Time (OFAT) Optimization
The various parameters that influence the enzyme production were optimized by using the OFAT method. Different parameters were standardized for maximal amylase and cellulase production. The parameters investigated were different substrates (wheat bran, rice bran, groundnut cake, coconut cake, soybean powder, bagasse, paddy husk), with a variable incubation period (3 to 8 days), initial pH (4 to 9), temperature (25, 30, 40 and 50 °C), salt concentration (1% to 4%), moisture content (5% to 20%), nitrogen sources (ammonium sulfate, ammonium nitrate, beef extract, and yeast extract), and carbon sources (starch, cellulose, glucose, and lactose). Each parameter was used individually for the optimization process to determine the best production medium [22]. Different parameters having optimum production of amylase and cellulase in selective isolates were then incorporated together to formulate a new fermentation medium for the enhanced production of enzymes.
Partial Purification
The partial purification of the extracted crude enzyme was achieved by ammonium sulfate precipitation followed by dialysis. In this process, the clear supernatant collected from the fermentation was subjected initially to 50% ammonium sulfate precipitation. The precipitate thus obtained was discarded as there was no enzyme activity and the supernatant was further subjected to 60–80% ammonium sulfate precipitation, kept for 1 h at 4 °C and centrifuged at 14,000×g for 20 min. The pellet was dissolved in 5 mL of 10 mM phosphate buffer (pH 7.0) and dialyzed in the same buffer several times at 4 °C. The dialyzed enzyme mixture was lyophilized for further use.
Molecular Characterization
The selective fungal isolates producing maximum amylase and cellulase activity were subjected to molecular characterization. The sequence analysis was performed at Genespy research service, Mysore. In brief, the fungal culture was transferred to cetyltrimethylammonium bromide (CTAB) buffer and the genomic DNA extraction was done according to the standard protocol [23]. The tissue was crushed and vortexed thoroughly in the mixture containing CTAB buffer (800 µL) (2% CTAB, 1.4 M NaCl, 20 mM EDTA, 100 mM Tris–HCl, pH 8.0, 0.2% Mercaptoethanol) at 60 °C. The entire mixture was held at 60 °C for 20 min and briefly vortexed several times. After the incubation, 600 µL of chloroform/octanol (24:1) was added by vortexing vigorously and centrifuged for 5 min. The supernatant was transferred to a microfuge tube containing an equal volume of ice-cold isopropanol and incubated on ice for 10 min. Later, it was centrifuged for 8 min and the pellet was rinsed with 80% ethanol, air-dried, suspended in 50–100 µL of water for Polymerase chain reaction (PCR) amplification.
The sequence analysis was performed by amplifying sequences of the internal transcribed spacer (ITS) regions 1 and 2 by PCR (Thermocycler, Biorad) using the fungal universal primers, 5′-TCCGTAGGTGAACCTGCGG-3′ (forward) and 5′-TCCTCCGCTTATTGTATGC-3′ (reverse). The PCR was carried out in 25 μL reaction mixture containing 4 μL of DNA template, 2.5 μL of 10× PCR buffer, 2 μL of MgCl2, 5 μL of dNTPs, 0.36 μL of Taq polymerase, and 0.5 μL of forward and reverse primer. The amplification was followed with initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 52 °C for 1 min, and 72 °C for 1 min with a final extension step of 72 °C for 5 min [24]. Sequence analysis was done using ClustalX and GENEDOC software. The analysis of ITS sequence similarity was performed using the basic local alignment search tool (BLAST) and comparisons to other microbial sequences were done with National Center for Biotechnology Information (NCBI) database [25]. The phylogenetic and molecular evolutionary analyses were conducted using the software MEGA by the maximum likelihood method [26].
Electrophoresis
The sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the standard method [27] using Bio-Rad mini protean II electrophoresis unit with 12% resolving gel and 4% stacking gel at 150 V for 65 min. The protein markers were run alongside the sample and protein bands were visualized by silver staining method [28].
Amylase Zymography
Amylase zymography was performed with slight modifications of SDS-PAGE containing 12% resolving gel and 1.5 mg/mL soluble starch, whereas the stacking gel was made with 5% acrylamide lacking starch. Enzyme samples were loaded on to the wells along with gel-loading dye. The gel was washed for 10 min each twice using a washing buffer with little agitation. The first washing buffer contained 2% Triton X-100 in 50 mM Tris–HCl buffer (pH 7.4) whereas the second buffer contained 50 mM Tris–HCl (pH 7.4). The gel was then incubated for 1 h in modified substrate buffer (0.3 g CaCl2, 0.01 g NaCl, 0.5 mL Triton X-100, 0.3 g NaNO3 in 50 mL of 50 mM Tris buffer pH 5.5) for substrate digestion. The gel was rinsed with distilled water and stained for 5 min by diluting 1 mL of Lugol’s iodine stock solution (0.05 g I2, 0.1 g KI/mL) in 50 mL distilled water. The gel was rinsed repeatedly with distilled water to remove excess stain and photographed for documentation [29].
Cellulase Zymography
The cellulase zymography was performed after SDS-PAGE as per the standard protocol with some modifications. A 12% separation gel containing 0.1% carboxymethyl cellulose was poured into the plates. The gel was dipped for 30 min in sodium citrate buffer (50 mM, pH 5.5) containing 1% Triton X-100 for zymogram analysis. This was followed by incubation of gel for 30 min in sodium citrate buffer (50 mM, pH 5.5) to allow for the enzymatic conversion of the substrate. Then the gel was stained for 30 min with 0.1% Congo red and 1 M NaCl solution was used to destain the gel [30].
