Abstract
Polyketide antibiotics are among the most important therapeutics used in human and animal health care. Type II polyketides are composed primarily of acetate-derived thioesters, and the subunits for the PKS are contained in a single module that includes a ketosynthase, acyl carrier protein, chain-length factor and sometimes a keto-reductase, aromatase, cyclase and modifying enzymes, such as glycosylases or hydroxylases. While the enzyme complexes that make up the PKS have been the focus of intense study (Khosla in Chem Rev 7:2577–2590, 1997), the pathways for precursor synthesis have not been established and predictions are complicated by the fact that acetate may be derived from a number of metabolic pathways. Here we show that 50% of the acetate for synthesis of the Type II polyketide, actinorhodin, in Streptomyces coelicolor, is derived from the catabolism of the branched amino acids by pathways that are nutrient dependent. The streptomycetes are apparently unique in that they contain two BCDH gene clusters, each of which is potentially capable of converting leucine, valine and isoleucine to the corresponding thioesters, and contain at least three different pathways for valine catabolism that are differentially used in response to nutrient availability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyketide antibiotics are produced on large multi-enzyme (PKS) complexes similar to those involved in fatty acid biosynthesis. The carbon backbones of the molecules are assembled by the successive condensation of small organic acids activated as thioesters. At each condensation, the backbone grows by two carbon units and additional carbons become side chains. Enzymes for Type I synthesis are contained in a series of modular polypeptides each encoding a separate PKS activity. Each condensation reaction is carried out by a different ketosynthase and each thioester is specified by a different acyltransferase similar to the Type II fatty acid synthases in fungi and vertebrates. This organization allows for synthesis of complex products as the starter unit, number of extender units, stereochemistry of the side chains, pattern of cyclization and programming of extender units vary from module to module. The PKS for Type II synthesis is contained in a single module that encodes a single complex used iteratively, similar in organization to bacterial and plant Type II fatty acid synthases. Most Type II polyketides use either malonyl-CoA or acetyl-CoA as starter units and acetate derived thioesters for extension [16]. While a great deal is known about the enzyme complexes that condense polyketides, relatively little is known about the metabolic pathways that produce the precursors for their synthesis.
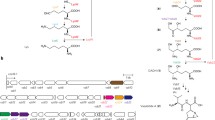
The branched-chain amino acids have been implicated in the generation of complex starter units for the Type I polyketides virginiamycin, tautomycin, mananumycin, and butyrolactols as well as for the Type II polyketide anthraquinones [16]. Type I polyketides are derived primarily from malonyl-CoA, methylmalonyl-CoA, propionyl-CoA and acetyl-CoA with more complex starter units that include short chain and often branched carbonyl-CoA molecules such as isobutyryl, isovaleryl and 2-methulbutyryl-CoA [16]. The most direct evidence for the role of branched-chain amino acids as precursors for these compounds comes from the fact that a deletion of the 5′ end of the Streptomyces avermitilis bkdFGH gene cluster (which corresponds to the bkdA1B1C1cluster in S. coelicolor, shown in Fig. 1a) eliminated production of the Type I polyketide antibiotic, avermectin [7]. This rather surprising result suggested that some of the precursors for avermectin biosynthesis derive exclusively from the degradation of branched-chain amino acids and that the bkdFGH cluster plays a key role in the generation of these precursors. Strains containing a deletion of the bkdFGH cluster may be fed alternative substrates to generate novel forms of avermectin [7].
Synthesis of actinorhodin, a Type II polyketide produced by S. coelicolor, begins with loading and decarboxylation of a malonyl-CoA starter unit (derived from acetyl-CoA by acetyl-CoA carboxylase during fatty acid biosynthesis) followed by the repetitive condensation of malonyl-CoA for elongation [4]. To determine if branched-chain amino acids were precursors for actinorhodin biosynthesis, a deletion mutant of the bkdA1B1C1 cluster was constructed in S. coelicolor. Here we show that the bkdA1B1C1 mutant cannot grow on branched-chain amino acids as sole carbon source, fails to produce branched-chain fatty acids (BCFAs), and shows a 50% reduction in actinorhodin production. Addition of isobutyrate (specific for valine catabolism), isovalerate (specific for leucine catabolism) or α-methylbutyrate (specific for isoleucine catabolism) restored actinorhodin production on rich media, but only isovalerate was able to rescue actinorhodin production in minimal medium. This report provides direct experimental evidence that branched-chain amino acids are a primary source of precursors for this acetate-primed Type II polyketide and that only one of the two BKD clusters participates in antibiotic biosynthesis.
