Abstract
ATP-dependent Lon protease plays important roles in different physiological processes, including cellular differentiation of the bacteria and is a part of an important stress response regulon (HspR/HAIR). In Streptomyces, biosynthesis of secondary metabolites starts with cellular differentiation and stress is one of the factor that affect metabolite production. To clarify the effect of Lon protease on secondary metabolite production, we constructed a recombinant strain of Streptomyces coelicolor A3(2) that has one extra copy of lon gene with its own promoter and transcriptional terminator in its genome. Expression of lon gene in the recombinant strain was determined by quantitative real time (RT-qPCR). Actinorhodin and undecylprodigiosin production of the recombinant cell was measured in liquid R2YE and it was found to produce about 34 times more actinorhodin and 9 times more undecylprodigiosin than the wild-type at 168 h of growth. Development of stable Streptomyces strains capable of producing high amounts of secondary metabolites is valuable for biotechnology industry. One extra copy of lon gene is enough to boost antibiotic production by S. coelicolor A3(2) and this change do not cause any metabolic burden in the cell.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
ATP-dependent Lon protease, which is present in all species from bacteria to human, is responsible for the degradation of unstable natural proteins and improperly folded proteins [1, 2]. Lon and two other proteases (ClpAP and ClpXP) are responsible for 70–80% of energy-dependent protein degradation in Escherichia coli [3]. This enzyme plays important roles in the control of physiological processes, such as cell division [4], DNA replication [5], adaptation to stress conditions [6] by degrading naturally folded regulator proteins.
Lon protease is also known to play a critical role in bacterial differentiation. For example, in Bacillus subtilis, Lon affects sporulation by degrading sigma factor σH [7]. The lon gene (lonD) has also been shown to be necessary for the developmental cycle of Myxococcus xanthus [8]. Similarly, cell differentiation in Vibrio parahaemolyticus is shown to be regulated by Lon protein [9]. In a recent study by Cerletti et al. [10], LonB protease was demonstrated to regulate carotenogenesis in the extremophilic archaeon Haloferax volcanii.
Lon protease also controls the expression of virulence genes in Brucella abortus and Salmonella enterica by regulating the degradation of transcription factor HilA [11]. In Pseudomonas aeruginosa, Lon mutants were shown to have reduced pathogenicity and also exhibit severe deficiency in swarming motility [12]. Recently, Lee et al. [13] determined that Lon protease modulates virulence traits in enterobacterial pathogen of apple, Erwinia amylovora.
Lon is not a vital enzyme for most bacterial species under normal growth conditions, but its absence results in a number of dysfunctions in the cells. Mutations in E. coli lon gene result in pleiotropic phenotypes, such as increased sensitivity to UV irradiation [14] and SOS-inducing agents [15], filament formation, mucoidy, and reduced degradation of various abnormal and normal proteins [16]. E. coli mutants deficient in either Lon or Clp proteases also show an extended growth lag after the downshift, which was more severe in the double mutant (lon, clp) [17]. In contrast to E. coli, one of the lon genes (lonV) in M. xanthus, is shown to be essential for viability, as attempts to construct a deletion mutant have failed [8].
Streptomycetes are gram positive, filamentous bacterium which produce a wide variety of bioactive compounds such as antibiotics, anticancer agents, immunosuppressant drugs, herbicides etc. They have a complex life-cycle with morphological differentiation [18]. In the liquid medium, the germinating spore cells form young vegetative mycelium. These young mycelium, also called “primary mycelium”, initiate pellet formation. Over time, the cell density increases and the cells in the pellet center begin to die (programmed cell death) and growth stops. The new mycelium with multi nucleoid (secondary mycelium) develops in and around the pellet and passes to the second growth phase. Secondary metabolites are produced by these new mycelium [19]. The expression of secondary metabolite biosynthesis genes are generally coordinated depending on the growth phase [20].