Statistical Analysis
The data recorded during experiments were subjected to significance testing using two-way ANOVA. GraphPad Prism software version 5.1 was used for the statistical analysis. Statistical significance was set at p < 0.05. Results were denoted as mean ± standard deviation (SD) of triplicate experiments.
Results and Discussion
Isolation, Screening, and Identification of Efficient Amylase and Cellulolytic Enzyme Producing Fungi
In the present study, after the primary screening on PDA medium, about 44 fungi were isolated from seven different soil samples of the Agumbe region by serial dilution method. Later, secondary screening was done to isolate amylase and cellulase producers by selective medium i.e., starch agar and CMC agar by point inoculation method. A total of 30 amylase and cellulase producing fungal isolates were obtained from secondary screening. These are represented as low (+), moderate (++), and high (+++) amylase and cellulase activity exhibiters as shown in Table 1.
Among these, seven best amylase and cellulase producing fungal isolates were considered for further work based on the zone of clearance on agar plate during primary screening. The selected species identified by morphotaxonomic features as, Cladosporium herbarum, Penicillium chrysogenum, Penicillium sp., Sarocladium mycophilum, Penicillium citrinum, Penicillium camemberti, and Aspergillus clavatus. These are further used for extracellular enzyme production by solid-state fermentation using wheat bran as substrate. The dialyzed enzyme extract obtained from each isolate after fermentation was quantitatively measured for amylase and cellulase activity and among these the best of two isolates, namely P. citrinum and A. clavatus, were selected for optimization and characterization of enzymes (Table 2).
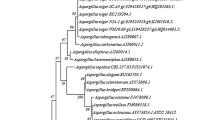
The phylogenetic tree was constructed for P. citrinum and A. clavatus (Fig. 1) by comparing with strains from GenBank with the highest similarities. The phylogenetic tree was constructed using the Maximum Likelihood method [31]. The neighbor-joining tree and subtree were generated using MEGA7 software [32]. The analysis of ITS sequences of P. citrinum and A. clavatus supported the results obtained in morphological identification. According to the NCBI blast search, the ITS region sequence of the isolated fungi had 100% homology with Penicillium citrinum and Aspergillus clavatus. Hence the ITS sequences of both the fungal strains, P. citrinum, and A. clavatus were submitted to GenBank and obtained accession numbers as MK271291 and MK271292, respectively.
Optimization of Amylase and Cellulase Production by Solid-State Fermentation
Effect of Different Substrates on the Production of Enzyme
The fermentation process includes the selection of appropriate substrate for SSF from various agro-industrial wastes for microbial growth and enzyme production [33]. Substrates such as rice bran, wheat bran, paddy husk, along with other starch-containing wastes have acquired importance as it supports the growth during enzyme production and wheat bran was reported as the utmost promising substrate [34]. The substrate optimization was achieved by SSF method using seven substrates, viz. wheat bran (Wb), rice bran (Rb), groundnut cake (Gc), coconut cake (Cc), soybean powder (Sp), bagasse (Ba) and paddy husk (Ph) for amylase and cellulase production by P. citrinum and A. clavatus. The maximal amylase and cellulase production was obtained in wheat bran by both P. citrinum (17.30 ± 0.6 U/mg; 14.88 ± 0.08 U/mg) and A. clavatus (47.98 ± 2.9 U/mg; 29.10 ± 0.4 U/mg). The sugarcane bagasse gave the lowest amylase (1.87 ± 0.2 U/mg) and cellulase (1.48 ± 0.08 U/mg) production in P. citrinum. Groundnut cake (20.88 ± 2.3 U/mg) and soybean powder (4.00 ± 0.3 U/mg) reduced amylase and cellulase production by A. clavatus (Fig. 2a). Though the production of enzyme from P. citrinum and A. clavatus is relatively low/similar when compared to earlier data (Table 3), further enrichment of production medium will enhance the enzyme production relatively with wheat bran substrate. Earlier studies have shown that the maximum amylase production was obtained from A. oryzae by using dry fermented wheat bran as substrate at pH 6.0 in SSF [20]. Similarly, with F. solani SY7 highest amylase production (141.18 U/g of dry substrate) was achieved under SSF using wheat bran after the 3rd day of incubation [35]. The wheat bran supplies appropriate nutrients and can remain loose even in moist conditions by providing huge surface area [36]. Hence, in the present study, wheat bran was used as a substrate for all the optimization criteria.