Methods and materials
Bacterial strains and growth conditions
The S. coelicolor strains used in these studies were M145 SCP1− SCP2− [10] and Δ bkdA1B1C1::aac(3)IV (constructed in this study). Media and protocols for genetic manipulation of Streptomyces [13] and Escherichia coli [18] as well as techniques for isolation and manipulation of nucleic acids were as described. Streptomyces strains were grown in YEME medium [13] for genomic DNA isolation and on MS agar [13] with the addition of 10 mM MgCl2 for conjugation experiments. Apramycin (50 μg/ml) was used as the selection and applied by overlay with soft nutrient agar. MYM agar medium [2] or MYM supplemented with isovaleric acid, α-methylbutyric acid or isobutyric acid (each 5 mM) were used in the analysis of actinohorodin production on agar medium. To test for growth on leucine, valine or isoleucine as sole carbon source, spores were innoculated into 50 ml of NMMP liquid medium without casamino acids and with 30 mM NH4SO4, and either 40 mM valine, leucine or isoleucine, or no carbon source and incubated for 6 days at 30°C shaking at 300 rpm. Samples were taken at 24-h intervals and growth was assayed spectrophotometrically at 450 nm.
Construction of the bkdA1B1C1 mutant
A deletion of the bkdA1B1C1 gene cluster in the S. coelicolor genome was constructed by replacing the coding sequence for the three genes of the cluster with the apramycin-resistance cassette (aac(3)IV) using the PCR-targeted gene replacement method described by Gust et al. [8]. The apramycin resistance replacement cassette was generated by PCR using primers with a 39 nt gene-specific extension and introduced into the S. coelicolor cosmid GC3, which contains a kanamycin resistance marker on the cosmid backbone, in E. coli BW25113. The mutagenized cosmid was introduced into E. coli ET12567 [15] and then transferred to S. coelicolor by conjugation. Exconjugants were selected by apramycin resistance and marker replacement events were identified by screening for kanamycin sensitivity. The deletion extended from nucleotide 54 of the bkdA1 gene (SCO3817), through the bkdA2 gene (SCO3818) to the end of the bkdA3 gene (SCO3819) leaving the last 69 bp of that gene at the end of the cassette. The 3.5 kb deletion in the S. coelicolor chromosome, as well as the fact that the bkdA2B2C2 gene cluster was intact and not affected, was confirmed by Southern hybridization.
Actinorhodin assays
Spores of S. coelicolor M145 and Δ bkdA1B1C1::aac(3)IV were grown for 6 h in 2 × YT medium [13] to allow germination, washed twice with modified NMMP medium [22], and then used to inoculate 500 ml of NMMP medium alone or supplemented with various compounds in the following concentrations: valine, leucine or isoleucine (each 15 mM), α-methylbutyric acid, isobutyric acid, isovaleric acid (each 0.5 mM). The cultures were grown at 30°C with shaking at 200 rpm, and samples were taken every 24 h for 192 h to assess growth and actinorhodin production [22] by spectrophotometric analysis at 450 and 608 nm, respectively.
Fatty acid analysis
Spores of S. coelicolor M145 and Δ bkdA1B1C1::aac(3)IV were incubated in 50 ml of NMMP for 3 days at 30°C shaking at 300 rpm. About 0.5 ml of this culture was used to inoculate 10 ml of NMMP and allowed to grow for 2 days. Cells were harvested by centrifugation and fatty acids from the cell pellet were analyzed by GC-MS as previously described [25]. An Agilent 19091 series gas chromatograph-mass selective detector equipped with a HP5 phenyl methyl siloxane capillary column with 0.25 μm film thickness was used for GC-MS analysis.