The regulation of secondary metabolite production is a highly complex process and involves many different regulatory protein families, extracellular and intracellular signaling molecules [21]. Internal signals such as ions [22], alarmones [23], or reactive oxygen species [24] are directing the regulation of secondary metabolites by influencing the transcription of the secondary metabolite gene cluster or by inducing a transcriptional activator of the target gene cluster [23, 25,26,27]. Stress conditions (shortage of nitrogen, phosphorus and carbon sources, amino acid starvation, lack of energy etc.) are also known to induce secondary metabolite biosynthesis in bacteria [21, 28,29,30,31]. In stress conditions protein degradation increase so that amino acids are provided in order to be used in the synthesis of proteins required for adaptation to new environmental conditions and the role of Lon protease in this process is critical [32]. Being an important part of stress response and regulating cellular differentiation, Lon seems to be a good candidate that will affect secondary metabolite production, however, there is no enough study showing the effect of Lon protease on secondary metabolism. To the best of our knowledge there is only one study that relate Lon with antibiotic production. Whistler et al. [33] demonstrated that Lon is a global regulator that negatively influences antibiotic pyoluteorin production in Pseudomonas fluorescens Pf-5. They proposed that Lon may represses antibiotic production by degrading a sigma factor required for pyoluteorin biosynthetic gene transcription or by degrading a positive regulator of antibiotic production.
Sobczyk et al. [34] cloned the lon gene of Streptomyces lividans and disrupted its expression by inserting the hygromycin resistance gene. Although the cell cycle has not changed drastically, it has been shown that the Lon mutant grows more slowly than the wild-type and that spore germination is affected. lon gene has been found to be a part of HspR/HAIR stress response regulon in the same study. Another study reported that S. lividans Lon protease could cause toxic effect when expressed in high copy [35].
In the present study, we determined the effect of one extra copy of lon gene on antibiotic production by Streptomyces coelicolor A3(2).
Materials and Methods
Bacterial Strains, Plasmids and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in LB liquid and solid medium at 37 °C. E. coli ET12567 containing the RP4 derivative pUZ8002 was used as the nonmethylating plasmid donor for intergeneric conjugation with S. coelicolor A3(2). E. coli DH5α was used as a host strain for cloning of lon gene into pRA (5769 bp) vector. pRA is derived from pSET152 and has the ability to integrate into attB site of the chromosomes. S. coelicolor strains were grown in TBO, TSB, YEME, MS and R2YE medium at 30 °C. Apramycin (50 µg/ml), chloramphenicol (25 µg/ml), kanamycin (50 µg/ml) and nalidixic acid (25 µg/ml) were added into growth media as required.
DNA Procedures
Isolation of plasmids, restriction enzyme digestions, agarose gel electrophoresis, Southern hybridization, competent cell preparation and transformation of E. coli cells were performed according to standard molecular biology techniques [36]. Isolation of Streptomyces total DNA were as described by Keiser [20]. Recombinant plasmid was transferred to S. coelicolor by intergeneric conjugation [37].
Southern Blot Hybridization
The Southern blot method was performed according to Sambrook et al. [36]. First, chromosomal DNA samples were digested with EcoRI and SphI restriction endonucleases. The transfer of DNA fragments from the gel to the membrane was achieved by the blotting system (Amersham) under vacuum with 40–50 mbar. The membrane was incubated first in prehibridization solution at 42 °C for 5 h, then in the hybridization solution containing lon gene probe overnight at 42 °C.
The lon gene fragment (1140 bp) to be used as a probe was obtained by PCR using Q5 polymerase enzyme and specific primers, lon probe F and lon probe R (Table 2). This DNA fragment was labelled with the DIG DNA labelling and detection kit (Roche). The same fragment was also used as a positive control in Southern Blot hybridization.
Plasmid Construction and Transformation
The 3397 bp genomic DNA region of S. coelicolor A3(2), which contains the lon coding sequence with its own putative promoter and transcriptional terminator, was amplified using forward (lon F) and reverse (lon R) lon primers (Table 2) and High Fidelity Q5 DNA polymerase (New England Biolabs). Blunt end PCR fragments were ligated into pRA vector, predigested with EcoRV, to obtain the recombinant plasmid pRAlon.