The enzymatic activity in the presence of different substrates [wheat bran (Wb), rice bran (Rb), groundnut cake (Gc), coconut cake (Cc), bagasse (Ba), paddy husk (Ph), and soybean powder (Sp)] on amylase [( ) A. clavatus, (
) A. clavatus, ( ) P. citrinum] and cellulase [(
) P. citrinum] and cellulase [( ) A. clavatus, (
) A. clavatus, ( ) P. citrinum] production (a), different incubation period and different carbon source on amylase (
) P. citrinum] production (a), different incubation period and different carbon source on amylase ( ), endoglucanase (
), endoglucanase ( ), FPase (
), FPase ( ), and β-glucosidase (
), and β-glucosidase ( ) production from A. clavatus (b, c), and P. citrinum (d, e) by SSF. The results are the mean of three independent experiments and the bars correspond to standard deviations
) production from A. clavatus (b, c), and P. citrinum (d, e) by SSF. The results are the mean of three independent experiments and the bars correspond to standard deviations
Effect of Incubation Time on the Production of Enzyme
The effect of incubation period on amylase and cellulase production by P. citrinum and A. clavatus was studied by incubating for 3 to 8 days. The production of amylase (64.00 ± 9.2 U/mg), endoglucanase (111.56 ± 4.9 U/mg), FPase (17.67 ± 2.5 U/mg), and β-glucosidase (10.99 ± 0.9 U/mg) was maximum on the 4th day of the incubation period and thereafter a decrease in the production was observed with P. citrinum (Fig. 2d). Increased production of amylase (155.84 ± 8.8 U/mg) and endoglucanase (15.47 ± 1.6 U/mg) was observed with A. clavatus on the 4th day of incubation. The FPase (7.42 ± 0.6 U/mg) and β-glucosidase (3.85 ± 1.0 U/mg) activity was maximum on the 6th day of incubation (Fig. 2b). Further, an increase in the incubation period resulted in decreased enzyme production in both cases. After a certain period, the reduced enzyme production may be due to the depleted nutrients, a pH change in the medium or depletion of more amorphous substrates [43]. This is inconsistent with an earlier observation that A. terreus GN1 isolated from the Jammu region compost soil showed a decrease in cellulase production with increased incubation time [44]. It was also found that the amylases production was maximum on the 5th day of the incubation at 30 °C by SSF from Aspergillus sp. MK07 [45]. Similarly, A. niger showed increased cellulase activity after 5–6 days of incubation in the presence of rice straw [46]. The depletion of oxygen in the medium due to static fermentation conditions may be another vital cause of the reduction in enzyme production.
Effect of Carbon Source on the Production of Enzyme
The effect of different carbon sources on amylase, FPase, β-glucosidase, and endoglucanase production by P. citrinum is shown in Fig. 2e. The data indicates that the maximum production of amylase (29.66 ± 0.5 U/mg) and FPase (14.57 ± 0.9 U/mg) were observed with 2% starch. The maximum β-glucosidase (3.13 ± 0.5 U/mg), and endoglucanase (14.57 ± 0.9 U/mg) production were observed with 2% glucose and 2% lactose. Further, 2% lactose reduced FPase (12.69 ± 1.2 U/mg) production and 2% cellulose as a supplement reduced amylase (17.02 ± 1.8 U/mg), endoglucanase (17.95 ± 2.7 U/mg), and β-glucosidase (1.62 ± 0.1 U/mg) production. The highest amylase (160.78 ± 10.9 U/mg) activity by A. clavatus was obtained when 2% lactose was used as a carbon source and 2% cellulose as carbon source reduced amylase (87.36 ± 1.9 U/mg) production but enhanced endoglucanase (19.88 ± 1.3 U/mg), FPase (7.34 ± 0.9 U/mg), and β-glucosidase (3.72 ± 0.39 U/mg) production. On the other hand, 2% lactose and 2% glucose lessened β-glucosidase (2.11 ± 0.1 U/mg), FPase (4.34 ± 0.6 U/mg), and endoglucanase (13.64 ± 1.0 U/mg) production (Fig. 2c). The starch was shown to be a suitable carbon source for the optimal production of amylase by A. niger and R. stolonifer [47]. All the reducing sugars provoke the secretion of cellulase and no non-reducing sugars stimulate cellulase production, and the reducing end of the oligosaccharides is required for the enzyme induction process [48].
Effect of Nitrogen Source on the Production of Enzyme
Both organic and inorganic nitrogen sources were added as supplements in the SSF medium. The maximal production of amylase (35.15 ± 3.2 U/mg), FPase (14.36 ± 1.6 U/mg), endoglucanase (39.26 ± 4.1 U/mg), and β-glucosidase (2.69 ± 0.3 U/mg) by P. citrinum was achieved with ammonium nitrate as the nitrogen source. But yeast extract as nitrogen source decreased amylase, endoglucanase, and FPase production in P. citrinum. The ammonium sulfate as nitrogen source increased the production of amylase (122.97 ± 11.7 U/mg), FPase (19.79 ± 1.6 U/mg), and β-glucosidase (6.29 ± 0.1 U/mg) production by A. clavatus but hindered β-glucosidase production in P. citrinum. Ammonium nitrate as nitrogen source also increased the production of endoglucanase (21.78 ± 2.6 U/mg) by A. clavatus but beef extract influenced the reduction of multienzyme production by A. clavatus considerably (Fig. 3a, b). It was observed that ammonium sulfate was the best inorganic nitrogen source for the maximum production of FPase and endoglucanase by A. flavus AT-2 and A. niger AT-3 strains [49]. Similar behavior has been observed in the case of C. asperatum for the production of amylase [50].
Effect of different nitrogen source, temperature, and pH on amylase ( ), Endoglucanase (
), Endoglucanase ( ), FPase (
), FPase ( ), and β-glucosidase (
), and β-glucosidase ( ) production from A. clavatus (a, c, e), and P. citrinum (b, d, f) by SSF respectively. The results are the mean of three independent experiments and the bars correspond to standard deviations
) production from A. clavatus (a, c, e), and P. citrinum (b, d, f) by SSF respectively. The results are the mean of three independent experiments and the bars correspond to standard deviations
Effect of Temperature and pH on the Production of Enzyme
In SSF, temperature acts as the essential physical variable altering the production [51]. The optimization of incubation temperature for the production of FPase (3.57 ± 0.2 U/mg), endoglucanase (13.62 ± 0.7 U/mg), and β-glucosidase (2.90 ± 0.2 U/mg) from A. clavatus under SSF conditions revealed that the enzyme production gradually increased from 25 to 40 °C and maximal enzyme production of all the three enzymes was observed at 40 °C. Any increase in temperature beyond 40 °C had a drastic adverse effect on the enzyme production but amylase (112.73 ± 8.2 U/mg) production was maximum at 30 °C and a further increase in temperature reduced enzyme production (Fig. 3c). In case of P. citrinum, the temperature of 25 °C was optimum for amylase (121.84 ± 8.8 U/mg) and FPase (4.34 ± 0.4 U/mg) production and 40 °C was found to be optimum for the maximum production of endoglucanase (13.59 ± 0.8 U/mg), whereas 30 °C was optimum for β-glucosidase (2.69 ± 0.3 U/mg) production (Fig. 3d). At 50 °C, both the organisms did not grow in the production medium. It has earlier been reported that an increased cellulase production was observed in A. niger (25 °C and 30 °C) and T. reesei (30 °C) up to 30 °C and later its production has declined [52, 53]. The optimum temperature of exactly 40ºC was needed for increased cellulase production from T. castaneum [53]. The best temperature for amylase production by A. niger was between 25 to 30 °C and incubation at higher temperatures affected the enzyme synthesis [45].
