Abstract
A Gram-reaction negative, aerobic, motile, non-pigmented and rod-shaped bacterium, designated as 168GH5-2-16T, was isolated from seawater Jeju island. Phylogenetic analysis based on 16S rRNA gene sequence comparison revealed that the strain formed a distinct lineage within the genus Vibrio and was most closely related to Vibrio variabilis R-40492T (96.0%). The DNA G+C content was 49.3 mol%. The major polar lipids were phosphatidylethanolamine (PE) and phosphatidylglycerol (PG). The predominant quinone was ubiquinone-8 (Q-8). The major fatty acids were C16:0, summed feature 3 (comprising C16:1 ω7c/C16:1 ω6c) and summed feature 8 (C18:1 ω7c/C18:1 ω6c) supported the affiliation of 168GH5-2-16T to the genus Vibrio. Moreover, the physiological, biochemical, and taxonomic analysis allowed the phenotypic and genotypic differentiation of strain 168GH5-2-16T from the recognized species of the genus Vibrio. Therefore, strain 168GH5-2-16T represents a novel species of the genus Vibrio, for which the name Vibrio hannami sp. nov. is proposed, with the type strain 168GH5-2-16T (=KACC 19277T = DSM105032T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Vibrio is on of the most diverse group of facultative anaerobes of the phylum Proteobacteria. Species of the genus Vibrio are commonly isolated from aquatic environments, usually from marine environments, both as free-living bacteria and as symbionts or parasites of fish, crustaceans, and molluscs [22]. Vibrios are Gram-negative, halophilic, usually motile rods, mesophilic and chemo-organotrophic, which have the facultatively fermentative metabolism [2]. Affiliates of the genus Vibrio obtained from shellfish and cultured fish around the world have provided more than 30% of the species currently recognized in the genus. Recently, based on multi-locus sequence analysis (MLSA) the species of genus Vibrio have been grouped in 23 clades [1]. At the time of writing, the genus Vibrio comprises more than 120 recognized species with validly published names (http://www.bacterio.net/vibrio.html).
In this study, we describe the taxonomic characterization of novel strain 168GH5-2-16T which belongs to the genus Vibrio.
Materials and Methods
Isolation of the Bacterial Strain
The water samples were collected from seawater (Goheung, South Korea), and after collection the samples were thoroughly suspended with 0.85% sterilized saline, following serial dilution, and then was spread onto marine agar medium (BD). The plates were incubated at 30 °C for 2 weeks. Single colonies were purified by subculture. Strain 168GH5-2-16T was seek out, and then it was routinely cultured on marine agar at 30 °C and maintained as a glycerol suspension (25%, v/v) at −80 °C.
Physiological, Morphological, and Biochemical Characteristics
The Gram-reaction was determined using the non-staining method, as described previously [3]. Cell motility was determined using the hanging drop method, while cell morphology was examined with the transmission electron microscope (SU-3500, Hitachi), using cells grown for 2 day at 30 °C on MA (Marine agar) medium. Oxidase activity was determined using 1% (w/v) N,N,N,N-tetramethyle-1,4-phenylenediamine reagent (bioMe′rieux). Catalase activity was determined by the production of bubbles from 3% (v/v) H2O2 solution. Hydrolysis activity was tested using the following substrates: starch, casein, DNase (DNase agar medium, Sharlau), Tween 80, and carboxyl methyl cellulose (CMC) [20]. All tests were performed and evaluated after 2 days of incubation at 30 °C. Biochemical tests in the commercial API kits [API ZYM, API 20NE, and API 32GN (bioMerieux)] were generally performed according to the manufacturer’s instructions. The API ZYM test strips were read after 4 h of incubation at 37 °C, and the other API streps were examined after 2 days of incubation at 30 °C. Growth at different temperatures (4, 10, 15, 25, 30, 37, and 40 °C) and various pH values (pH 4–10 at intervals of 0.5 and 1 pH units) was assessed after 3 days of incubation at 30 °C. The following buffers (final concentration, 50 mM) were used to adjust the pH of nutrient broth. Acetate buffer was used for pH 4.0–5.5; phosphate buffer was used for pH 6.0–8.0; and Tris buffer was used for pH 8.5–10.0. Salt tolerance test was evaluated on marine agar medium supplemented with 2–8% (w/v at intervals of 0.5 and 1% unit) NaCl and growth was assessed after 7 days of incubation at 30 °C. Growth on different media was also tested by using nutrient agar (NA, Difco), trypticase soy agar (TSA, Difco), lysogeny broth (LB) agar (Difco), and MacConkey agar (Difco) at 30 °C for 1 week.
