Abstract
A non-motile, pleomorphic rod-shaped or oval, red-pigmented (nearly scarlet), extremely halophilic archaeon, strain Y78T, was isolated from a salt deposit of Yunnan salt mine, China. Analysis of the 16S rRNA gene sequence showed that it was phylogenetically related to species of the genus Halorubrum, with a close relationship to Halorubrum rutilum YJ-18-S1T (98.6%), Halorubrum yunnanense Q85T (98.3%), and Halorubrum lipolyticum 9-3T (98.1%). The temperature, NaCl, and pH ranges for growth were 25–50 °C, 12–30% (w/v), and 6.5–9.0, respectively. Mg2+ was required for growth. The polar lipids of strain Y78T were phosphatidylglycerol, phosphatidylglycerol phosphate methyl ester, phosphatidylglycerol sulfate, and a sulfated diglycosyl diether. The DNA G+C content was 66.6 mol%. DNA–DNA hybridization values between strain Y78T and two closely related species of the genus Halorubrum were far below 70%. Based on the data presented in this study, strain Y78T represents a novel species for which the name Halorubrum depositum sp. nov. is proposed; the type strain is Y78T (= CGMCC 1.15456T = JCM 31272T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Species of genus Halorubrum established by McGenity and Grant in 1995 [16] have always been found in salt mines [3, 29]. At the time of writing, there are 35 validly named Halorubrum species. Many species of halotolerant bacterium or haloarchaea were isolated from the salt mine or salt crystal [22, 27, 28]. Hypersaline environments such as salt lakes, salt mine, and evaporation ponds for the production of commercial salts including edible and industrial salts are found on all continents. They are inhabited by a great diversity of microorganisms adapted to life at high salt concentrations [19]. Salt mine, the fixed hypersaline environment, is one kind of unique habitats for keeping the old microorganisms alive in it [12]. The discovery of many new microbial groups, and their isolation has recently turned our limited view of the hypersaline environment in new directions [26]. In this study, we searched for haloalkaliphiles able to grow at pH 8.0, and isolated a novel slightly alkaliphilic haloarchaeon strain Y78T that belongs to the genus Halorubrum. Based on the phylogenetic and phenotypic features, we aim to propose a novel species within the genus Halorubrum.
Materials and Methods
Strain Isolation and Cultivation
The salt deposits were collected from Yunnan salt mine (China) in Sep. 2013 (99°46′42.05″E, 26°06′04.05″N). The sample (100 g) was washed with 100 ml sterile water twice for 1 min for each time, and then the rest of the salt sediment was dissolved in 100 ml of sterile 5% (w/v) NaCl solution. And 500 μl of the sample dissolving liquid was spread on JCM 168 medium agar plates. The JCM 168 medium used for cultivating halophilic bacteria or archaea constituted of (per liter): casamino acids (BD-Difco, 5.0 g), yeast extract (BD-Difco, 5.0 g), sodium glutamate (1.0 g), trisodium citrate (3.0 g), MgSO4·7H2O (20.0 g), KCl (2.0 g), NaCl (200.0 g), FeCl2·4H2O (36.0 mg), and MnCl2·4H2O (0.36 mg) (http://jcm.brc.riken.jp/en/). The pH of the medium was adjusted to 8.0 with 1 M KOH.
The inoculated agar plates were sealed by plastic preservative film and incubated in a sealed plastic bags at 38 °C for 2 weeks. Red-colored (nearly scarlet) colonies occurred on the agar plates. The colonies that occurred on the agar plates were transferred to fresh agar plates, and pure cultures were obtained after successive streaking. All the colonies were purified for the further searches.
Preliminary Identification by 16S rRNA Gene
The pure cultures obtained via successive streaking were re-suspended in sterile water and the lysates were taken as the PCR template for preliminary identification. PCR amplification of the 16S rRNA gene was performed by using the primer pair F8 (5′-TTGATCCTGCCGGAGGCCATTG-3′) and R1462 (5′-ATCCAGCGCAGATTCCCCTAC-3′) [13]. The PCR products verified by electrophoresis were sent to a biotechnological company for direct DNA sequencing.