Sequencing of DNA fragment containing lon coding region in pRAlon was carried out at Iontek Company (Istanbul, Turkey) by the dideoxy chain-termination method using an ABI Prism 310 instrument (Perkin–Elmer) and DYEnamic ET Terminator Cycle Sequencing Kit (Amersham).
Actinorhodin and Undecylprodigiosin Determination
Actinorhodin and undecylprodigiosin production were determined spectrophotometrically as described by Keiser [20]. 108 spores from each strain were inoculated into R2YE medium and incubated at 30 °C, 200 rpm for 48 h. Pellets with equal wet weight from this preculture were used to inoculate fresh R2YE medium and grown at 30 °C, 200 rpm. For antibiotic and dry weight measurements, samples were taken at every 24 h of fermentation. 1 ml of culture was treated overnight with 250 μL of KOH (5 M) at 4 °C. Then the sample was centrifuged at 13000 rpm for 5 min and the A640 value of the supernatant was measured to determine actinorhodin concentration. Molarity was quantified by the formula: Absorbance = ε × [M], where ε640 = 25230 for the pure compound. To determine undecylprodigiosin concentration (a mixture of at least four prodiginines), the mycelium (pellet from actinorhodin measurement) was washed two times with 0.5 M HCl. Then, the pellet was dissolved in 1 ml of methanol-HCl (0.5 M) mixture and incubated at room temperature for 2 h. Then, the A530 of the supernatant was determined (ε530 = 100500).
For the dry weight measurement, the pellets from 24, 48, 72, 96, 120, 144, and 168 h samples were washed twice with dH2O, dried at 80 °C for 5 days and then the weights were measured.
Gene Expression Quantification
For RNA extraction, the cell pellets were resuspended in TE solution with lysozyme and were incubated at 37 °C for 60 min. Total RNA isolation was performed with RNA Isolation Kit (Macherey–Nagel, Germany) according to manufacturer’s instructions with small modifications. Possible DNA contamination was eliminated using Deoxyribonuclease I (Thermo Scientific, USA). The absence of contaminated DNA in the RNA samples was confirmed by PCR reactions without reverse transcriptase. The RNA concentration and purity was determined by spectrophotometric measurement. RNA samples were aliquoted then stored at − 80 °C for future use.
cDNA was synthesized by the Iscript Advanced cDNA Synthesis KIT (Bio-rad Laboratories, USA), using 300 ng of total RNA. Reaction mixtures, prepared according to manufacturer’s recommendations, were first incubated at 70 °C for 5 min, followed by 2–3 min of incubation in ice to eliminate hairpin formation and primer dimer formation. For the first-strand cDNA synthesis conditions were as follows: incubation at 25 °C for 5 min, 46 °C for 60 min, followed by 85 °C for 5 min. For quantitative RT-PCR StepOnePlus™ Real-Time PCR System (Applied Biosystems) and the SYBR Green qPCR master mix (Bio-rad Laboratories, USA), including 500 nM primers (each) and 25 ng cDNA in a total volume of 20 μl were used. The thermal profile consisted of 10 min initial denaturation at 98 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Each cDNA sample was quantified three times in independent experiments. The primers used for transcriptional analysis were designed via a web-based tool, primer 3. Sequences of primer pairs were given in Table 2. The target gene expression level was normalized internally to the level of the gyrA and hrdB genes. Relative gene expression levels were calculated using the 2−ΔΔCt formula [38] and the error bars represent the standard deviations obtained from three independent biological replicates.