The pH is also a significant factor where the concentration of hydrogen ions in the medium determines the development and morphology of microorganisms in the enzyme production [54]. The pH is condition which play a vital role in enzyme production. The maximum productivity of FPase (7.99 ± 1.2 U/mg), β-glucosidase (4.20 ± 0.7 U/mg), and endoglucanase (46.94 ± 6.6 U/mg) were recorded at pH 7.0 but amylase (29.20 ± 3.1 U/mg) production was highest at pH 5.0 by P. citrinum (Fig. 3e). The optimum pH for the production of endoglucanase (8.03 ± 0.7 U/mg), and β-glucosidase (2.05 ± 0.2 U/mg) was at pH 7.0 whereas amylase (50.44 ± 4.5 U/mg), and FPase (3.20 ± 0.3 U/mg) production was increased at pH 4.0 by A. clavatus (Fig. 3f). The cellulase production by Fomitopsis sp. RCK2010 was also maximum at medium pH 5.5. The optimum pH 5.0 increased the cellulase production by P. chrysogenum in SSF with pea pod as a substrate [40]. The amount of α-amylase increased significantly in A. niger by increasing the pH value up to 6.5 [55].
Effect of Salt Concentration and Moisture Content on the Production of Enzyme
To understand the role of salt, different concentrations of NaCl (1–4%) were employed to check the effect of concentration of NaCl on amylase and cellulase production by P. citrinum and A. clavatus. The results indicated that P. citrinum showed maximum amylase (39.26 ± 9.0 U/mg), FPase (17.63 ± 1.6 U/mg), β-glucosidase (4.73 ± 0.3 U/mg), and endoglucanase (45.63 ± 6.7 U/mg) production in the medium containing 4% NaCl. But 1% NaCl concentration decreased enzyme production and 4% NaCl concentration enhanced multienzyme production in P. citrinum (Fig. 4a). A. clavatus gave maximum titer of amylase (162.71 ± 17.9 U/mg) at 2% NaCl, whereas 3% NaCl increased FPase (19.17 ± 0.7 U/mg), β-glucosidase (4.52 ± 0.6 U/mg), and endoglucanase (24.25 ± 4.5 U/mg) activity. A further increase in NaCl concentration reduced enzyme production (Fig. 4c). The optimized NaCl concentration for maximum production of amylase by A. niger-ML-17 and R. oligosporus-ML-10 was 0.5% and 0.75%, respectively [56]. In another study, however, the purified and characterized amylase enzyme produced by A. oryzae in SSF was with 0.1% concentration of NaCl [38].
Effect of salt concentration and moisture content on amylase ( ), endoglucanase (
), endoglucanase ( ), FPase (
), FPase ( ), and β-glucosidase (
), and β-glucosidase ( ) production from A. clavatus (a, b), and P. citrinum (c, d) by SSF respectively. The results are the mean of three independent experiments and the bars correspond to standard deviations
) production from A. clavatus (a, b), and P. citrinum (c, d) by SSF respectively. The results are the mean of three independent experiments and the bars correspond to standard deviations
The appropriate moisture content of the substrate is one of the critical factors influencing SSF. Our results revealed that, as moisture content increased, FPase (17.67 ± 2.2 U/mg; 14.07 ± 1.7 U/mg) and endoglucanase (29.54 ± 4.3 U/mg; 18.29 ± 2.9 U/mg) production also increased in both P. citrinum and A. clavatus. The maximum production was found at 15% moisture content in P. citrinum and A. clavatus, respectively. A 5% moisture content increased the amylase (43.37 ± 6.1 U/mg; 142.32 ± 9.2 U/mg) production by P. citrinum and A. clavatus and further alteration in moisture content had an adverse effect on enzyme production. The β-glucosidase (3.56 ± 0.4 U/mg; 4.63 ± 0.8 U/mg) production was increased in presence of 15% and 20% moisture content in A. clavatus and P. citrinum, respectively (Fig. 4b, d). It was earlier reported that the highest amylase production was observed at 25% of moisture content by A. niger DTO: H5 using wheat bran as substrate [57]. Further, 70% of moisture content was found to be optimum for cellulase production when banana peel was used as the substrate [58]. The moisture enabled better utilization of the substrate by the microorganisms and the effective mass transfer in the solid phase particles depends on the characteristics of the substrate and suitable moisture content. However, the increased moisture content influenced enzyme production negatively. It reduced the surface area of the particles and made the water film thicker, which lessened the accessibility of the air to the particles [59].