Phylogenetic Tree Construction and Determination of DNA G+C Content (mol%)
For phylogenetic analysis, the genomic DNA was extracted with a commercial genomic DNA extraction kit (Solgent). The bacterial universal primer sets 27F, 800R, 518F, and 1492R were used to amplify the 16S rRNA gene sequence [13]. The purified PCR product was sequenced by Genotech according to Kim et al. [10]. Nearly full-length sequence of the 16S rRNA gene was compiled using SeqMan software (DNASTAR). The 16S rRNA gene sequences of related taxa were obtained from GenBank database (http://www.ncbi.nlm.nih.gov/genbank) or http://www.ezbiocloud.net/eztaxon, [11]. Multiple sequence alignments were performed via the Clustal_X program [23]. Gaps were edited in the BioEdit program [8] and 1370 nucleotides were used for phylogenetic tree construction. For maximum-likelihood tree analysis, evolutionary distances were calculated using Kimura two-parameter model [12] and the gapes were treated by complete deletion. The neighbor-joining tree was constructed by using the same model with complete deletion of gaps [18]. Similarly, maximum-parsimony tree was made with Subtree–Pruning–Regrafting (SPR) heuristic method and the gaps were edited with complete deletion [7] using MEGA6 program [21] with bootstrap values based on 1000 replications [6].
To analyze the DNA G+C content, the genomic DNA was extracted and purified as previously described [17], degraded enzymatically into nucleosides, and the DNA G+C content was determined as described by Mesbah et al. [15] using a reverse-phase HPLC.
Chemotaxonomic Analysis
The novel isolate was examined for their polar lipid contents as described by Minnikin et al. [16] and the polar lipids were developed in the first direction by using the chloroform/methanol/water (65:25:4, by v/v), while in the second direction, it was developed by chloroform/acetic acid/methanol/water (80:15:12:4, by v/v). Isoprenoid quinone of the isolate was extracted with chloroform/methanol (2:1, v/v), evaporated under vacuum condition, and re-extracted in n-hexane/water (1:1, v/v). The crude n-hexane-quinone solution was purified using Sep-Pak Vac cartridges silica (Waters) and subsequently analyzed by HPLC as previously described [9]. Cellular fatty acid profiles were determined after 48 h of growth at 30 °C on R2A agar medium. The cellular fatty acids were saponified, methylated, and extracted according to the described method of Sherlock Microbial Identification System (MIDI). The fatty acids analyzed by a gas chromatograph (Hewlett Packard 6890) were identified by the Microbial Identification software package based on Sherlock Aerobic Bacterial Database (TSBA60) [19].
Results and Discussion
Morphological and Phenotypic Characteristics
Cells of strain 168GH5-2-16T were Gram-reaction-negative, aerobic, motile, and rod-shaped Fig. S1. Colonies of strain 168GH5-2-16T grown on marine agar were circular, convex, opaque, and non-pigmented after 48 h of incubation at 30 °C. The isolate did not grow on LB (BD), NA (BD), and MacConkey agar (Difco), whereas weakly grew on R2A (BD) and TSA (Difco) at 30 °C. Strain 168GH5-2-16T negative for the hydrolysis of starch, casein, cellulose, and DNase. Furthermore, physiological characteristics of strain 168GH5-2-16T is summarized in the species description and a comparison of selective characteristics of the isolated strain and related type strain is given in Table 1.