Phenotypic and Chemotaxonomic Characterization
Colony morphology was observed on the agar plate after incubation for 2–3 weeks at 38 °C. Gram-staining was performed according to method described by Dussault [8]. Cell morphology and motility were examined using a phase contrast microscope (OLYMPUS BX51 equipped with OLYMPUS DP72) and scanning electron microscope (HITACHI SU8010). The range of salinity for growth was determined by using the JCM 168 medium containing various concentrations of NaCl 5–30% (w/v) with intervals of 5% (w/v). The pH range for growth was assayed from pH 5.0–9.5 at intervals of 0.5 in liquid media containing 50 mM pH buffers (MES for pH 5.0–6.0, PIPES for pH 6.5–7.0, HEPES for pH 7.5–8.0, tricine for pH 8.5, or CHES for pH 9.0–9.5). The temperature range for the growth was determined at 10, 15, 20, 25, 30, 35, 38, 40, 42, 45, 50, and 55 °C in a medium of pH 8.0 with 20% (w/v) NaCl. The concentrations of Mg2+ in the media were set to 0, 0.005, 0.01, 0.03, 0.05, 0.1, 0.3, 0.5, 0.7, and 1.0 M, for detecting the requirement of Mg2+ for the growth. In this test, the MgSO4·7H2O was replaced by MgCl2·6H2O to avoid the impacts of SO42−. Cell lysis was performed by re-suspending the cell pellets in distilled water and centrifugation.
The similarity search of 16S rRNA gene sequence showed that strain Y78T was affiliated to the genus Halorubrum. The following phenotypic tests were performed under the optimal growth condition, according to the proposed minimal standards for the description of new taxa in the order Halobacteriales [20]. Tests for catalase and oxidase activities and for the hydrolysis of starch, gelatin, skim milk, Tween 80, Tween 60, Tween 40, and Tween 20 were performed as described by González et al. [9]. H2S formation from l-cysteine was detected using a filter-paper strip impregnated with lead acetate (10%, w/v) [5]. Indole production from tryptophan was assessed as described by Oren et al. [20]. To determine the utilization of different organic substrates such as carbohydrates, alcohols, amino acids, and organic acids as the only source of carbon, nitrogen, and energy, a medium containing 0.01% (w/v) yeast extract and supplemented with 1% (w/v) of the tested substrate (filtration sterilization) was assessed as described by Oren et al. [20]. Reduction of nitrate and nitrite was detected by using the sulfanilic acid and α-naphthylamine reagent [23]. Anaerobic growth was tested with l-arginine, KNO3 and DMSO with the final concentration of 5 g l− 1 in screw-topped sealed vials.
Susceptibility to Antibiotics
Antibiotic sensitivity tests were performed by spreading cell suspensions on culture plates and then placing discs impregnated with antibiotics. Antibiotics and amounts (μg per disc, unless indicated) were shown as follows; ampicillin (10), bacitracin (0.04 IU), chloramphenicol (30), ciprofloxacin (5), erythromycin (15), kanamycin (10), neomycin (30), norfloxacin (10), novobiocin (30), penicillin G (10 IU), rifampicin (5), streptomycin (10), tetracycline (30), and vancomycin (30).
Polar Lipids Profile
Polar lipids were extracted using the widely used chloroform–methanol system and detected using two-dimensional thin layer chromatography (2D-TLC) on an aluminum-backed silica gel 60 plates (20 × 20 cm, Merck) [18]. Glycolipids and phospholipids were detected by spraying with 0.5% (w/v) α-naphthol in methanol:water (1:1, by vol), after drying, with sulfuric acid and ethanol (1:1, by vol), and followed by heating at 120 °C for 10 min. The plate was scanned immediately for recording the results.