Results and discussion
There are two branched-chain amino acid dehydrogenase complexes in S. coelicolor but only one allows growth on valine or leucine as sole carbon source. The streptomycetes are apparently unique among bacteria and even other actinomycetes in that they contain two branched-chain α-keto acid dehydrogenase (BCDH) gene clusters (Fig. 1a), each of which encodes E1α (a dehydrogenase), E1β (a decarboxylase), E2 (a dihydrolipoamide acyltransferase) and E3 (a dihydrolipoamide dehydrogenase) subunits. In S. coelicolor each cluster has been shown to be transcribed [21] and homologues of these gene clusters have been found in the genomes of all of the streptomycete species that have been sequenced, including S. avermitilis (http://avermitilis.ls.kitusato-u.ac.jp), S. griseus and S. scabies (http://www.sanger.ac.uk). Other bacteria and even other actinomycetes such as mycobacteria (http://www.sanger.ac.uk), corynebacteria (http://www.sanger.ac.uk) or nocardia species (http://nocardia.nih.go.jp/blast) have a single cluster. In the streptomycetes, the E3 subunit, a flavin containing protein that participates in the donation of electrons, is not linked to this cluster and likely resides elsewhere in the genome [20]. While a search of the S. coelicolor genome database revealed several possibilities, including a dehydrogenase associated with the pyruvate dehydrogenase complex, it is not clear which might function in the BCDH enzyme complex.
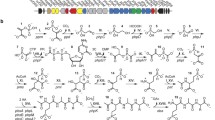
Branched-chain amino acid catabolism begins with three reactions that employ common enzymes (Fig. 1b): transamination by valine dehydrogenase to the corresponding α-keto acids; oxidative decarboxylation by the branched-chain dehydrogenase complex to the corresponding acetyl-CoA derivatives, α-ketoisovalerate from valine, α-keto-β-methyvalerate from isoleucine and α-ketoisocaproate from leucine; and dehydrogenation by FAD to form a double bond. The remainder of the isoleucine degradation pathway is the same as for fatty acid oxidation, common to most bacteria, yielding one mole of acetyl-CoA and one mole of propionyl-CoA, which is subsequently converted into succinyl-CoA. Three moles of acetyl-CoA are generated per mole of leucine and valine degradation may proceed by three different pathways yielding propionyl-CoA, methylmalonyl-CoA and acetyl-CoA. Streptomyces is unusual in its ability to convert valine to acetyl-CoA by the intramolecular conversion of isobutyryl-CoA to butyryl-CoA and subsequent β-oxidation to form acetyl-CoA [19], allowing the generation of acetyl-CoA as an end product of the catabolism of each branched-chain amino acid.
A mutation of the bkdFGH cluster in S. avermitilis (which corresponds to the bkdA1B1C1 cluster in S. coelicolor) was previously reported to affect branched-chain amino acid catabolism [7] but interpretations about the phenotypes of this mutant were compromised by the fact that the mutant contained a deletion of only the E1α subunit and extended into an upstream open reading frame. To further investigate the role of the bkdA1B1C1 cluster in amino acid catabolism, a deletion of this cluster was constructed in S. coelicolor by replacing the E1α, E1β and E2 open reading frames with an apramycin resistance cassette (Fig. 1a) using the PCR-targeted gene replacement method of Gust et al. [8]. The resulting strain, Δ bkdA1B1C1::aac(3)IV was tested for growth on valine, leucine, or isoleucine as sole carbon source, with growth on alanine as a control, as well as on a medium containing no carbon source. S. coelicolor spores store carbon in the form of trehalose and glycogen [1, 9] and are capable of germinating in liquid medium without a carbon source. As shown in Fig. 2, both the wild type M145 strain and the Δ bkdA1B1C1::aac(3)IV mutant strain germinate in medium without a carbon source but growth ceases once the storage carbohydrate is exhausted. Both the mutant and wild type grow relatively well on alanine as sole carbon source and to the same cell densities, that are significantly above growth with no carbon source (Fig. 2a). Growth yield of the wild type on valine is relatively poor compared to that observed on alanine. Growth of the Δ bkdA1B1C1::aac(3)IV mutant on valine is reduced and indistinguishable from the wild type without a carbon source, suggesting that the mutant cannot use valine as sole carbon source (Fig. 2b). Growth of the wild type on leucine is also relatively poor but is detectable above the no carbon source control culture while growth of the Δ bkdA1B1C1::aac(3)IV mutant is less than the wild type without a carbon source (Fig. 2c), suggesting that the mutant is unable to use leucine as sole carbon source. Growth on isoleucine for both the wild type and the mutant strain is indistinguishable from growth without a carbon source, suggesting that even the wild type S. coelicolor strain cannot use isoleucine as sole carbon source (Fig. 2d). We conclude from these data that the bkdA1B1C1 gene cluster alone is responsible for the level of catabolism required for the ability of S. coelicolor to grow on leucine or valine as sole carbon source. Interestingly, wild type cultures with leucine as sole carbon source produced more of the blue pigment associated with actinorhodin production under these conditions.