Phylogenetic and DNA G+C Content Analysis
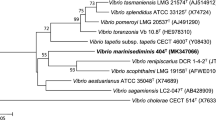
The almost complete 16S rRNA gene sequence of strain 168GH5-2-16T (1,427 nt) was determined and subjected to comparative analysis. Phylogenetic analysis using the maximum-likelihood method based on 16S rRNA gene sequences indicated that strain clustered within the genus Vibrio (Fig. 1) and form a monophyletic clad with Vibrio aestivus KCTC 23860T. Moreover, this relationship was also evident in phylogenetic trees based on the neighbor-joining and maximum-parsimony methods. Vibrio variabilis LMG 25438T showed the highest sequence similarity (96%) to the new isolate.
Phylogenetic relationship between strain 168GH5-2-16T and other related species of the genus Vibrio. The tree was constructed using the maximum-likelihood method based on 16S rRNA gene sequences. Bootstrap values (expressed as percentages of 1000 replications) greater than 60% are shown at branch points. Filled circles indicate that the corresponding nodes were also recovered in the tree generated with maximum-parsimony and neighbor-joining algorithms. Photobacterium aqua AE6T (JQ948040) was used as an outgroup. Scale bar, 0.005 substitutions per nucleotide position
Based on 16S rRNA gene sequence and phylogenetic trees analysis, Vibrio variabilis LMG25438T, Vibrio aestivus KCTC 23860T, and Vibrio maritimus LMG 25439T were selected as the closest recognized neighbor of strain 168GH5-2-16T and used as reference strains in most of the subsequent phenotypic analysis.
DNA G+C contents of the strain 168GH5-2-16T were 49.3 mol%, which was similar to those of the described species of the genus Vibrio (Table 1).
Chemotaxonomic Characteristics
The main polar lipids of strain 168GH5-2-16T were phosphatidylethanolamine (PE) and phosphatidylglycerol (PG); the minor polar lipids were two unknown polar lipids (L1, L2), one unknown phospholipid (PL) three unknown aminolipids (AL1–AL3) and one unknown amino phospholipid (APL) (Fig. S2). Based on the polar lipid analysis, strain 168GH5-2-16T share similar major polar lipid PE and PG with the recently described species of the genus Vibrio [5]. The major respiratory quinone was Q-8. The major fatty acids of strain 168GH5-2-16T were C16:0 (11.7%), summed feature 3 [(comprising C16:1 ω7c and/or C16:1 ω6c) 37.4%] and summed feature 8 [(comprising C18:1 ω7c/C18:1 ω6c) 28.6%], which is a typical profile of members of the genus Vibrio. However, some qualitative and quantitative differences in the fatty acids distinguished strain 168GH5-2-16T from the closely related strains Vibrio aestivus KCTC 23860T, Vibrio variabilis LMG 25438T, and Vibrio maritimus LMG 25439T (Table 2).
Taxonomic Conclusions
In summary, the characteristics of strain 168GH5-2-16T are consistent with descriptions of the genus Vibrio with regard to morphological, biochemical, and chemotaxonomic properties. However, on the basis of phylogenetic distance from known Vibrio species indicated by 16S rRNA gene sequence similarities and the combination of unique phenotypic characteristics (Table 1), strain 168GH5-2-16T represents a novel species, for which the name Vibrio hannami sp. nov. is proposed.
Description of Vibrio hannami sp. nov.
Vibrio hannami (han.nam’i. N.L. masc. gen. n. hannami of Hannam University Republic of Korea, where the strain was taxonomically characterized).
The novel isolate is positive for catalase and oxidase activities. Growth occurs at 20–42 °C and pH 6.0–10.0 with 1.0–8.0% NaCl (w/v). Optimum growth occurs at 25 °C and pH 6.0 with 2.0% NaCl (w/v) supplement. In the API kits (API 20NE and 32GN API ZYM) system, positive for alkaline phosphatase, esterase, esterase lipase, leucine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-glucosidase, indole production, glucose acidification, arginine dihydrolase, β-glucosidase, β-galactosidase, d-glucose, l-arabinose, N-acetyl-glucosamine, d-maltose, gluconate, adipate, malate, citrate, d-glucose, salicin, d-melibiose, l-fucose, d-sorbitol, l-arabinose, propionate, citrate, l-histidine, 2-ketogluconate, 4-hydroxy-benzoate, l-proline, l-rhamnose, N-acetyl-glucosamine, d-ribose, inositol, d-sucrose, d-maltose, itaconate, malonate, acetate, l-alanine, 5-ketogluconate, and l-serine. List of all negative traits of commercial kits is shown in Table S1. Ubiquinone Q-8 is the predominant respiratory quinone and C16:0, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c) and summed feature 8 (C18:1 ω7c/C18:1 ω6c) are the major cellular fatty acids. The major polar lipids were PE and PG. The DNA of the type strain is 49.3 mol%.