Results
Cloning and Expression of Streptomyces coelicolor A3(2) lon gene
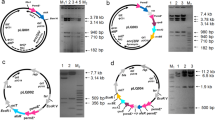
A 3397 bp genomic DNA region of S. coelicolor, that contain ATP-dependent Lon protease gene (SCO5285, GeneBank accession number: AL939123.1) was amplified by PCR with its own putative promoter and transcriptional terminator. HAIR motif with a “ATTGAGT-cgatgta-ACTCAAC” sequence at the upstream of the lon gene [34] was included in the promoter region (Fig. 1). lon gene was cloned into EcoRV site of pRA (derived from pSET152) and after verification by DNA sequencing the recombinant plasmid (pRAlon) was transferred to S. coelicolor cells by conjugation. Possible recombinant strains were first selected according to apramycin resistance, followed by Southern hybridization. Hybridization sites of lon probe on pRAlon plasmid and in the genome of S. coelicolor was shown in Fig. 2. The presence of second lon gene in the genome of recombinant strain (Sco-pRAlon) was confirmed by Southern hybridization (Fig. 3a and b). S. coelicolor with empty pRA (Sco-pRA) was used as the control strain. A hybridization band of 4195 bp was obtained for the wild-type, Sco-pRA and Sco-pRAlon with a lon gene probe. Second lon copy (a hybridization band of 6455 bp) was present only in Sco-pRAlon strain (Fig. 3b). The relative expression of lon gene in the Sco-pRAlon strain was found to be about 16, 24 and 12-fold higher compared to the wild-type at 60th, 72th and 96th hour of growth, respectively (Fig. 3c).
Determination of the presence of an extra copy of lon gene in S. coelicolor genome by Southern Blot. a Red safe-stained agarose gel and b Southern blots results where lon probe was used in hybridization. 1140 bp lon gene fragment is positive control (lane 1), hybridization pattern of the SphI-EcoRI-digested chromosomal DNA of the wild-type S. coelicolor (lane 2), SphI-EcoRI digested chromosomal DNA of the Sco-pRA (lanes 3), SphI-EcoRI digested chromosomal DNA of the Sco-pRAlon (lane 4) probed with the lon gene fragment. M: 1 kb DNA marker (Fermentas). c Relative gene expression of the lon gene in the Sco-pRAlon strain compared with the wild-type (WT) grown in R2YE medium for 60th, 72 th and 96th hour. Fold changes were calculated using the 2−ΔΔCt method. The statistical analyses performed with GraphPad Prism 8.0.2 software. P values were calculated using the Mann–Whitney test. P < 0.05 was considered as statistically significant. Asterisks (*) represents P < 0.05. Vertical bars indicate standard deviation from the mean value
Growth of Recombinant Strain and Antibiotic Production
Growth of Sco-pRAlon, Sco-pRA and wild-type strains were compared on R2YE liquid culture and quite similar growth profile were obtained for all strains until 96 h of growth, then the growth of Sco-pRAlon was impaired compared to others (Fig. 4a). Recombinant strains were shown to sporulate efficiently on TBO medium (Fig. 4b).
Actinorhodin and undecylprodigiosin production of the cells were measured in liquid R2YE for 168 h. Sco-pRAlon strain produced about 20–34 times more actinorhodin (Fig. 5a) and 5–9 times more undecylprodigiosin (Fig. 5b) than the wild-type and control strain Sco-pRA at 168 h of growth depending on the amount and age of the inoculum. These results were consistent in repeated experiments.
Comparison of a specific actinorhodin and b undecylprodigiosin production (nmol/mg dry weight) by wild-type, Sco-pRA and Sco-pRAlon strains grown in R2YE. Vertical bars indicate standard deviation from the mean value. P values < 0.05 were considered to be statistically significant. One asterisks (*) represents P < 0.05 and double asterisks represents (**) P < 0.005 according to the t test (Microsoft Office Excel). Data are the average of six independent experiments
Expression of sigma factor sigB, which is responsible from general stress response in the cell and that of actIIORF4 and redK, which encodes regulatory protein for actinorhodin biyosynthetic gene cluster and one of gene in undecylprodigiosin biyosynthetic gene cluster, respectively, were measured by RT-qPCR. sigB, actIIORF4 and redK expressions in Sco-pRAlon strain were found to be increased significantly compared to the wild-type (Fig. 6).