Growth of S. coelicolor M145 and the Δ bkdA1B1C1::aac(3)IV mutant in NMMP liquid minimal medium replacing casamino acids with 30 mM NH4SO4 and 40 mM a alanine (Ala), b valine (Val), c leucine (Leu), or d isoleucine (Ile), as sole carbon source or no carbon source. Samples were taken from liquid cultures incubated at 30°C shaking at 300 rpm, at 24-h intervals for 6 days. Growth was measured spectrophotometrically at 450 nm. Error bars indicate the standard deviation of two independent trials
A deletion of the bkdA1B1C1 cluster in S. coelicolor eliminated the ability to grow on valine or leucine as sole carbon source even in the presence of a wild type copy of the bkdA2B2C2 cluster. This observation might be explained, in part, by the expression patterns of the two clusters [21]. RNA transcribed from the bkdA1B21C1 cluster was readily detected throughout growth and development while transcript from the bkdA2B2C2 cluster is detected at relatively low levels and increases only at the initiation of development. Virtually no transcript from the bkdA2B2C2 cluster was detected during the vegetative growth phase [21].
Deletion of the bkdA1B1C1 gene cluster in S. coelicolor resulted in defects in actinorhodin production on rich solid medium that were rescued by products of valine, leucine or isoleucine catabolism. On relatively rich media (MYM) with maltose as carbon source, the wild type strain produced the blue pigment associated with actinorhodin production while the Δ bkdA1B1C1::aac(3)IV mutant did not (Fig. 3a). The mutant does produce the red pigment associated with the peptide antibiotic undecylprodigiosin suggesting that it is not pleotropically defective in antibiotic production.
To test whether the defects in actinorhodin production in the Δ bkdA1B1C1::aac(3)IV mutant resulted from defects in valine, leucine or isoleucine catabolism on rich medium, the mutant was grown on MYM agar plates supplemented with isobutyrate (specific for valine catabolism), isovalerate (specific for leucine catabolism) or α-methylbutyrate (specific for isoleucine catabolism). As shown in Fig. 3, the addition of each of these compounds to the growth medium restored actinorhodin production in the Δ bkdA1B1C1::aac(3)IV mutant suggesting that the defect in actinorhodin synthesis by cells grown on rich medium results from the failure of the mutant to catabolize these amino acids. Similar results were obtained on MYM medium containing glucose as carbon source (data not shown). While isoleucine is not catabolized at levels required for growth, it apparently plays a role in antibiotic biosynthesis on solid rich medium.
Deletion of the bkdA1B1C1 gene cluster in S. coelicolor results in defects in actinorhodin production in liquid minimal medium that are rescued only by products of leucine catabolism. While color intensity on agar medium is a good indication of actinorhodin production, S. coelicolor grows relatively poorly on minimal medium making comparisons of the wild type and mutant strains less reliable. To test whether the defects in actinorhodin production in the Δ bkdA1B1C1::aac(3)IV mutant resulted from defects in valine, leucine or isoleucine catabolism on minimal medium quantitative assays for actinorhodin production were performed on cultures grown in liquid medium with glucose as carbon source. As shown in Fig. 4, growth rates and growth yields of the mutant were the same as those of the wild type (shown as insets in each panel). The Δ bkdA1B1C1::aac(3)IV mutant, however, produced approximately 50% less actinorhodin than wild type S. coelicolor (Fig. 4a). Production of actinorhodin by wild type supplemented with 15 mM valine (Fig. 4b) or leucine (Fig. 4c) increased actinorhodin production by 7.7% (from 96.2 to 104.2 μg/ml) and 13.3% (from 96.2 to 111 μg/ml), respectively. Isoleucine (Fig. 4d) had virtually no effect on actinorhodin in the wild type cultures. Actinorhodin production in the Δ bkdA1B1C1::aac(3)IV mutant (Fig. 4b, d, f) decreased by approximately 25% when cells were supplemented with either valine (from 53.5 to 40.3 μg/ml), leucine (from 53.5 to 39.7 μg/ml) or isoleucine (from 53.5 to 42.1 μg/ml). One explanation for this observation is that the inability to catabolize these amino acids might lead to the accumulation of these compounds in the growth medium and result in feedback inhibition of antibiotic production. In fact, addition of valine to valine dehydrogenase (vdh) mutants of S. fradiae and S. ambofacians, that are defective in valine catabolism, partially restored antibiotic production at low (10 mM) concentrations but decreased production of tylosin and spiramycin, respectively, at high (25 mM) concentrations [23].
