Abstract
In this study, a bacterial strain P13 capable of degrading pendimethalin was isolated from the soil of a fruit garden. Based on observed cellular morphology and physiology characteristics and a phylogenetic analysis of 16S rRNA gene sequences, strain P13 was identified as a member of the genus Paracoccus. Strain P13 grew on pendimethalin as the sole carbon source, and could degrade 100 mg/L pendimethalin within 2 days and 200 mg/L pendimethalin within 5 days. Pendimethalin degradation was proposed to be initiated by oxidation ring cleavage to yield 1,3-dinitro-2-(pentan-3-ylamino)butane-1,4-diol, an alkane organic compound that was identified by ultra-high performance liquid chromatography coupled to tandem mass spectrometry (UHPLC–MS/MS), which then underwent a series of enzymatic reactions to produce CO2 and H2O. The optimal pH and temperature for pendimethalin degradation by strain P13 were 7.0 and 30 °C, respectively. This study identified the bacterial strain Paracoccus sp. P13, which degraded pendimethalin with a relatively high efficiency, and presents a previously unreported microbial pendimethalin degradation pathway.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Currently, increases in food quantity and quality rely heavily on the use of pesticides. Pendimethalin [N-(1-ethylpropyl)-2,6-dinitro-3,4-xylidine], is a selective preemergence dinitroaniline herbicide, which is commonly used to control most annual grasses and certain broadleaf weeds for many crops, such as cotton, soybeans, wheat, rice, and maize. Its mode of action is inhibiting plant cell division and cell wall formation [9, 20]. Pendimethalin has a relatively low volatility [16, 21], and is moderately persistent with a field half-life of approximately 90 days and does not undergo rapid microbial degradation [23]. Due to its widespread use, pendimethalin has been detected as a contaminant in soil, ground water, and surface water [14, 24]. Pendimethalin is a possible human carcinogen and shows high toxicity to terrestrial and aquatic invertebrates [1, 3]. Furthermore, pendimethalin can bioaccumulate and concentrate to 7000 times its initial concentration [17], and the US Environmental Protection Agency (EPA) has classified pendimethalin as a persistent, bioaccumulative, and toxic substance [18]. Thus, a better understanding of the degradative fate of pendimethalin in soil is needed.
Biodegradation is the most important route by which pendimethalin is degraded in the environment [20, 22, 25]. Numerous pendimethalin-degrading microorganisms have been isolated, including Bacillus sp. Y3 [12], Bacillus lehensis XJU [11], Pseudomonas aeruginosa, Bacillus mycoides, Bacillus cereus [19], Pseudomonas putida E15 [2], Bacillus circulans [10], Fusarium oxysporum [8], and Azotobacter chroococcum [7]. Generally, pendimethalin degradation is initiated by three different mechanisms, nitroreduction, oxidative N-dealkylation, and cyclization [7]. In this study, we described a Paracoccus sp. bacterium P13 that was observed to degrade pendimethalin, which was determined to be initiated through oxidation ring cleavage on the basis of metabolite identification.
Materials and Methods
Chemicals and Medium
Pendimethalin (97% purity) was provided by Rosi Chemical Co., Ltd. (Zhejiang Province, China). Methanol and acetonitrile were of chromatographic grade (Sigma-Aldrich, Shanghai, China), dichloromethanol and other organic reagents were of analytical grade (Shou-De, Nanjing, China). Pendimethalin was first dissolved in methanol and then filtered through a 0.22 µm Millipore membrane filter before use.
Mineral salts medium (MSM) consisted of (in g/L) 1.0 NH4NO3, 0.5 NaCl, 1.5 K2HPO4, 0.5 KH2PO4, and 0.2 MgSO4·7H2O, supplemented with pendimethalin as the sole carbon source. The Luria–Bertani (LB) broth contained (in g/L) 10.0 tryptone, 5.0 yeast extract, and 10.0 NaCl. Agar was added at 15.0 g/L for solid medium. Both media were sterilized at 121 °C for 30 min before use.