Comparison of relative gene expression levels of asigB, bredK and cactIIORF4 in Sco-pRAlon and wild-type strains grown in R2YE medium. Fold changes were calculated using the 2−ΔΔCt method. The statistical analyses performed with GraphPad Prism 8.0.2 software. P values were calculated using the Mann–Whitney test. P < 0.05 was considered as statistically significant. Asterisks (*) represents P < 0.05. Vertical bars indicate standard deviation from the mean value
Discussion
From bacteria to human, ATP-dependent Lon protease is one of the important enzyme in protein quality control [39, 40]. In this study, the effect of Lon on antibiotic production by S. coelicolor A3(2) was determined. For this, a recombinant strain (Sco-pRAlon), which have one extra copy of lon gene (with its own putative promoter and transcriptional terminator) was prepared. Actinorhodin concentration was dramatically increased in recombinant strain and it was found to be around 34 times more than the wild-type strain.
The elicitation mechanism of secondary metabolism is a complex process and many different factors play role in this process. No significant increase was observed in antibiotic production in the control strain obtained by transferring the empty pRA vector into S. coelicolor compared to the wild-type. It seems that the excessive increase in antibiotic production by Sco-pRAlon recombinant strain is due to the expression of extra lon gene. pRA and other integrative vectors enter to an attB site in a gene (SCAC2.o6c; SCO3798) encoding “putative chromosome condensation protein” in S. coelicolor A3(2) genome. This gene has been shown not to be essential for cell survival [41]. Although the vector integrate to this attB site with an efficiency of 300-fold, it is also known that there are other pseudo-attB sites in the genome that the vector might enter [41]. Three of these sites (pseB1: inside putative dihydropteroate synthase gene, pseB2: inside hypothetic protein gene and pseB3: inside aspartate aminotransferase gene) were shown to be active phage-integration sites. Neither attB nor pseudo-attB sites are inside the genes that may affect secondary metabolism.
“Duplicating a gene would double the gene’s transcript” idea was not true for this study, since expression of lon gene in the wild-type and Sco-pRAlon strains was quite different. Understanding of the quantitative factors that govern gene expression is not very well understood. In the literature, there are different studies which show that transcription of a gene increase more than two times when it is integrated into the chromosome as an extra copy. For example, Schmid, Roth [42] have translocated the first four genes of the his operon (hisGDCB) of Salmonella typhimurium, along with the his promoter, to 16 chromosomal sites. They measured the level of hisD expression at these sites under different growth conditions and they have found that different chromosomal sites express the his genes at different levels. Other studies also demonstrates that expression of the genes varies depending on its position on the chromosome [43, 44]. Position effect on gene expression is also seen in eukaryotic organisms [45].
Stress is one of the important factor that triggers secondary metabolite biosynthesis in bacteria [21, 31]. In the case of amino acid starvation and energy shortage, bacteria produce stringent response alarmons (ppGpp and pppGpp), which are produced by “RelA” and “SpoT” proteins, cause cessation of tRNA and rRNA synthesis together with protein synthesis and increase protein degradation processes. By this way amino acids are used in the synthesis of proteins required for adaptation to new environmental conditions. The role of Lon protease in this process is critical [32]. Presence of one extra copy of lon in the genome of S. coelicolor could cause production of more precursors for antibiotic biosynthesis by direct protein degradation. Besides, ppGpp and pppGpp were also shown to be responsible for direct stimulation of transcription of several amino acid biosynthetic genes [46] which are precursors of some secondary metabolites including actinorhodin [47]. Traxler et al. [48] determined that starvation triggers induction of genes involved in the branched chain amino acid pathways as well as genes in pathways which generate precursors for branched chain amino acid biosynthesis. Branched-chain amino acid catabolism, in turn, are known to provide precursors for the Type II polyketide antibiotic, actinorhodin, via pathways that are nutrient dependent [49]. Expression of sigma factor that is responsible from general stress response in bacteria, sigB, was shown to be increased in the recombinant strain. This result shows that with one extra copy of lon gene Sco-pRAlon is under stress. So it is possible that recombinant strain produce more branched chain amino acids which resulted in more actinorhodin production. Detailed experiments are underway to verify this possibility.