Actinorhodin production by the wild type M145 and the Δ bkdA1B1C1::aac(3)IV mutant in modified liquid minimal (NMMP) media (a), NMMP supplemented with valine (b), isobutyric acid (c), isoleucine (d), α-methylbutyrate (e), leucine (f), or isovaleric acid (g). Growth (shown in insets) and actinorhodin production were measured spectrophotometrically at 450 and 608 nm, respectively. Pre-germinated spores [3], prepared in 2 × YT medium [13] were used as inoculum. Amino acids were added at a concentration of 15 mM, the keto acids at 0.5 mM. The cultures were grown at 30°C with shaking at 200 rpm. Bars indicate the standard deviation of three independent trials
To test whether defects in actinorhodin production resulted directly from defects in the ability to catabolize valine, leucine or isoleucine, cells were grown in liquid minimal medium supplemented with isobutyric acid (specific for valine catabolism), isovaleric acid (specific for leucine catabolism) or α-methylbutyric acid (specific for isoleucine catabolism). Because excess amounts of these compounds had been shown to cause inhibition of antibiotic production in some strains, various concentrations were used to supplement the medium [23]. The plate assays shown in Fig. 3 were supplemented with 5 mM of the indicated keto-acid. The addition of various concentrations of either isobutyric acid or α-methylbutyric acid, 0.5 mM (Fig. 4c, e, respectively), 1, 5, or 10 mM, had no effect on actinorhodin production by the Δ bkdA1B1C1::aac(3)IV mutant suggesting that the breakdown of valine or isoleucine does not play a major role in providing the precursors for actinorhodin production in liquid minimal medium with glucose as carbon source. Addition of 0.5 mM isovaleric acid to the mutant, however, resulted in a 27.4% increase in actinorhodin production over that on minimal medium with glucose alone (Fig. 4g), suggesting that leucine catabolism provides some of the precursors for actinorhodin production under these conditions.
These data are in agreement with the observation that wild type cultures grown in minimal medium supplemented with leucine but not valine or isoleucine as sole carbon source produced the blue pigment associated with actinorhodin production and suggest that rescue of leucine catabolism in the Δ bkdA1B1C1::aac(3)IV mutant also rescues actinorhodin production. Leucine was identified as a source of precursors for actinorhodin production on rich medium agar plates as well as liquid minimal medium. The fact that experiments using solid rich medium identified valine, leucine and isoleucine as precursors while supplementation of liquid minimal medium identified only leucine suggests that use of these amino acids for actinorhodin production is medium dependent.
Differences in antibiotic production on solid versus liquid medium, and rich versus poor media have been reported for a number of streptomycete species suggesting different pathways may be used under different growth conditions. In addition, different species of streptomycetes apparently use different pathways even under the same conditions. Valine catabolism, for example, may be accomplished by at least three different pathways (Fig. 1b) whose activities are medium dependent. The most common pathway in microorganisms and mammals converts valine to propionyl-CoA with 3-hydroxybutyryl-CoA and 2-methylmalonic acid semialdehyde as intermediates. Evidence for the existence of this pathway in S. coelicolor comes from the fact that mutations in methylmalonic acid semialdeyde dehydrogenase, encoded by the msdA gene of S. coelicolor, cannot use valine as sole carbon source on minimal medium [27]. An msdA mutant of S. cinnamonensis, however, was able to utilize both valine and isobutyrate as sole carbon source and showed no decrease in production of the polyketide antibiotic monensin suggesting that this pathway is not important for valine catabolism or anitbiotic production on minimal medium in this species [14].