The type strain, isolated from seawater, Jeju Island, South Korea, Republic of Korea, is 168GH5-2-16T (=KACC 19277T = DSM105032T).
References
Al-Saari N, Gao F, Rohul AA, Sato K, Sato K, Sato K, Mino S, Suda W, Oshima K, Hattori M, Ohkuma M et al (2015) Advanced microbial taxonomy combined with genome-based-approaches reveals that Vibrio astriarenae sp. nov., an Agarolytic Marine Bacterium, Forms a New Clade in Vibrionaceae. PLoS ONE 10(8):e0136279
Baumann P, Furniss AL, Lee JV (1984) Genus I. Vibrio Pacini 1854. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology. Williams & Wilkins, Baltimore, pp 518–538
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Chimetto LA, Cleenwerck I, Moreira APB, Brocchi M, Willems A, De Vos P (2001) Thompson FL Vibrio variabilis sp. nov. and Vibrio maritimus sp. nov., isolated from Palythoa caribaeorum. Int J Syst Evol Microbiol 61:3009–3015
Doi H, Chinen A, Fukuda H, Usuda Y (2016) Vibrio algivorus sp. nov., an alginate- and agarose-assimilating bacterium isolated from the gut flora of a turban shell marine snail. Int J Syst Bacteriol 66:3164–3169
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution Int J org Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: Minimum change for a specified tree topology. Syst Zool 20:406–416
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Kim JK, Kang MS, Park SC, Kim KM, Choi K, Yoon MH, Im WT (2015) Sphingosinicella ginsenosidimutans sp. nov., with ginsenoside converting activity. J Microbiol 53:435–441
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi Het al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press Cambridge, New York, Cambridge
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt EM (ed) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–176
Lucena T, Ruvira MA, Arahal DR, Macián MC, Pujalte MJ (2012)) Vibrio aestivus sp. nov. and Vibrio quintilis sp. nov., related to Marisflavi and Gazogenes clades, respectively. Int J Syst Syst Appl Microbiol 35:427–431
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Moore DD, Dowhan D (1995) Preparation and analysis of DNA. In: Ausubel FW, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Wiley, New York, pp 2–11
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc, Newark
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, pp 607–655
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson FL, Iida T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol Rev 68:403–431
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Acknowledgements
This research was supported by a grant from the Marine Biotechnology Program (20170431) funded by the Ministry of Oceans and Fisheries, Korea, and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A3B03933067), Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
The digital protologue database (DPD) number of Current Microbiology for the strain 168GH5-2-16T is TA00219.
The GenBank accession number for the 16S rRNA gene sequence of strain 168GH5-2-16T is KY451770.
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2017_1376_MOESM2_ESM.pptx
Fig. S2. Two-dimensional TLC of the total polar lipids of strain 168GH5-2-16T. The TLC plate was stained for total polar lipids with 5 % ethanolic molybdophosphoric acid. Abbreviations: PE, Phosphatidylethanolamine; PG, phosphatidylglycerol; PL, unknown phospholipid; APL, unknown aminophospholipids; ALs, unknown aminolipids and Ls, unknown polar lipids. (PPTX 1202 KB)
Rights and permissions
About this article
Cite this article
Lee, GE., Im, WT. & Park, JS. Vibrio hannami sp. nov., Isolated from Seawater. Curr Microbiol 75, 278–283 (2018). https://doi.org/10.1007/s00284-017-1376-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1376-x