Phylogenetic Analysis
An single colony of strain Y78T was picked with sterile toothpick from the agar plate (2–3 weeks incubation) and re-suspended in 40 μl sterile distilled water. Cells lysed in distilled water were taken as the PCR template. The 16S rRNA gene was amplified by using the primer pair F8 and R1462 [13]. The PCR products were purified with a DNA gel extraction kit (Axygen) and inserted into the cloning vector pMD-18T (TaKaRa) for transformation. At least five PCR-verified recombinants were picked for the DNA sequencing. The almost complete 16S rRNA gene sequence was taken as the query to search the public database via the online BLAST searching tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the EzBioCloud, a public and professional website focusing on the prokaryotic taxonomy [31].
The rpoB’ gene was amplified with the pair of degenerated primers 1420F (5′-TGTGGGCTNGTGAAGAACTT-3′) and 153R (5′-GGGTCCATCAGCCCCATGTC-3′) [17], while the ef-2 gene was amplified with another pair of degenerated primers EF-2f (5′-ATGGGYMGACGHAAGAA-3′) and EF-2r (5′-GCBGGRCCRCGGTGGAT-3′) [11]. The procedure of the cloning and sequencing of the rpoB’ and ef-2 genes was identical to that of 16S rRNA gene.
Multiple sequence alignments were performed using the Clustal W program implemented in the BioEdit software [10] prior to the phylogenetic analysis. DNA sequences used for the reconstruction of phylogenetic trees were retrieved from the public database. Phylogenetic trees were reconstructed using maximum-likelihood algorithms and neighbor-joining algorithms in the MEGA 5.0 software [25]. Multiple sequence alignments were created after taking into account the amino acid alignments for the rpoB′ protein-encoding gene generated by translating this gene to protein sequences. To evaluate the robustness of the phylogenetic tree, a bootstrap analysis (1000 replications) was performed.
Determination of the DNA G+C Content and DNA–DNA Relatedness
Genomic DNA of strain Y78T was obtained by the method of Marmur [15]. The G+C content of genomic DNA was inferred from the mid-point (Tm) of the thermal denaturation profile [14] using the equation of Owen and Hill [21]. DNA–DNA hybridization was performed between strain Y78T and its closely related species (Halorubrum rutilum YJ-18-S1T and Halorubrum yunnanense Q85T) on a Perkin-Elmer Lambda 35 spectrophotometer equipped with a high-performance temperature controller (PTP-6 Peltier system, Perkin-Elmer, USA) in accordance with the thermal denaturation and renaturation approach [7].
Results and Discussion
A red-pigmented (nearly scarlet) colony, entitled strain Y78T, was isolated from the deposit sample of Yunnan Salt Mine (China) for further analysis. Similarity search of the almost complete 16S rRNA gene (KX376712, 1439 bp) sequence showed that strain Y78T was closely related to Halorubrum rutilum YJ-18-S1T, Halorubrum yunnanense Q85T, and Halorubrum lipolyticum 9-3T with the similarities of 98.6, 98.3, and 98.1%, respectively. Lower similarities were obtained with the type strains of other species of genus Halorubrum and other haloarchaeal genera. Based on the preliminary identification of the 16S rRNA gene, strain Y78T was closely related to species of the genus Halorubrum in family Halorubraceae [1].
Strain Y78T was pleomorphic rod-shaped or oval (Fig. S1). It differed from all the closely related species in genus Halorubrum on the mobility and the optimum pH for growth. The mobility of strain Y78T was weak, nearly non-motile, while others were motile (Table 1). The optimum pH for growth of strain Y78T was pH 8.5, which obviously was higher than others (Table 1). Strain Y78T was capable of growing in the range of 10–30% (w/v) NaCl, pH 6.5–9.0, and 25–50 °C. Weak growth also observed with the temperature between 15 and 25 °C when the duration of incubation prolonged (> 4 weeks). Strain Y78T was resistant to bacitracin under the indicated amount (0.04 IU), while other closely related species were sensitive to it.