Valine can also be converted to methylmalonyl-CoA by streptomycetes that utilize methylmalonyl-CoA as extender units in polyketide synthesis via methylmalonyl-CoA semialdehyde as the intermediate [14]. Precursor labeling studies in S. cinnamonensis have shown that methylmalonyl-CoA from labeled valine is incorporated into the monensin carbon framework and is produced via the direct oxidation of β-hydroxybutyric acid to methylmalonyl-CoA [14]. Unlike the growth studies with msdA mutants, the labeling studies were done in rich medium that promoted increased production of the Type I polyketide, monensin. It was suggested by the authors of these studies that pathways for precursors identified on minimal media may or may not be important for antibiotic production in complex rich medium [14].
A third potential pathway allows valine catabolism to acetyl-CoA via the reversible intramolecular conversion of isobutyryl-CoA to butyryl-CoA by isobutyryl-CoA mutase (ICM), encoded by the icmA gene [17] followed by the β-oxidation of butyryl-CoA to form acetyl-CoA [19]. While ICM activity has not been studied in S. coelicolor specifically, an icmA gene homologous to those found in other actinomycetes is present in the S. coelicolor genome. An icmA mutant of S. cinnamonensis was unable to utilize valine or isobutyrate as sole carbon source but could utilize butyrate suggesting that this pathway allows valine catabolism in minimal medium [24]. The phenotypes of these mutants as well as precursor labeling studies suggest that different pathways for valine catabolism are used in rich versus minimal medium and provide a possible explanation for the differences observed for actinorhodin production. It is possible, if not likely, that S. coelicolor uses multiple pathways for the breakdown of valine on rich medium but is limited to a single pathway in minimal medium. We speculate that the difference in rescue of actinohorodin production in the Δ bkdA1B1C1::aac(3)IV mutant with valine catabolites may be due to the different activities of the pathways for valine catabolism on different media and under different conditions.
Leucine, however, is apparently a precursor for actinorhodin production under all conditions. Leucine degradation yields one mole of acetyl-CoA and one mole of acetoacetate, which may be converted stoichiometrically to acetoacetyl-CoA by 3-oxoadipate-CoA transferase (SCO6702), which is converted to two moles of acetyl-CoA by acetoacetyl-CoA thiolase (SCO5399) yielding a total of three moles of acetyl-CoA per mole of leucine. Isoleucine is apparently degraded poorly in minimal medium (Fig. 2) and one mole yields one mole of acetyl-CoA and one mole of propionyl-CoA. Catabolism of one mole of valine in most organisms yields one mole of propionyl-CoA but in Streptomyces sp. valine can be converted to two moles of acetyl-CoA through β-oxidation of butyryl-CoA. The pathway for valine degradation acetyl-CoA is not used by S. coelicolor on minimal medium [27] and taken together these data suggest that leucine degradation is the best source of precursors for actinorhodin biosynthesis in minimal medium.
The Δ bkdA1B1C1mutant is defective in branched-chain fatty acid production. Streptomycetes contain primarily BCFAs [5, 25]. Most bacteria that contain predominately BCFAs use them to maintain membrane fluidity even when unsaturated fatty acids (UFAs) are also present [11]. In Bacillus, branched-chain fatty acids are essential for membrane fluidity and, therefore, cell viability [26]. Since branched-chain fatty acids are derived from branched-chain amino acid catabolism, if the bkdA1B1C1 cluster is the primary pathway for catabolism in S. coelicolor, deletion of this cluster should also result in changes in fatty acid metabolism. An analysis of the fatty acid content of the Δ bkdA1B1C1::aac(3)IV mutant was performed using GC-MS. As shown in Fig. 5, the mutant showed a dramatic change in branched-chain fatty acid content. Branched-chain fatty acids decreased from 61 to 3% of the total fatty acid content with an increase in straight chain fatty acids, from 36% of the total in the wild type to 90% in the mutant. UFAs also increased from 6% of the total in wild type to 34% in the mutant, perhaps allowing the mutant to maintain membrane fluidity and grow without BCFA supplementation. These observations are consistent with previous reports from fatty acid analysis of bkdFGH mutants of S. avermitilis, which were reported to contain less than 1% of the total fatty acids as branched-chain derivatives [5], and suggest that the bkdA1B1C1 cluster is the primary cluster involved in branched-chain fatty acid production in S. coelicolor.