Recently, Bucca et al. [50] studied the genome-wide transcriptional and translational changes following heat stress exposure in S. coelicolor and find out little correlation between the “transcriptome” and ‘translatome’ data. Although key proteins like major molecular chaperones and proteases were found to be highly induced at both the transcriptional and translational level, many other transcripts were observed to be not induced at the transcriptional level but they were more highly polysome-associated following heat stress. Based on this study, a detailed transcriptomic and translatomic analysis of the recombinant strain would explain the reason why one extra copy of lon gene-boosted antibiotic production in S. coelicolor.
Although our attempts still continue, many trials to delete the lon gene by using REDIRECT method [37] were unsuccessful suggesting that this gene may be essential in S. coelicolor. To our knowledge there is no Streptomyces mutant strain with lon gene deletion. Sobczyk et al. [34] disrupted expression of lon gene in S. lividans and the mutant was shown to grow more slowly than the wild-type. However, it is known that disruption and deletion mutants may have different characteristics. Although Lon protease is not a vital enzyme for most bacterial species, there is at least one example where the lon gene is essential for viability [8]. Our preliminary results suggest that lon may be an essential gene for S. coelicolor A3(2). Ching et al. [51] observe a dramatic upregulation of surface antigen gene, surA1, in the lon− mutant, which could be caused by an energetic burden (phosphorylation sink) or by disturbance of the functions of the cell, thus having a role in inducing cell death. Although it remains to be elucidated, it is possible that SurA1 protein concentration increase to critical level in S. coelicolor so that we were not able to obtain lon deletion mutant of this strain.
The development of stable Streptomyces strains that produce high amount of secondary metabolites is highly attractive in biotechnology. Although Mak, Nodwell [52] concluded that actinorhodin is a pH-sensitive, strong bacteriostatic antibiotic, to our knowledge it is not used medically. However, it is known that actinorhodin and other colored metabolites produced by Streptomyces have been tested in some industrial areas as an alternative to toxic synthetic dyes and effective results have been obtained (https://www.asm.org/index.php/general-science-blog/item/6929bacterialdyesinfashion;https://www.ted.com/talks/natsai_audrey_chieza_fashion_has_a_pollution_problem_can_biology_fix_it). In this respect, our recombinant strain has the potential to be used in industry. But the subject we really wonder is “Does the presence of one extra copy of lon gene in the genome of other Streptomyces, which produce secondary metabolites with high medical and biotechnological value, will result the same increase in the production efficiency?” Our experiments are ongoing to find an answer to this question.
References
Chung CH, Goldberg AL (1982) DNA stimulates ATP-dependent proteolysis and protein-dependent ATPase activity of protease La from Escherichia coli. Proc Natl Acad Sci 79(3):795–799
Goldberg AL, Rock KL (1992) Proteolysis, proteasomes and antigen presentation. Nature 357(6377):375
Bellier A, Gominet M, Mazodier P (2006) Post-translational control of the Streptomyces lividans ClgR regulon by ClpP. Microbiology 152(4):1021–1027
Schoemaker J, Gayda R, Markovitz A (1984) Regulation of cell division in Escherichia coli: SOS induction and cellular location of the sulA protein, a key to lon-associated filamentation and death. J Bacteriol 158(2):551–561
Mascarenhas J, Volkov AV, Rinn C, Schiener J, Guckenberger R, Graumann PL (2005) Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC Cell Biology 6(1):28
Tsilibaris V, Maenhaut-Michel G, Van Melderen L (2006) Biological roles of the Lon ATP-dependent protease. Res Microbiol 157(8):701–713
Liu J, Cosby WM, Zuber P (1999) Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol Microbiol 33(2):415–428