Enrichment, Isolation, and Identification of Pendimethalin-Degrading Strains
A conventional enrichment culture method was performed to isolate pendimethalin-degrading bacteria. Ten gram soil samples, collected from the surface layer (0–10 cm) of a fruit garden in Shandong Province, China, was added to an Erlenmeyer flask with 100 mL of sterile deionized water and shaken for 1 h at 30 °C and 150 r/min. Next, 5 mL of the soil suspension was added to a 250-mL Erlenmeyer flask containing 95 mL fresh MSM supplemented with 100 mg/L pendimethalin as the sole carbon source. The mixture was incubated on a rotary shaker at 30 °C and 150 r/min in the dark for 5 days. Subsequently, 5 mL of the initial enrichment culture was transferred into 95 mL of fresh MSM with 100 mg/L pendimethalin, and incubated for another 5 days under the same conditions described above. The transfer was repeated three times.
Serial gradient dilutions (1.0 × 10−1–1.0 × 10−7) of the final enrichment culture were prepared, and then 100 µL of each dilution was spread on MSM agar plates supplemented with 200 mg/L pendimethalin. The plates were incubated at 30 °C for 5 days and monitored for the appearance of bacterial colonies. Single colonies were picked and further purified by repeated streaking onto LB broth agar plates supplemented with 200 mg/L pendimethalin.
Each isolate was then tested for the ability to degrade pendimethalin. Isolates that were effective at degrading pendimethalin were characterized and identified by morphological, physiological, and biochemical characteristics as well as by 16S rRNA gene sequences analysis according to the method described by Nie et al. [13].
Cell Growth and Pendimethalin Degradation
The isolates were pregrown in baffled Erlenmeyer flasks containing fresh LB broth, which were incubated overnight at 30 °C on a rotary shaker at 150 r/min. The contents were harvested by centrifugation at 5000 r/min for 10 min and then washed twice with fresh MSM. The cell density was adjusted to 1.0 × 108 CFU/mL and served as seed cultures. An aliquot of seed culture (1%, V/V) was transferred into 20 mL MSM supplemented with 200 mg/L pendimethalin in a 50-mL Erlenmeyer flask. The degradation of pendimethalin by free-living cells of strain P13 was carried out aerobically at 30 °C on a rotary shaker at 150 r/min in the dark. Control experiments without free cells or pendimethalin were performed under the same conditions. Each treatment was performed three times. The enrichment cultures were sampled at 24 h intervals to calculate cell growth and residual pendimethalin. Bacteria growth was monitored by measuring the colony-forming units (CFU/mL), and the residual concentration of pendimethalin was confirmed by ultra-high performance liquid chromatography (UHPLC) as described below.
Chemicals Analysis
The culture medium samples were partitioned with an equal volume of dichloromethanol. The organic extracts were passed through anhydrous sodium sulfate and evaporated to dryness under a gentle nitrogen stream (40 °C). Finally, all the samples were redissolved in methanol, filtered through 0.22 µm Millipore membrane filters, and then used for the following analysis.
The structural identification of pendimethalin metabolites in samples was performed by LC–MS. The LC–MS system consisted of a UHPLC (Dionex, Thermo, USA) coupled with an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The UHPLC column utilized was a Hypersil GOLD C18 column (100 mm × 2.10 mm, 3 µm particle sizes, Thermo Fisher Scientific). The UHPLC mobile phase consisted of 0.02% of formic acid in water (buffer A) and 100% acetonitrile (buffer B). Separation was carried out over 30 min at a flow rate of 0.2 mL/min under the following conditions: 0–4 min, 30% B; 4–20 min, 30–95% B; 20–24.5 min, 95% B; 24.5–25 min, 95–30% B; and 25–30 min, 30% B. The injection volume was 5 µL, and the detection wavelength was set at 240 nm for pendimethalin (UV-900 wavelength absorption detector). An LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with an electrospray ionization (ESI) probe, was used for the mass spectral analysis of pendimethalin and its metabolites. The capillary temperature was set at 300 °C, and the source voltage was set at 4 kV. The sheath gas was set at 35 arb, and the Aux Gas was set at 10 arb. The data analysis was performed under positive mode ESI conditions [15]. Data were processed using Xcalibur.
Effect of pH and Temperature on Pendimethalin Degradation by Strain P13
To determine the effect of pH and temperature on pendimethalin degradation by strain P13, 20 mL MSM containing 200 mg/L pendimethalin was used as the pendimethalin-degrading medium, and inoculated by a bacterial seed culture to an initial cell density of 1.0 × 106 CFU/mL. To determine the optimum pH, experiments were performed at pH values of 4.0–9.0. To determine the effect of temperature, the pendimethalin-degrading medium with the optimum pH was incubated at 20–45 °C. The effect of pH and temperature on pendimethalin degradation by strain P13 was examined using an incubation period of 5 days on a rotary shaker at 30 °C and 150 r/min. Residual pendimethalin under different conditions was measured by UHPLC, and the amount of degraded pendimethalin was calculated.