GCMS profile of fatty-acid content in a wild type and b the Δ bkdA1B1C1::aac(3)IV mutant. Spores of S. coelicolor M145 and Δ bkdA1B1C1::aac(3)IV were incubated in NMMP medium for 3 days at 30°C shaking at 300 rpm. Cells were harvested by centrifugation and fatty acids from the cell pellet were analyzed by GC-MS as previously described [25]. An Agilent 19091 series gas chromatograph-mass selective detector equipped with and HP5 phenyl methyl siloxane capillary column with 0.25 μm film thickness was used for GC-MS analysis. Branch-chain fatty acids are indicated: iC14:0, isomyristate; iC15:0, isopentadecanoate; iC16:0, isopalmitate; iC17:0, isoheptadecanoate; aiC17:0, anteisoheptadecanoate. Straight-chain fatty acids: C14:0, myristate; C15:0, pentadecanoate; C16:1 palmitoleate; C16:0, palmitate; C17:1, heptadecenoate; CFA, cyclopropyl fatty acid; C17:0, heptadecanoate; C18:1, octadecenoate; C18:0, octadecanoate
Conclusions
Here we show that a mutant containing a constructed deletion of one of the branched-chain amino acid dehydrogenase complexes of S. coelicolor, Δ bkdA1B1C1::aac(3)IV, was unable to grow on branched-chain amino acids as sole carbon source and produced 50% less actinorhodin that the wild type. This observation suggests that this gene cluster alone is responsible for the levels of catabolism needed for growth on these substrates and the fact that the defects in actinorhodin production were rescued by the addition of products specific to branched-chain amino acid catabolism suggests that branched-chain amino acids serve as precursors for this Type II polyketide. Amino acid catabolism pathways are perhaps the most relevant for antibiotic production in streptomycetes given the metabolic activity of the organism at this stage in the life cycle. When vegetative growth ceases, the cells enter stationary phase and rely on nutrients supplied by the lysing substrate mycelium. Protein is the primary source of carbon and branched-chain amino acids are the most abundant amino acids in protein.
Another important conclusion from this and other work is that different pathways for precursor synthesis may be used under different growth conditions and results from the analysis of antibiotic production using minimal media may differ from those obtained using rich media. Moreover, different pathways are apparently used by species even under the same growth conditions. An understanding of carbon flow from substrates such as the branched-chain amino acids to the production of a class of antibiotics, such as polyketides, will be critical for the metabolic engineering of antibiotic production pathways in this important genus.
References
Brana AF, Manzanal MB, Hardisson C (1982) Characterization of intracellular polysaccharides of Streptomyces. Can J Microbiol 28:1320–1323
Brawner ME, Auerbach JI, Fornwald JA, Rosenberg M, Taylor DP (1985) Characterization of Streptomyces promoter sequences using the Escherichia coli galactokinase gene. Gene 40:191–201. doi:10.1016/0378-1119(85)90042-3
Burke J, Schneider D, Westpheling J (2001) Generalized transduction in Streptomyces coelicolor. Proc Natl Acad Sci USA 98:6289–6294. doi:10.1073/pnas.101589398
Carreras CW, Khosla C (1998) Purification and in vitro reconstitution of the essential protein components of an aromatic polyketide synthase. Biochemistry 37:2084–2088. doi:10.1021/bi972919+
Cropp TA, Smogowicz AA, Hafner EW, Denoya CD, McArthur HA, Reynolds KA (2000) Fatty-acid biosynthesis in a branched-chain α-keto acid dehydrogenase mutant of Streptomyces avermitilis. Can J Microbiol 46:506–514. doi:10.1139/cjm-46-6-506
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi:10.1073/pnas.120163297
Denoya CD, Fedechko RW, Hafner EW, McArthur HA, Morgenstern MR, Skinner DD, Stutzman-Engwall K, Wax RG, Wernau WC (1995) A second branched-chain alpha-keto acid dehydrogenase gene cluster (bkdFGH) from Streptomyces avermitilis: its relationship to avermectin biosynthesis and the construction of a bkdF mutant suitable for the production of novel antiparasitic avermectins. J Bacteriol 177:3504–3511
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546. doi:10.1073/pnas.0337542100