The profiles of the major glycolipids and phospholipids were similar to those of Halorubrum trueperi Y73T [4]. Strain Y78T contained sulfated diglycosyl diether (S-DGD-1) as the sole glycolipid and phosphatidylglycerol phosphate methyl ester (PGP-Me) and phosphatidylglycerol sulfate (PGS) as the major phospholipids (Fig. S2).
The sequences of rpoB’ gene (KX530069, 1830 bp) and ef-2 gene (KX530068, 1692 bp) were deposited in the GenBank. The relatives of strain Y78T deduced from the sequence similarity search of 16S rRNA gene were the same as the rpoB’ gene. The similarity search of rpoB’ gene sequence showed that strain Y78T was closely related to Halorubrum yunnanense Q85T, Halorubrum rutilum YJ-18-S1T, and Halorubrum saccharovorum JCM 8865T with the similarities of 96.5, 96.5, and 96.0%, respectively. However, the relatives deduced from the ef-2 gene sequence were different from 16S rRNA gene and rpoB’ gene. The similarity search of ef-2 gene sequence showed that strain Y78T was closely related Halorubrum rubrum YC87T, Halorubrum xinjiangense CGMCC 1.3527T, and Halorubrum trapanicum JCM 10477T with the similarities of 95.4, 95.4, and 95.3%.
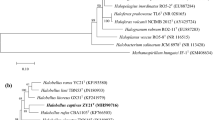
The phylogenetic tree based on the 16S rRNA gene reconstructed by the maximum-likelihood method confirmed that strain Y78T was closely related to Halorubrum rutilum YJ-18-S1T and Halorubrum yunnanense Q85T and that they formed a independent lineage (Fig. 1). Topologies of phylogenetic trees inferred by using the neighbor-joining were highly similar to that of the tree reconstructed by the maximum-likelihood method (Fig. S3).
Maximum-likelihood phylogenetic trees based on 16S rRNA gene (a), rpoB′ gene (b) sequences, showed the relationships between strain Y78T and other members of the genus Halorubrum and other related groups. Species Halococcus morrhuae JCM 8876T and Halobacterium salinarum JCM 8978T were taken as outgroups for the 16S rRNA gene and the rpoB’ gene tree. Bootstrap values (expressed as percentages of 1000 replications) greater than 50% are shown. GenBank accession numbers are shown in parentheses. Bar: 2% substitution
The calculated DNA G+C content of strain Y78T was 66.6 mol%. The DNA–DNA relatedness between strain Y78T and Halorubrum rutilum YJ-18-S1T was 27.3 ± 0.5%, while 38.6 ± 0.9% for strain Y78T and Halorubrum yunnanense Q85T, which are far below the threshold value (70%) for the separation of two different species [24].
The phylogeny based on the rpoB′ gene was assessed after recovering gene sequences from the GenBank database. Topology of phylogenetic tree inferred by rpoB′ gene was coincident with that inferred by the 16S rRNA gene (Fig. 1), which strengthens the phylogenetic position and relationship of strain Y78T. However the topology of phylogenetic tree deduced from the ef-2 gene was different from 16S rRNA gene and rpoB′ gene, which reflects the divergence of the evolutionary rate (data not shown). The similar results had happened in the Halorubrum species, Halorubrum yunnanense [2]. Horizontal gene transfer may attribute to the fact that different collinear genes share different evolutionary rates.
Other detailed results of the physiological tests are shown in the species description and Table 1.
Conclusion
A polyphasic approach including phylogenetic analyses using 16S rRNA and rpoB′ gene sequence comparisons (Fig. 1), polar lipid profiles (Fig. S2), DNA–DNA hybridization, and detailed phenotypic characterization (Table 1) confirms that the strain Y78T represents a novel species of the genus Halorubrum, for which the name Halorubrum depositum sp. nov. is proposed.
Description of Halorubrum depositum sp. nov
Halorubrum depositum (de.po′si.tum. L. neut. part. adj. depositum deposited).