Strain Deposition and Nucleotide Sequence Accession Numbers
Strain P13 was deposited at the China Center for Type Culture Collection (CCTCC) under the deposition number CCTCC AB 2015071. The GenBank accession number for the strain P13 16S rRNA gene sequence is KP215280.
Results and Discussion
Strain Isolation and Identification
After four rounds of selective enrichment, the enrichment mixture degraded approximately 99.9% of the initial 100 mg/L pendimethalin within 5 days, and 13 morphologically different microorganisms were isolated from the enrichment samples. All 13 strains were individually cultured in MSM and LB broth supplemented with 200 mg/L pendimethalin as the carbon source to assess pendimethalin degradation. The results suggested that only 3 of the 13 strains were able to grow in both media and degrade pendimethalin. One strain (P13) exhibited the highest degradation rate (99.8%) compared with the other two strains (degradation rates of 57 and 46%) and was chosen for further study.
Strain P13 was identified by morphological and physiological characteristics and a phylogenetic analysis of 16S rRNA gene sequences. Strain P13 was determined to be a gram-negative, non-spore-forming, and non-motile bacterium. Colonies grown on LB and MSM agar media were pale yellow, wet, convex, rod-shaped, and glossy with a smooth surface and neat edges. In addition, this microorganism tested positive for nitrate reduction, oxidase and catalase, and for the acidification of l-arabinose, d-glucose, d-mannose, and d-fructose. Strain P13 tested negative for urease, indole reaction, methyl red, and Voges–Proskauer tests, hydrolysis of gelatin and starch, and for the acidification of maltose, sucrose, lactose, and trehalose. The obtained 16S rRNA gene sequence of strain P13, nearly complete, was 1336 bp in length. The sequence was deposited under the accession number KP215280 in GenBank. Phylogenetic analysis of 16S rRNA gene sequences suggested that strain P13 grouped among Paracoccus species and was most closely related to Paracoccus huijuniae strain FLN-7T (similarity = 100%), forming a subclade with P. huijuniae FLN-7T (100% similarity) and P. aminovorans DSM 8537 (99.18% similarity) (Fig. S1). Thus, in light of our results, strain P13 was identified as a Paracoccus sp. and was deposited at the China Center for Type Culture Collection (CCTCC) under the deposition number CCTCC AB 2015071.
Enrichment culturing is a useful and efficient method to isolate bacterial strains that are able to degrade different pesticides with high efficiency, and the addition of pendimethalin provides a selective pressure for the evolution of microorganisms that could degrade this xenobiotic compound. Paracoccus species are widely distributed in soils and involved in the degradation of various xenobiotics and pollutants. Numerous Paracoccus sp. have been isolated, for example, Paracoccus sp. FLN-7 has been reported to degrade diflubenzuron [26], Paracoccus sp. M-1 has been shown to degrade monocrotophos [5], and Paracoccus sp. QCT6 has been reported to degrade metribuzin [4]. This is the first report of a Paracoccus sp. that can use pendimethalin as the carbon source for cell growth and degrade it. Further study of this genus may provide a better understanding of the ecological diversity of these organisms and their usefulness for biodegradation.
Utilization and Degradation of Pendimethalin by Paracoccus sp. P13
The observed growth of Paracoccus sp. P13 in liquid MSM containing 200 mg/L pendimethalin and its capacity to degrade pendimethalin are presented in Fig. 1. The growth curve showed a steady increase in P13 population, and the cell density increased from 1.0 × 106 to 3.42 × 107 CFU/mL within 5 days. In the meantime, UHPLC analysis showed a substantial reduction of the pendimethalin concentration in the medium, as 50% of the initial 200 mg/L pendimethalin was degraded by strain P13 within 2 days, and the degradation rate reached 99.8% within 5 days. No cell growth was observed when strain P13 was incubated in the cultures without pendimethalin, and no significant change in the pendimethalin concentration was observed in uninoculated medium. The consumption of pendimethalin supported the cell growth of strain P13, suggesting that this microorganism could degrade pendimethalin and utilize it as a carbon source for growth.