As a result of optimization, the enzyme production was maximum for each factor depending upon the provided nutrient. By combining all the best optimized parameters to each enzyme, one new medium was formulated to obtain maximum production of amylase and cellulase by P. citrinum and A. clavatus through SSF which gave promising results. The optimized medium influenced the increased production of all the four enzymes in both the isolates when compared to individual optimization criteria. In P. citrinum, the amylase (121.84 ± 8.8 U/mg) production was maximum followed by endoglucanase (66.11 ± 5.2 U/mg), FPase (46.94 ± 6.6 U/mg) and β-glucosidase (12.96 ± 1.0 U/mg) production. The increased production of amylase (131.40 ± 7.9 U/mg) endoglucanase (25.59 ± 1.5 U/mg), FPase (14.88 ± 0.08 U/mg), and β-glucosidase (9.49 ± 0.5 U/mg) was observed in A. clavatus (Table 4). Taken together, our results indicate that the newly formulated optimal production medium yields the highest production of multienzymes in both P. citrinum and A. clavatus.
Partial Purification of Amylase and Cellulase
The crude enzyme extract was used for the purification of amylase and cellulase by ammonium sulfate precipitation followed by dialysis. The protein was precipitated by using various ammonium sulfate concentrations in the range of 20 to 90% (w/v). The maximum activity was observed between 50 and 80% ammonium sulfate fractions as compared to the rest of fractions. Accordingly, the crude enzyme extract was precipitated using 50–80% ammonium sulfate concentration for both the enzymes. The fractions were then subjected to dialysis and the activity was measured. In the case of A. clavatus, the partially purified amylase enzyme fraction showed 2219 U of total activity, 55.47/mg of specific activity with a recovery of 30% and 21.01-fold of purification. Further, the partially purified cellulase fraction showed a total activity of 410 U, specific activity of 13.66 U/mg with 19% yield and 11.38-fold purification. On the other hand, the partially purified amylase fraction from P. citrinum showed a total activity of 1080 U, 30.85 U/mg of specific activity with 30% of recovery and 8.69-fold purification. In the case of cellulase, the total activity was 462 U, the specific activity was 23.1 U/mg with a recovery of 33% and 4.16-fold purification (Table 5). Earlier studies have reported that the 80% ammonium sulfate fraction had a protein content of 21.54 mg/ml, specific activity of 14,864.94 U/mg with 83.8% recovery and 4.7-fold purification for cellulase from B. vallismortis RG-07 [60]. The endocellulase produced from endophyte F. oxysporum was reported to have achieved 15.9-fold purification with a yield of 27.7% [61]. Similarly, the cell-free dialyzed α-amylase from A. terreus NCFT4269 displayed 381 U of total activity and 15.24 U/mg protein specific activity with 36.95% yield and 2.305-fold purification [62].
SDS-PAGE and Zymographic Analysis
The partially purified amylase and cellulase from A. clavatus and P. citrinum were analyzed for their purity by SDS-PAGE and zymography technique. The molecular mass of partially purified P. citrinum amylase was found to be ~ 75 kDa and that of cellulase was ~ 155 kDa, ~ 130 kDa, and ~ 115 kDa, respectively, as observed by the zymographic analysis (Fig. 5a). In case of A. clavatus, the zymography revealed the presence of three bands of ~ 135 kDa, ~ 75 kDa, ~ 62 kDa with amylase activity and three bands with cellulase activity having molecular weight (MW) of ~ 130 kDa, ~ 69 kDa, and ~ 35 kDa (Fig. 5b). The samples loaded for zymographic analysis were neither boiled nor treated with any denaturing agents. Hence, the proteins were in their native state and not denatured during gel separation. This confirms that the P. citrinum and A. clavatus produce three isoforms of cellulase and A. clavatus secretes three isoforms of amylase. However, in the case of P. citrinum, only one type of amylase was observed.
The electrophoretic analysis of partially purified enzymes from P. citrinum (a) and A. clavatus (b). M: Prestained protein ladder (245–10 kDa); 1: Partially purified enzyme obtained after ammonium sulfate precipitation and dialysis; 2: Enzymatic activity of partially purified amylase. 3: Zymogram of partially purified cellulase. Protein was detected by silver staining, a portion of the gel was separated and stained by iodine and Congo red stains for amylase and cellulase activity later the appeared zymogram was compared with marker proteins
According to the available literature, to date, there are no reports about the production of amylase and cellulase isoforms both in A. clavatus and P. citrinum. In contrast, ours is the first report of the production of various isoforms of amylase as well as cellulase in A. clavatus and P. citrinum. However, the multifunctional and multienzymes that are helpful in degrading cellulose and polysaccharide exist in many microorganisms including filamentous fungi [63]. The amylases and cellulases can cohabit in multiple forms as isoenzymes and the production of these isoforms can be accomplished by controlling numerous extracellular parameters [64]. The two purified α-amylase isoforms from A. oryzae strain S2 was reported to have the MW of 50 kDa and 42 kDa [65]. An earlier study has reported the presence of two CM-Cellulase isozymes in the ammonium sulfate precipitate fraction of S. fredii CCRC15769 as indicated by zymography which are shown to possess the MW of 19.6 kDa and 30 kDa [66]. In another study, the endoglucanase zymography of A. japonicus PJ01 revealed that the multienzyme complexes consisted of different isozymes that are produced in submerged and solid-state fermentation [67].
Conclusion
Among the several isolates screened, A. clavatus and P. citrinum yielded maximum multienzymes (amylase, endoglucanase, and FPase) via solid-state fermentation using wheat bran as the substrate as compared to other substrates studied. The productivity and yield of the enzyme were found to be appreciably influenced by various parameters such as temperature, pH, carbon source, and nitrogen source. The optimized production medium resulted in the increased production of amylase, endoglucanase, β-glucosidase, and FPase enzymes. The zymographic analysis demonstrated that the partially purified enzymes from both the isolates constituted different isoforms. Overall, our results indicated that the fungi from this unique Western Ghat region, which has not been explored so far, could be a potential source of commercially and industrially important enzymes. However, further characterization of these enzymes is necessary to understand their unique properties for industrial applications.