Cells stain Gram-negative. Colonies on agar medium containing 20% (w/v) NaCl are 1–2 mm in diameter, translucent, red-pigmented (nearly scarlet), circular, slightly raised, and smooth. Cells are non-motile and pleomorphic rods or oval-shaped (approximately 0.6–0.8 × 0.8–1.2 μm). Cell lysed in distilled water. Chemo-organotrophic, aerobic growth occurs at 10–30% (w/v) NaCl, pH 6.5–9.0, and 25–50 °C. Optimum NaCl concentration, pH, and temperature for growth are 20% (w/v), pH 8.5, and 38 °C, respectively. Mg2+ is required for growth (at least 0.005 M) with an optimum at 0.3 M. They are catalase- and oxidase-positive. Anaerobic growth with nitrate, DMSO, or l-arginine does not occur. Nitrate is not reduced to nitrite. Indole is not produced. Gelatin is not liquefied. Starch, aesculin, Tween 60, Tween 40, and Tween 20 are not hydrolysed; however, Tween 80 is hydrolysed. The following substrates are utilized for growth as sole sources of carbon and energy: d-glucose, d-mannose, maltose, sucrose, glycerol, d-sorbitol, acetate, pyruvate, lactate, succinate, malate, fumarate, and citrate. The strain can produce acid from d-glucose, d-mannose, maltose, and sucrose. d-galactose, d-fructose, l-sorbose, d-ribose, d-xylose, lactose, starch, and d-mannitol are not used as sole sources of carbon and energy. The following amino acids are used as sole sources of carbon, nitrogen, and energy: l-arginine, l-glutamate, l-ornithine. Glycine, l-alanine, l-asparagine, and l-lysine are not used as sole sources of carbon, nitrogen, and energy. They are sensitive to rifampicin and novobiocin, and resistant to ampicillin, bacitracin, chloramphenicol, ciprofloxacin, erythromycin, kanamycin, neomycin, norfloxacin, penicillin G, streptomycin, tetracycline, and vancomycin. The major components of the polar lipids are sulfated diglycosyl diether, phosphatidylglycerol, phosphatidylglycerol phosphate methyl ester, and phosphatidylglycerol sulfate. The DNA G+C content is 66.6 mol% (Tm).
The type strain, Y78T (= CGMCC 1.15456T = JCM 31272T), was isolated from a salt deposit of Yunnan salt mine, China (99°46′42.05″E, 26°06′04.05″N).
References
Amoozegar MA, Siroosi M, Atashgahi S, Smidt H, Ventosa A (2017) Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology 163:623–645
Chen S, Liu HC, Zhao D, Yang J, Zhou J, Xiang H (2015) Halorubrum yunnanense sp. nov., isolated from a subterranean salt mine. Int J Syst Evol Microbiol 65:4526–4532
Chen S, Liu HC, Zhou J, Xiang H (2016) Halorubrum pallidum sp. nov., an extremely halophilic archaeon isolated from a subterranean rock salt. Int J Syst Evol Microbiol 66:2980–2986
Chen S, Xu Y, Ke LX (2017) Halorubrum trueperi sp. nov., a halophilic archaeon isolated from a salt mine. Int J Syst Evol Microbiol 67:1564–15705
Cui HL, Lin ZY, Dong Y, Zhou PJ, Liu SJ (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:2204–2206
Cui HL, Tohty D, Zhou PJ, Liu SJ (2006) Halorubrum lipolyticum sp. nov. and Halorubrum aidingense sp. nov., isolated from two salt lakes in Xin-Jiang, China. Int J Syst Evol Microbiol 56:1631–1634
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Dussault HP (1955)) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485
González C, Gutiérrez C, Ramírez C (1978) Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Han D, Cui HL (2015) Halorubrum laminariae sp. nov., isolated from the brine of salted brown alga Laminaria. Antonie Van Leeuwenhoek 107:217–223
Jaakkola ST, Ravantti JJ, Oksanen HM, Bamford DH (2016) Buried alive: microbes from ancient halite. Trends Microbiol 24:148–160
Kharroub K, Quesada T, Ferrer R, Fuentes S, Aguilera M, Boulahrouf A, Ramos-Cormenzana A, Monteoliva-Sánchez M (2006) Halorubrum ezzemoulense sp. nov., a halophilic archaeon isolated from Ezzemoul sabkha, Algeria. Int J Syst Evol Microbiol 56:1583–1588