In this study, Paracoccus sp. P13 was observed to degrade 100 mg/L pendimethalin within 2 days and 200 mg/L pendimethalin within 5 days. A. chroococcum was previously shown to degrade 45 and 55% of 25 µg/mL pendimethalin within 10 and 20 days [7], respectively. Lecanicillium saksenae degraded 99.5% of 25 mg/L pendimethalin within 10 days [14]. P. aeruginosa, B. mycoides, and B. cereus could degrade 80.75, 71.75, and 80.25% of an initial 400 mg/L pendimethalin within 30 days, respectively [19]. Bacillus sp. Y3 degraded 99.5% of 100 mg/L pendimethalin within 2.5 days [12]. Compared with these pendimethalin-degrading strains, strain P13 possessed a relatively high degradation efficiency, suggesting that this microorganism may have a greater potential in eliminating pendimethalin residue pollution.
Metabolites of Pendimethalin Degradation by Strain P13
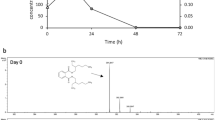
Identification of the target compound in samples was based on determining HPLC retention times (RTs) compared to that of a standard, and the identification of pendimethalin metabolites was performed by UHPLC–MS/MS. By injecting a standard sample and comparing the RT, a peak with a RT of 18.64 min was detected as pendimethalin, which had a molecular peak [M + H]+ at m/z 282.1448 and a base peak [M + H]+ at m/z 212.1212 due to oxidation N-dealkylation. In the tested sample, only one metabolite was detected with a RT of 3.29 min (Fig. 2). The MS/MS spectrum of this metabolite indicated that it had a molecular peak [M + H]+ at m/z 266.1354, in agreement with the molecular formula of C9H19N3O6. The fragmentation pattern showed the base peak [M + H]+ was at m/z 166.0024, which was due to the removal of N-alkyl group and nitroreduction of one nitro group. According to the above analysis, the metabolite was identified as 1,3-dinitro-2-(pentan-3-ylamino)butane-1,4-diol. Therefore, the proposed biochemical pathway of pendimethalin degradation in strain P13 was figured out (Fig. 3): pendimethalin first underwent oxidative ring cleavage to form 1,3-dinitro-2-(pentan-3-ylamino)butane-1,4-diol, with a subsequent series of enzymatic reactions resulting in the production of CO2 and H2O.
UHPLC–MS/MS analysis of the metabolite of pendimethalin degradation by strain P13. a UHPLC–MS/MS analysis of pendimethalin, RT = 18.64 min, the molecular peak [M + H]+ was at m/z 282.1448, and the base peak [M + H]+ was at m/z 212.1212; b UHPLC–MS/MS analysis of the metabolic product, RT = 3.29 min, the molecular peak [M + H]+ was at m/z 266.1354, and the base peak [M + H]+ at m/z 166.0024
Previous reports have indicated that some microorganisms can degrade pendimethalin, however the degradation pathway of pendimethalin was diverse. In A. chroococcum, three mechanisms were involved in pendimethalin degradation: nitroreduction to form 6-aminopendimethalin, oxidation N-dealkylation to produce 3,4-dimethyl-2,6-dinitroaniline, and arylmethyl group oxidation to yield [6-methyl-3,4-dinitro-3-(pentanyl-3-amino)phenyl]methanol [7]. 6-Aminopendimethalin and 3,4-dimethyl-2,6-dinitroaniline were identified as pendimethalin degradation products by B. circulans and B. lehensis XJU [10, 11]. Pendimethalin was observed to be initially degraded by nitroreduction, and three metabolites, 6-aminopendimethalin, 5-amino-2-methyl-3-nitroso-4-(pentan-3-ylamino) benzoic acid, and 8-amino-2-ethyl-5-(hydroxymethyl)-1,2-dihydroquinoxaline-6-carboxylic acid, were identified [12]. Pendimethalin degradation involved nitroreduction of both nitro groups and demethylation of one methyl group, and the metabolite was identified as N-(1-ethylpropyl)-3-methyl-2,6-diaminobenzine in P. aeruginosa [19]. N-dealkylation and nitroreduction are the very common pathways of microbial transformation of pendimethalin and other dinitroaniline compounds, which directed to detoxify their herbicidal activities [7]. Sequentially, the metabolites are cleaved by the ortho- or meta-cleavage pathway as that used for catechol in microorganisms [6]. In this study, Paracoccus sp. P13 was determined to initially degrade pendimethalin by oxidative ring cleavage to produce the alkane metabolite 1,3-dinitro-2-(pentan-3-ylamino)butane-1,4-diol, whereas previously identified metabolites of pendimethalin biodegradation were all aromatic products. This indicated that a different process involving unique degrading enzymes is responsible for the biodegradation of this dinitroaniline herbicide in Paracoccus sp. P13. Further studies with additional molecular biological techniques are needed to better understand the details of pendimethalin degradation by strain P13.