References
Ugoh SC, Ijigbade B (2013) Production and characterization of amylase by fungi isolated from soil samples at Gwagwalada, FCT, Abuja-Nigeria. Rep Opin 5:44–53
Conesa A, Punt PJ, van Luijk N, van den Hondel CA (2001) The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet Biol 33:155–171. https://doi.org/10.1006/fgbi.2001.1276
Oliveira PC, de Brito AR, Pimentel AB, Soares GA, Pacheco CSV, Santana NB, da Silva EGP, Fernandes A, Ferreira MLO, Oliveira JR, Franco M (2019) Cocoa shell for the production of endoglucanase by Penicillium roqueforti ATCC 10110 in solid state fermentation and biochemical properties. Rev Mexicana Ingeniería Quím 18:777–787. https://doi.org/10.24275/uam/izt/dcbi/revmexingquim/2019v18n3/Oliveira
Santos TC, Reis NS, Silva TP, Bonomo RC, Aguiar-Oliveira E, Oliveira JR, Franco M (2017) Production, optimisation and partial characterisation of enzymes from filamentous fungi using dried forage cactus pear as substrate. Waste Biomass Valoriz. https://doi.org/10.1007/s12649-016-9810-z
Sajith S, Priji P, Sreedevi S, Benjamin S (2016) An overview on fungal cellulases with an industrial perspective. J Nutr Food Sci 6:1–13. https://doi.org/10.4172/2155-9600.1000461
Banakar PS, Thippeswamy B, Thirumalesh BV, Naveenkumar KJ (2012) Isolation, production and partial purification of fungal amylase from forest soils of Bhadra Wildlife Sanctuary. Western Ghats. Inventi Rapid Pharm Biotechnol Microbiol 3:1–7
Manivasagan P, Gnanam S, Sivakumar K, Thangaradjou T, Vijayalakshmi S, Balasubramanian T (2010) Isolation, identification and characterization of multiple enzyme producing actinobacteria from sediment samples of Kodiyakarai Coast, the Bay of Bengal. Afr J Microbiol Res 4:1550–1559
Pereira AS, Fontan RI, Franco M, Júnior EC, Veloso CM, Sampaio VS, Bonomo P, Bonomo RC (2018) Study of alpha-amylase obtained by solid state fermentation of cassava residue in aqueous two-phase systems. Braz J Chem Eng 35:1141–1152. https://doi.org/10.1590/0104-6632.20180353s20170003
Mukunda S, Onkarappa R, Prashith Kekuda TR (2012) Isolation and screening of industrially important fungi from the soils of Western Ghats of Agumbe and Koppa, Karnataka, India. Star J 7522:27–32. https://doi.org/10.4314/star.v1i4.98816
Garlapati VK, Maheswari N, Gupta A (2015) Isolation and screening of fungal isolates for multienzyme production through submerged and solid state fermentations. J Bioprocess Biotechnol 5:249. https://doi.org/10.4172/2155-9821.1000249
Bagewadi ZK, Ninnekar HZ (2015) Production, purification and characterization of endoglucanase from Aspergillus fumigatus and enzymatic hydrolysis of lignocellulosic waste. Int J Biotechnol Biomed Sci 1:25–32
Iqbal NHM, Ahmed I, Zia MA, Irfan M (2011) Purification and characterization of the kinetic parameters of cellulase produced from wheat straw by Trichoderma Viride under SSF and its detergent compatibility. Adv Biosci Biotechnol 2:149–156. https://doi.org/10.4236/abb.2011.23024
Elad Y, Chet I, Henis YA (1981) Selective medium for improving quantitative isolation of Trichoderma spp. from soil. Phytoparasitica 9:59–67. https://doi.org/10.1007/BF03158330
Upadhyay MK, Sharma R, Pandey KA, Rajak RC (2005) An improved zymographic method for detection of amylolytic enzymes of fungi on polyacrylamide gels. Mycologist 19:138–140. https://doi.org/10.1017/S0269915X05004015
Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A (2008) A rapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr Microbiol 57:503–507. https://doi.org/10.1007/s00284-008-9276-8
Ogbonna CN, Okpokwu NM, Okafor CU, Onyia CE (2014) Isolation and screening of amylase producing fungi obtained from garri processing site. Int J Biotechnol Food Sci 2:88–93
Muthulakshmi C, Gomathi D, Kumar DG, Ravikumar G, Kalaiselvi M, Uma C (2011) Production, purification and characterization of protease by Aspergillus flavus under solid state fermentation. Jordan J Biol Sci 4:137–148
Bernfeld P (1955) Amylases alpha and beta. Methods Enzymol 1:149–158. https://doi.org/10.1016/0076-6879(55)01021-5
Mandels M, Andreotti R, Roche C (1976) Measurement of saccharifying cellulase. Biotechnol Bioeng Symp 6:21–33
Zambare V (2010) Solid state fermentation of Aspergillus oryzae for glucoamylase production on agro residues. Int J Life Sci 4:16–25. https://doi.org/10.3126/ijls.v4i0.2892
Soni R, Nazir A, Chadha BS (2010) Optimization of cellulase production by a versatile Aspergillus fumigatus Fresenius strain (AMA) capable of efficient deinking and enzymatic hydrolysis of Solka floc and bagasse. Ind Crops Prod 31:277–283. https://doi.org/10.1016/j.indcrop.2009.11.007
Singh AK, Maharana AK, Masih H, Kumar Y, Mishra SK (2012) Production, optimization and purification of bacterial cellulase by solid state bio-processing of agro biomass. Res J Pharm Biol Chem Sci 3:977–989
Zhang YP, Uyemoto KBC (1998) A small scale procedure for extracting nucleic acids from woody plants infected with various phytopathogens for PCR assay. J Virol Methods 71:45–50. https://doi.org/10.1016/s0166-0934(97)00190-0