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3:208–218
McGenity TJ, Grant WD (1995) Transfer of Halobacterium saccharovorum, Halobacterium sodomense, Halobacterium trapanicum NRC 34021 and Halobacterium lacusprofundi to the genus Halorubrum gen. nov. as Halorubrum saccharovorum comb. nov., Halorubrum sodomense comb. nov., Halorubrum trapanicum comb. nov., and Halorubrum lacusprofundi comb. nov. Syst Appl Microbiol 18:237–243
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B’ (rpoB’) gene. Int J Syst Evol Microbiol 60:2398–2408
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Oren A (2015) Halophilic microbial communities and their environments. Curr Opin Biotechnol 33:119–124
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238
Owen RJ, Hill LR (1979) The estimation of base compositions, base pairing and genome size of bacterial deoxyribonucleic acids. In: Skinner FA, Lovelock DW (eds) Identification methods for microbiologists, 2nd edn. Academic Press, London, pp 217–296
Schubert BA, Lowenstein TK, Timofeeff MN, Parker MA (2010) Halophilic Archaea cultured from ancient halite, Death Valley, California. Environ Microbiol 12:440–454
Smibert RM, Krieg NR (1981) General characterization. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 409–443
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Ventosa A, de la Haba RR, Sánchez-Porro C, Papke RT (2015) Microbial diversity of hypersaline environments: a metagenomic approach. Curr Opin Microbiol 25:80–87
Vreeland RH, Rosenzweig WD, Powers DW (2000) Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature 407:897–900
Wang YX, Liu JH, Xiao W, Ma XL, Lai YH, Li ZY, Ji KY, Wen ML, Cui XL (2013) Aliifodinibius roseus gen. nov., sp. nov., and Aliifodinibius sediminis sp. nov., two moderately halophilic bacteria isolated from salt mine samples. Int J Syst Evol Microbiol 63:2907–2913
Xiao W, Wang ZG, Wang YX, Schneegurt MA, Li ZY, Lai YH, Zhang SY, Wen ML, Cui XL (2013) Comparative molecular analysis of the prokaryotic diversity of two salt mine soils in southwest China. J Basic Microbiol 53:942–952
Yin S, Wang Z, Xu JQ, Xu WM, Yuan PP, Cui HL (2015) Halorubrum rutilum sp. nov. isolated from a marine solar saltern. Arch Microbiol 197:1159–116430
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Acknowledgements
We thank Prof. Zhu L. Yang from the Kunming Institute of Botany, CAS, for the help in sample collection, and thank Prof. Hua Xiang from the Institute of Microbiology, CAS, for technical assistance. This work was supported by grants from the National Natural Science Foundation of China (31460003), the Anhui Provincial Key Lab. of the Conservation and Exploitation of Biological Resources (591601), the Anhui Provincial Natural Science Research Project (KJ2017A318), and the Education Department of Anhui province (gxyqZD2017011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research Involving Animal Participants
This article does not contain any studies with animals performed by any of the authors.
Additional information
The GenBank/EMBL/DDBJ accession numbers of the 16S rRNA, rpoB′ and ef-2 sequences of strain Y78T are KX376712, KX530069, and KX530068, respectively. The protologue has been submitted to the Digital Protologue database (http://imedea.uib-csic.es/dprotologue/) under the Taxon Number TA00336.
Electronic Supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, S., Sun, S., Xu, Y. et al. Halorubrum depositum sp. nov., a Novel Halophilic Archaeon Isolated from a Salt Deposit. Curr Microbiol 75, 677–683 (2018). https://doi.org/10.1007/s00284-018-1432-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1432-1