Effect of pH and Temperature on Pendimethalin Degradation by Strain P13
Factors such as pH and temperature can influence the growth of microorganisms, and hence influence their ability to degrade pesticides. The effect of pH on pendimethalin degradation by strain P13 is shown in Fig. 4. The maximum degradation rate was observed at pH 7.0, and strain P13 could degrade >75% of 200 mg/L pendimethalin at pH values of 6.0–8.0. The degradation rate was significantly decreased under strong acid conditions (pH values below 4.0). The optimum temperature for pendimethalin degradation was determined to be 30 °C (Fig. 5). More than 80% of 200 mg/L pendimethalin was degraded at 25–35 °C, and pendimethalin degradation was inhibited at 20 and 40 °C.
Microorganisms are a major component of the natural environment and play a considerable role in the biodegradation of herbicides. Additional studies of the biodegradation characteristics in Paracoccus sp. P13 will further our understanding of how microorganisms are involved in the biodegradation of dinitroaniline herbicides.
Conclusion
Pendimethalin is a widely used herbicide with a high herbicidal efficiency. The dinitroaniline herbicide pendimethalin is very stable and persists in the environment. Pendimethalin exhibits high toxicity to terrestrial and aquatic invertebrates and is a possible human carcinogen. Microbial metabolism is the most important factor in the degradation of pendimethalin in soil. Strain P13 was isolated from the soil of a fruit garden and was characterized as a Paracoccus sp. that could degrade approximately 100 and 200 mg/L pendimethalin after 2 and 5 days of incubation, respectively. An alkane metabolite, 1,3-dinitro-2-(pentan-3-ylamino)butane-1,4-diol, was identified as a product of P13-mediated pendimethalin degradation by UHPLC–MS/MS. This finding suggested that there may be a new degradation mechanism of pendimethalin in strain P13 that differs from those that are commonly observed, the details of which will need to be further studied.
References
Das N, Ray S, Jena S, Mohanty P (1998) Effect of certain herbicides on weeds and population of root-knot nematode (Meloidogyne incognita) in mustard. Crop Res 16:156–158
Elsayed B, El-Nady MF (2013) Bioremediation of pendimethalin-contaminated soil. Afr J Microbiol Res 7(21):2574–2588
Fliedner A (1997) Ecotoxicity of poorly water-soluble substances. Chemosphere 35:295–305
Huang X, Zhang H, Chen F, Song M (2018) Colonization of Paracoccus sp. QCT6 and enhancement of metribuzin degradation in maize rhizosphere soil. Curr Microbiol 75(2):156–162
Jia KZ, Cui ZL, He J, Guo P, Li SP (2006) Isolation and characterization of a denitrifying monocrotophos-degrading Paracoccus sp. M-1. FEMS Microbiol Lett 263(2):155–162
Ju KS, Parales RE (2010) Nitroaromatic compounds, from synthesis to biodegradation. Microbiol Mol Biol Rev 74(2):250–272
Kole RK, Saha J, Pal S, Chaudhuri S, Chowdhury A (1994) Bacterial degradation of the herbicide pendimethalin and activity evaluation of its metabolites. Bull Environ Contam Toxicol 52:779–786
Kulshrestha G, Singh SB, Lal SP, Yaduraju NT (2000) Effect of long-term field application of pendimethalin: enhanced degradation in soil. Pest Manag Sci 56:202–206
Ma JY, Wang SF, Wang PW, Ma LJ, Chen XL, Xu RF (2006) Toxicity assessment of 40 herbicides to the green alga Raphidocelis subcapitata. Ecotoxicol Environ Safe 63:456–462
Megadi VB, Tallur PN, Hoskeri RS, Mulla SI, Ninnekar HZ (2010) Biodegradation of pendimethalin by Bacillus circulans. Indian J Biotechnol 9:173–177