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA 109:6241–6246. https://doi.org/10.1073/pnas.1117018109
Altschul SF, Maddan TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306. https://doi.org/10.1093/bib/bbn017
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Celis JE, Carte N, Hunter T, Simons K, Small JV, Shotton D (2006) Cell biology: a laboratory handbook. Elsevier, Academic Press, New York
Wood TM, Bhat KM (1988) Methods for measuring cellulase activities. Methods Enzymol 160:87–112. https://doi.org/10.1016/0076-6879(88)60109-1
Geib SM, Tien M, Hoover K (2010) Identification of proteins involved in lignocelluloses degradation using in gel zymogram analysis combined with mass spectroscopy-based peptide analysis of gut proteins from larval Asian longhorned beetles, Anoplophora glabripennis. Insect Sci 17:253–264. https://doi.org/10.1111/j.1744-7917.2010.01323.x
Amura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kunamneni A, Permaul K, Singh S (2005) Amylase production in solid state fermentation by the thermophilic fungus Thermomyces lanuginosus. J Biosci Bioeng 2:168–171. https://doi.org/10.1263/jbb.100.168
Anto H, Trivedi UP, Patel KC (2006) Glucoamylase production by solid-state fermentation using rice flake manufacturing waste products as substrate. Bioresour Technol 97:1161–1166. https://doi.org/10.1016/j.biortech.2005.05.007
Akatin MY (2019) An overview of amylase production by solid state fermentation (SSF) since 2010. J Technol Soc Sci 9:1–7
Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandey A (2007) Alpha amylase production by Aspergillus oryzae employing solid-state fermentation. J Sci Ind Res 66:621–626
Tallapragada P, Dikshit R, Jadhav A, Sarah U (2017) Partial purification and characterization of amylase enzyme under solid state fermentation from Monascus sanguineus. J Genet Eng Biotechnol 15:95–101. https://doi.org/10.1016/j.jgeb.2017.02.003
Irfan M, Nadeem M, Syed Q (2012) Media optimization for amylase production in solid state fermentation of wheat bran by fungal strains. J Cell Mol Biol 10:55–64
Sibi G, Rajan C, Grover S, Biswas R, Pyne O, Tirkey SR, Veeramallegowda VPC, Gonsalves SM (2019) Amylase production by Aspergillus niger using agroindustrial residues under temperature mediated solid state fermentation. Enliven Microb Microbial Tech 6:1–5
Khattab MSA, Azzaz HH, Tawab AM, Murad HA (2019) Production optimization of fungal cellulase and its impact on ruminal degradability and fermentation of diet. Int J Dairy Sci 14:61–68. https://doi.org/10.3923/ijds.2019.61.68
Ortiz GE, Guitart ME, Cavalitto SF, Albertó EO, Fernández-Lahore M, Blasco M (2015) Characterization, optimization, and scale-up of cellulases production by Trichoderma reesei cbs 836.91 in solid-state fermentation using agro-industrial products. Bioprocess Biosyst Eng 38:2117–2128. https://doi.org/10.1007/s00449-015-1451-2
Pandey AK, Edgard G, Negi S (2016) Optimization of concomitant production of cellulase and xylanase from Rhizopus oryzae SN5 through EVOP-factorial design technique and application in Sorghum Stover based bioethanol production. Renew Energy 98:51–56. https://doi.org/10.1016/j.renene.2016.05.071
Ghose TK (1987) Measurement of cellulase activities. Pure Appl Chem 59:257–268. https://doi.org/10.1351/pac198759020257
Nazir A, Soni R, Saini HS, Kaur A, Chadha BS (2010) Profiling differential expression of cellulases and metabolite footprints in Aspergillus terreus. Appl Biochem Biotechnol 162:538–547. https://doi.org/10.1007/s12010-009-8775-9
Chimata MK, Sasidhar P, Challa S (2010) Production of extracellular amylase from agricultural residue by a newly isolated Aspergillus species in solid state fermentation. Afr J Biotechnol 9:5162–5169
Kang SW, Park YS, Lee JS, Hong SI, Kim SW (2004) Production of cellulases and hemicellulases by Aspergillus niger KK2 from lignocellulosic biomass. Bioresour Technol 91:153–156. https://doi.org/10.1016/S0960-8524(03)00172-X
Saleem A, Ebrahim MKH (2014) Production of amylase by fungi isolated from legume seeds collected in Almadinah Almunawwarah, Saudi Arabia. J Taibah Univ Sci 8:90–97. https://doi.org/10.1016/j.jtusci.2013.09.002
Korish M (2003) Production, purification, properties and application of the cellulases from a wild type strain of a yeast isolate. PhD, Institute of Microbiology and Wine Research, Johannes Gutenberg University, Mainz
Dutt D, Kumar A (2014) Optimization of cellulase production under solid-state fermentation by Aspergillus flavus (At-2) and Aspergillus niger (At-3) and Its Impact on stickies and ink particle size of sorted office paper. Cell Chem Technol 48:3–4
Sanghvi GV, Rina DK, Kishore SR (2011) Isolation, optimization, and partial purification of amylase from Chrysosporium asperatum by submerged fermentation. J Microbiol Biotechnol 21:470–476. https://doi.org/10.4014/jmb.0910.10014