Hey-Ferguson A, Mitchell M, Elbein AD (1973) Trehalose metabolism in germinating spores of Streptomyces hygroscopicus. J Bacteriol 116:1084–1085
Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H (1985) genetic manipulation of Streptomyces: a laboratory manual, 1st edn. The John Innes Foundation, Norwich
Kaneda T (1991) Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev 55:288–302
Khosla C (1997) Harnessing the biosynthetic potential of modular polyketide synthases. Chem Rev 7:2577–2590. doi:10.1021/cr960027u
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich
Li C, Akopiants K, Reynolds KA (2006) Identification and disruptional analysis of the Streptomyces cinnamonensis msdA gene, encoding methylmalonic acid semialdehyde dehydrogenase. J Ind Microbiol Biotechnol 33:75–83. doi:10.1007/s10295-005-0053-4
MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T (1992) Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68. doi:10.1016/0378-1119(92)90603-M
Moore BS, Hertweck C (2002) Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat Prod Rep 19:70–99. doi:10.1039/b003939j
Ratnatilleke A, Vrijbloed JW, Robinson JA (1999) Cloning and sequencing of the coenzyme B12-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J Biol Chem 274:31679–31685. doi:10.1074/jbc.274.44.31679
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sherman MM, Yue S, Hutchinson CR (1986) Biosynthesis of lasalocid A. Metabolic interrelationships of carboxylic acid precursors and polyether antibiotics. J Antibiot (Tokyo) 39:1135–1143
Skinner DD, Morgenstern MR, Fedechko RW, Denoya CD (1995) Cloning and sequencing of a cluster of genes encoding branched-chain alpha-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1 [αβ] component in Escherichia coli. J Bacteriol 177:183–190
Sprusansky O, Stirrett K, Skinner D, Denoya C, Westpheling J (2005) The bkdR gene of Streptomyces coelicolor is required for morphogenesis and antibiotic production and encodes a transcriptional regulator of a branched-chain amino acid dehydrogenase complex. J Bacteriol 187:664–671. doi:10.1128/JB.187.2.664-671.2005
Strauch E, Takano E, Baylis HA, Bibb MJ (1991) The stringent response in Streptomyces coelicolor A3(2). Mol Microbiol 5:289–298. doi:10.1111/j.1365-2958.1991.tb02109.x
Tang L, Zhang YX, Hutchinson CR (1994) Amino acid catabolism and antibiotic synthesis: valine is a source of precursors for macrolide biosynthesis in Streptomyces ambofaciens and Streptomyces fradiae. J Bacteriol 176:6107–6119
Vrijbloed JW, Zerbe-Burkhardt K, Ratnatilleke A, Grubelnik-Leiser A, Robinson JA (1999) Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: Influence of polyketide antibiotic biosynthesis. J Bacteriol 181:5600–5605
Wallace KK, Zhao B, McArthur HA, Reynolds KA (1995) In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol Lett 131:227–234. doi:10.1111/j.1574-6968.1995.tb07781.x
Willecke K, Pardee AB (1971) Fatty acid-requiring mutant of Bacillus subtilis defective in branched-chain α-keto acid dehydrogenase. J Biol Chem 246:5264–5272
Zhang YX, Tang L, Hutchinson CR (1996) Cloning and characterization of a gene (msdA) encoding methylmalonic acid semialdehyde dehydrogenase from Streptomyces coelicolor. J Bacteriol 178:490–495
Acknowledgments
We thank Kevin Reynolds and Galina Florova for performing the fatty acid analysis and for assistance in interpreting the data, Timothy Dore for his help in interpretation of the fatty acid analysis, Larry Shimketts and Mike Adams for advice throughout the course of the work and for critical review of the manuscript, and David Brown for help in preparation of the figures. K. S. was supported by a predoctoral training grant from the National Institute for General Medical Sciences GM07103 to the Genetics Department of the University of Georgia, an ARCS Foundation Scholarship, as well as a grant from Pfizer, Inc. to J. W.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Stirrett, K., Denoya, C. & Westpheling, J. Branched-chain amino acid catabolism provides precursors for the Type II polyketide antibiotic, actinorhodin, via pathways that are nutrient dependent. J Ind Microbiol Biotechnol 36, 129–137 (2009). https://doi.org/10.1007/s10295-008-0480-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0480-0