Tojo N, Inouye S, Komano T (1993) The lonD gene is homologous to the lon gene encoding an ATP-dependent protease and is essential for the development of Myxococcus xanthus. J Bacteriol 175(14):4545–4549
Stewart BJ, Enos-Berlage JL, McCarter LL (1997) The lonS gene regulates swarmer cell differentiation of Vibrio parahaemolyticus. J Bacteriol 179(1):107–114
Cerletti M, Paggi R, Troetschel C, Ferrari MC, Guevara CR, Albaum S, Poetsch A, De Castro R (2018) LonB protease ıs a novel regulator of carotenogenesis controlling Degradation of phytoene synthase in Haloferax volcanii. J Proteome Res 17(3):1158–1171
Brazas MD, Breidenstein EB, Overhage J, Hancock RE (2007) Role of lon, an ATP-dependent protease homolog, in resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob Agents Chemother 51(12):4276–4283
Breidenstein EB, Janot L, Strehmel J, Fernandez L, Taylor PK, Kukavica-Ibrulj I, Gellatly SL, Levesque RC, Overhage J, Hancock RE (2012) The Lon protease is essential for full virulence in Pseudomonas aeruginosa. PLoS ONE 7(11):e49123
Lee JH, Ancona V, Zhao Y (2018) Lon protease modulates virulence traits in Erwinia amylovora by direct monitoring of major regulators and indirectly through the Rcs and Gac-Csr regulatory systems. Mol Plant Pathol 19(4):827–840
Mizusawa S, Gottesman S (1983) Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc Natl Acad Sci 80(2):358–362
Gonzalez M, Frank EG, Levine AS, Woodgate R (1998) Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: in vitro degradation and identification of residues required for proteolysis. Genes Dev 12(24):3889–3899
Torres-Cabassa A, Gottesman S (1987) Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol 169(3):981–989
Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A (2001) Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293(5530):705–708
Garrity GM, Bell JA, Lilburn T (2004) Taxonomic outline of the prokaryotes. Bergey’s manual of systematic bacteriology. Springer, New York
Manteca A, Alvarez R, Salazar N, Yagüe P, Sanchez J (2008) Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl Environ Microbiol 74(12):3877–3886
Keiser T (2000) Practical streptomyces genetics. The John Innes Foundation, Norwich
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8(2):208–215
Murphy TM, Nilsson AY, Roy I, Harrop A, Dixon K, Keshavarz T (2011) Enhanced intracellular Ca2+ concentrations in Escherichia coli and Bacillus subtilis after addition of oligosaccharide elicitors. Biotech Lett 33(5):985–991
Kawai K, Wang G, Okamoto S, Ochi K (2007) The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol Lett 274(2):311–315
Radman R, Bland EJ, Sangworachat N, Bucke C, Keshavarz T (2006) Effects of oligosaccharides and polysaccharides on the generation of reactive oxygen species in different biological systems. Biotechnol Appl Biochem 44(3):129–133
Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, Van Wezel GP (2008) Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9(7):670–675
Tanaka A, Takano Y, Ohnishi Y, Horinouchi S (2007) AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369(2):322–333
Nair R, Roy I, Bucke C, Keshavarz T (2009) Quantitative PCR study on the mode of action of oligosaccharide elicitors on penicillin G production by Penicillium chrysogenum. J Appl Microbiol 107(4):1131–1139
Martín JF, Demain AL (1980) Control of antibiotic biosynthesis. Microbiol Rev 44(2):230
Ghorbel S, Kormanec J, Artus A, Virolle M-J (2006) Transcriptional studies and regulatory interactions between the phoR-phoP operon and the phoU, mtpA, and ppk genes of Streptomyces lividans TK24. J Bacteriol 188(2):677–686
Lounes A, Lebrihi A, Benslimane C, Lefebvre G, Germain P (1996) Regulation of spiramycin synthesis in Streptomyces ambofaciens: effects of glucose and inorganic phosphate. Appl Microbiol Biotechnol 45(1–2):204–211
Yeşilirmak F, Doruk T, Yilmaz Ş, Gedik ST (2018) Comparative proteomic analysis of Bacillus thuringiensis wild-type and two mutant strains disturbed in polyphosphate homeostasis. Turk J Biol 42(1):87–102
Kuroda A, Tanaka S, Ikeda T, Kato J, Takiguchi N, Ohtake H (1999) Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. Proc Natl Acad Sci 96(25):14264–14269