Krishna C (2005) Solid state fermentation systems-an overview. Crit Rev Biotechnol 25:1–30. https://doi.org/10.1080/07388550590925383
Hanif A, Yasmeen A, Rajoka MJ (2004) Induction, production, repression and derepression of exoglucanase synthesis in Aspergillus niger. Bioresour Technol 94:311–319. https://doi.org/10.1016/j.biortech.2003.12.013
Abdullah JJ, Greetham D, Pensupa N, Tucker GA, Du C (2016) Optimizing cellulase production from municipal solid waste (MSW) using solid state fermentation (SSF). J Fundam Renew Energy Appl 6:206. https://doi.org/10.4172/2090-4541.1000206
Rehman FU, Aslam M, Tariq MI, Shaheen A, Sami AJ, Naveed NH, Batool AI (2009) Isolation of cellulolytic activities from Tribolium castaneum (red flour beetle). Afr J Biotechnol 8:6710–6715. https://doi.org/10.4314/ajb.v8i23.66387
Rizk MA, El-Kholany EA, Abo-Mosalum EMR (2019) Production of α-amylase by Aspergillus niger isolated from mango kernel. Middle East J Appl Sci 9:134–141
Deswal D, Khasa YP, Kuhad RC (2011) Optimization of cellulase production by a brown rot fungus Fomitopsis sp. RCK2010 under solid state fermentation. Bioresour Technol 102:6065–6072. https://doi.org/10.1016/j.biortech.2011.03.032
Patel AK, Nampoothiri MK, Ramachandaran S (2005) Partial purification and characterization of α-amylase produced by Aspergillus oryzae using spent brewing grains. Indian J Biotechnol 4:336–341
Ire FS, Eruteya OC, Amaechi V (2017) Optimization of culture conditions using one-factor-at-time methodology and partial purification of amylase from Aspergillus niger of DTO: H5 under solid state fermentation. Int J Curr Microbiol Appl Sci 6:307–325. https://doi.org/10.20546/ijcmas.2017.605.035
Krishna C (1999) Production of bacterial cellulases by solid state bio-processing of banana wastes. Biores Technol 69:231–239. https://doi.org/10.1016/S0960-8524(98)00193-X
Raimbault M, Alazard D (1980) Culture method to study fungal growth in solid fermentation. Eur J Appl Microbiol Biotechnol 9:199. https://doi.org/10.1007/BF00504486
Gaur R, Tiwari S (2015) Isolation, production, purification and characterization of an organic-solvent-thermostable alkalophilic cellulase from Bacillus vallismortis RG-07. BMC Biotechnol 15:1–12. https://doi.org/10.1186/s12896-015-0129-9
Dar RA, Saba I, Shahnawaz M, Sangale MK, Ade AB, Rather SA, Qazi PH (2013) Isolation, purification and characterization of carboxymethyl cellulase (CMCase) from endophytic Fusarium oxysporum producing podophyllotoxin. Adv Enzyme Res 1:91–96. https://doi.org/10.4236/aer.2013.14010
Sethi BK, Jana A, Nanda PK, Das Mohapatra PK, Sahoo SL, Patra JK (2016) Production of α-amylase by Aspergillus terreus NCFT4269.10 using pearl millet and its structural characterization. Front Plant Sci 7:1–13. https://doi.org/10.3389/fpls.2016.00639
Ohtsuki T, Suyanto YS, Ui S, Mimura A (2005) Production of large multienzyme complex by aerobic thermophilic fungus Chaetomium sp. nov. MS-017 grown on palm oil mill fibre. Lett Appl Microbiol 40:111–116. https://doi.org/10.1111/j.1472-765X.2004.01644.x
Sahnouna M, Bejara S, Sayaria A, Triki MA, Kriaaa M, Kammouna R (2012) Production, purification and characterization of two α-amylase isoforms from a newly isolated Aspergillus oryzae strain S2. Process Biochem 47:18–25. https://doi.org/10.1016/j.procbio.2011.09.016
Hu C-Y, Lin L-P (2003) Characterization and purification of hydrolytic enzymes in Sinorhizobium fredii CCRC15769. World J Microbiol Biotechnol 19:515–522. https://doi.org/10.1023/A:102512271
Li P, Xia J, Shan Y, Nie Z (2015) Comparative study of multi-enzyme production from typical agro-industrial residues and ultrasound-assisted extraction of crude enzyme in fermentation with Aspergillus japonicus PJ01. Bioprocess Biosyst Eng 38:2013–2022. https://doi.org/10.1007/s00449-015-1442-3
Acknowledgements
The authors gratefully acknowledge Kuvempu University, Karnataka, India, for providing research facility, Mr. Santhosh K N for suggesting comments that greatly improved the manuscript, and Mr. Sanjay Yalashetti for assisting in phylogenetic tree construction.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
BRS performed all the experiments, wrote the manuscript and analyzed data. RNHA carried out the computational analysis, results interpretation and reviewed the final draft. NBT designed the experiment, carried out results interpretation and manuscript correction.
Corresponding author
Ethics declarations
Conflict of interest
Authors have declared as they have no conflict of interest.
Research Involving Human and Animal Rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shruthi, B.R., Achur, R.N.H. & Nayaka Boramuthi, T. Optimized Solid-State Fermentation Medium Enhances the Multienzymes Production from Penicillium citrinum and Aspergillus clavatus. Curr Microbiol 77, 2192–2206 (2020). https://doi.org/10.1007/s00284-020-02036-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02036-w