More VS, Tallur PN, Niyonzima FN, More SS (2015) Enhanced degradation of pendimethalin by immobilized cells of Bacillus lehensis XJU. 3 Biotech 5(6):967–974
Ni HY, Yao L, Li N, Cao Q, Dai C, Zhang J, He Q, He J (2016) Biodegradation of pendimethalin by Bacillus subtilis Y3. J Environ Sci 41:121–127
Nie ZJ, Hang BJ, Cai S, Xie XT, He J, Li SP (2011) Degradation of cyhalofop-butyl (Cyb) by Pseudomonas azotoformans strain qdz-1 and cloning of a novel gene encoding cyb-hydrolyzing esterase. J Agric Food Chem 59(11):6040–6046
Pinto A, Serrano C, Pires T, Mestrinho E, Dias L, Teixeira DM, Caldeira A (2012) Degradation of terbuthylazine, difenoconazole and pendimethalin pesticides by selected fungi cultures. Sci Total Environ 435:402–410
Ramakrishna M, Mohan SV, Shailaja S, Narashima R, Sarma PN (2008) Identification of metabolites during biodegradation of pendimethalin in bioslurry reactor. J Hazard Mater 151:658–661
Richardson ML, Gangolli SE (eds) (1994) The dictionary of substances and their effects, vol 6. The Royal Society of Chemistry, Clays Ltd., Cambridge, pp 431–432
Ritter L, Solomon K, Forget J, Stemeroff M, O’Leary C (1995) Persistent organic pollutants: an assessment report on DDT. Aldrin, dieldrin, endrin, chlordane, heptachlor, hexachlorobenzene, mirex, toxaphene, polychlorinated biphenyls, dioxins, and furans. International Programme on Chemical Safety (IPCS)
Roca E, D’Errico E, Izzo A, Strumia S, Esposito A, Fiorentino A (2009) In vitro saprotrophic basidiomycetes tolerance to pendimethalin. Int Biodeterior Biodegrad 63:182–186
Sharef IB, Abdelbagi A, Elsheikh EA, Ahmed AES, Elsaid OEG (2013) Biodegradation of pendimethalin by three strains of bacteria isolated from pesticide-polluted soils. U K J Agric Sci 21(2):233–252
Strandberg M, Scott-Fordsmand JJ (2004) Effects of pendimethalin at lower trophic levels—a review. Ecotoxicol Environ Safe 57:190–201
Tomlin C (1997) Pesticide manual, 11th edn. British Crop Protection Council, Farnham, ISBN 1-901396-11-8
Venkata Mohan S, Rama Krishna M, Muralikrishna P, Shailaja S, Sarma P (2007) Solid phase bioremediation of pendimethalin in contaminated soil and evaluation of leaching potential. Bioresour Technol 98(15):2905–2910
Wauchope RD, Buttler T, Hornsby A, Augustijn-Beckers P, Burt J (1992) The SCS/ARS/CES pesticide properties database for environmental decision-making. Springer, New York, pp 1–155
Younes M, Galal-Gorchev H (2000) Pesticides in drinking water—a case study. Food Chem Toxicol 38:S87–S90
Zhang C, Hughes JB, Nishino SF, Spain JC (2000) Slurry-phase biological treatment of 2,4-dinitrotoluene and 2,6-dinitrotoluene: role of bioaugmentation and effects of high dinitrotoluene concentrations. Environ Sci Technol 34:2810–2816
Zhang J, Yin JG, Hang BJ, Cai S, He J, Zhou SG, Li SP (2012) Cloning of a novel arylamidase gene from Paracoccus sp. strain FLN-7 that hydrolyzes amide pesticides. Appl Environ Microbiol 78(14):4848–4855
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31760031 and 31600080), the Natural Science Foundation of Jiangxi Province (20171BAB214002), and the Natural Science Foundation of Shandong Province (ZR2016CB29).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Haiyan Ni and Na Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ni, H., Li, N., Qiu, J. et al. Biodegradation of Pendimethalin by Paracoccus sp. P13. Curr Microbiol 75, 1077–1083 (2018). https://doi.org/10.1007/s00284-018-1494-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1494-0