Whistler CA, Stockwell VO, Loper JE (2000) Lon protease influences antibiotic production and UV tolerance of Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 66(7):2718–2725
Sobczyk A, Bellier A, Viala J, Mazodier P (2002) The lon gene, encoding an ATP-dependent protease, is a novel member of the HAIR/HspR stress-response regulon in actinomycetes. Microbiology 148(6):1931–1937
Bucca G, Brassington AM, Hotchkiss G, Mersinias V, Smith CP (2003) Negative feedback regulation of dnaK, clpB and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol Microbiol 50(1):153–166
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci 100(4):1541–1546
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods 25(4):402–408
Wickner S, Maurizi MR (1999) Here’s the hook: similar substrate binding sites in the chaperone domains of Clp and Lon. Proc Natl Acad Sci 96(15):8318–8320
Gottesman S, Wickner S, Maurizi MR (1997) Protein quality control: triage by chaperones and proteases. Genes Dev 11(7):815–823
Combes P, Till R, Bee S, Smith MC (2002) The Streptomyces genome contains multiple pseudo-attB sites for the φC31-encoded site-specific recombination system. J Bacteriol 184(20):5746–5752
Schmid MB, Roth JR (1987) Gene location affects expression level in Salmonella typhimurium. J Bacteriol 169(6):2872–2875
Thompson A, Gasson MJ (2001) Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactisand Saccharomyces cerevisiae. Appl Environ Microbiol 67(8):3434–3439
Bryant JA, Sellars LE, Busby SJ, Lee DJ (2014) Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res 42(18):11383–11392
Loehlin DW, Carroll SB (2016) Expression of tandem gene duplicates is often greater than twofold. Proc Natl Acad Sci 113(21):5988–5992
Paul BJ, Berkmen MB, Gourse RL (2005) DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci 102(22):7823–7828
Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M (2007) The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol 8(8):R161
Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T (2008) The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68(5):1128–1148
Stirrett K, Denoya C, Westpheling J (2009) Branched-chain amino acid catabolism provides precursors for the Type II polyketide antibiotic, actinorhodin, via pathways that are nutrient dependent. J Ind Microbiol Biotechnol 36(1):129–137
Bucca G, Pothi R, Hesketh A, Möller-Levet C, Hodgson DA, Laing EE, Stewart GR, Smith CP (2018) Translational control plays an important role in the adaptive heat-shock response of Streptomyces coelicolor. Nucleic Acids Res 46(11):5692–5703
Ching C, Yang B, Onwubueke C, Lazinski D, Camilli A, Godoy VG (2019) Lon protease has multifaceted biological functions in Acinetobacter baumannii. J Bacteriol 201(2):e00536
Mak S, Nodwell JR (2017) Actinorhodin is a redox-active antibiotic with a complex mode of action against G ram-positive cells. Mol Microbiol 106(4):597–613
Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ (1999) Evidence that the extracytoplasmic function sigma factor ςE is required for normal cell wall structure in Streptomyces coelicolor A3 (2). J Bacteriol 181(1):204–211
Perez-Redondo R, Santamarta I, Bovenberg R, Martin JF, Liras P (2010) The enigmatic lack of glucose utilization in Streptomyces clavuligerus is due to inefficient expression of the glucose permease gene. Microbiology 156(5):1527–1537
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4):557–580
MacNeil DJ, Occi JL, Gewain KM, MacNeil T, Gibbons PH, Ruby CL, Danis SJ (1992) Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115(1):119–125
Hopwood DA (1999) Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145(9):2183–2202
Acknowledgements
This research was supported by Gebze Technical University, Grant Number 2018-A-102-16.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Demir, Z., Bayraktar, A. & Tunca, S. One Extra Copy of lon Gene Causes a Dramatic Increase in Actinorhodin Production by Streptomyces coelicolor A3(2). Curr Microbiol 76, 1045–1054 (2019). https://doi.org/10.1007/s00284-019-01719-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01719-3