Abstract
Helicobacter pylori infection plays an important role in the etiology of various gastroduodenal diseases. However, pathogenic mechanism of H. pylori is not clear. More potential pathogenic factors need to be further discovered and studied. In this study, two vectors for generating double-crossover recombination gene knockout plasmids in H. pylori were designed based on the backbone of plasmid pLYL03. Genes on plasmid pLYL03 were rearranged, and the redundant sequences were reduced. Erythromycin-resistant gene on pLYL03 was replaced by aphA or catGC to generate the kanamycin-resistant plasmid pSJHK or the chloramphenicol-resistant plasmid pSJHC. The sizes of pSJHK and pSJHC are 4371 and 3949 bp, respectively. Based on plasmids pSJHK and pSJHC, double-crossover recombination gene knockout plasmids targeting hp0169 and hp0788 were constructed, and deletion mutants were achieved by electroporation of the gene-targeting plasmids. The results indicated that plasmids pSJHK and pSJHC are efficient for gene deletion in H. pylori, and the transformation efficiency of pSJHC is slightly higher than pSJHK. These plasmids provide convenient genetic tools for further research of novel pathogenic factors in H. pylori. Derivative plasmids developed by changing antibiotic-resistant genes would also provide valuable tools for the study of functional genes in other bacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Helicobacter pylori is recognized as a pathogen that chronically infects more than half of the world’s population. H. pylori plays an important role in the development of peptic ulcers, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue lymphoma (MALToma) [14, 17, 22]. However, only approximately 20 % of infected individuals developed severe disease [20]. The mechanism of different outcomes of an infection is not clear. It is thought to involve an interplay between the virulence of the infecting strain, host genetics, and environmental factors [4, 23, 26].
Many virulence factors of H. pylori have been reported to play important roles in pathogenesis, which are generally classified into two categories. One category contains flagellum [15], urease [13], and outer-membrane proteins (oipA, sabA, sabB, babB, babC, and hopZ) [8, 21, 28], which are thought to participate in the adhesion and colonization (first step in pathogenesis). The other category contains vacA [1], CagA [2, 3, 6], and peptidoglycan [10], which directly take part in pathogenesis. The studies of these pathogenic virulences enrich the understanding of the H. pylori pathogenesis; however, the mechanism of different outcomes of an infection has not been solved.
Experiences on other bacterial pathogens studies suggest that H. pylori-specific factors may exist and influence the pathogenicity of H. pylori. Several novel pathogenic factors have been reported to be associated with increased risks of special clinical outcome, involving duodenal ulcers and gastric cancers [11, 12, 15]. In order to better understand the pathogenic mechanism and explain the different outcomes of an infection in detail, more potential pathogenic factors need to be further discovered and studied.
Construction of gene deletion mutant is an effective method for gene functional analysis, among which double-crossover recombination was the common method to construct the gene deletion. As the establishment of genetic manipulation, gene knockout was widely used in the studies of various virulence genes of H. pylori [5, 13, 15, 25]. Plasmid pBluescript II SK was usually used in generation double-crossover recombination gene knockout cassette in knockout studies [27], and PCR products-based homologous recombination has also been used to create gene knockout in H. pylori [24]. However, the construction of specific gene knockout cassette was usually time-consuming. In this study, two vectors (pSJHK, pSJHC) for generating double-crossover recombination gene knockout plasmids in H. pylori were established. Based on these vectors, the construction of gene-targeting plasmid was convenient, and gene deletion in H. pylori was easily achieved. The construction of these plasmids was valuable for further research of novel pathogenic factors in H. pylori.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains, plasmids, and primers used in this study are listed in Table 1 and Table S1. Helicobacter strains were cultivated under microaerobic conditions (85 % N2, 10 % CO2, 5 % O2) at 37 °C as described previously [9]. For selection of H. pylori mutant strains carrying the catGC resistance marker or the aphA cassette, serum plates were supplemented with chloramphenicol (10 mg/l) or kanamycin (15 mg/l). All cloning procedures were performed in Escherichia coli, and E. coli strains were grown at 37 °C in Luria-Bertani medium supplemented with chloramphenicol (30 mg/l) or ampicillin (100 mg/l) when needed.
Construction of the Double-Crossover Template Plasmids pSJHK and pSJHC
Plasmid pLYL03, which could not replicate in H. pylori, was used as the backbone for generating double-crossover recombination template plasmid. Plasmid pLYL03 was digested with ScaI and EcoRI, and the 2.5-kbp fragment containing oriT and a partial bla gene was collected. The remaining portion of bla was amplified using primers bla-1 and bla-2, digested with ScaI and EcoRI, and then ligated with the 2.5-kbp fragment, yielding plasmid pSJH. The DNA fragment containing erythromycin-resistant gene (ermf) was amplified from pLYL03 by PCR with primers ermf-1 and ermf-2, digested with BamHI and SphI, and inserted into the corresponding sites of pSJH to create pSJHE. Meanwhile, several restriction sites (multiple cloning site) were designed on the flanks of ermf for insertion of the homologous arms.
Kanamycin-resistant gene (aphA) amplified from pHimarEm1 by PCR with primers km-1 and km-2 was digested with KpnI and SacI. The digested fragment replaced ermf on pSJHE to generate kanamycin-resistant plasmid pSJHK. Chloramphenicol-resistant plasmid pSJHC was constructed from the same method, and chloramphenicol-resistant gene (catGC) was amplified from pTnMax9 by PCR with primers cm-1 and cm-2.
Construction of Gene-Targeting Plasmids pSJHK-1069, pSJHC-0169, pSJHK-0788, and pSJHC-0788
A 844-bp fragment spanning the last 69 bp of hp0169 and downstream sequence was amplified with primers 0169-3 and 0169-4, which was used as the downstream homologous arm for double-crossover recombination. The fragment was ligated into the corresponding sites of pSJHK or pSJHC after digested by SacI and XbaI. Then, upstream homologous arm (a 898-bp fragment spanning the first 130 bp of hp0169 and upstream sequence) amplified with primers 0169-1 and 0169-2 was also inserted into plasmids, yielding plasmids pSJHK-0169 or pSJHC-0169, respectively.
Plasmids pSJHK-0788 and pSJHC-0788 were constructed with the same procedure. Upstream homologous arm was amplified with primers 0788-1 and 0788-2, and downstream homologous arm was amplified with primers 0788-3 and 0788-4.
Electrotransformation of H. pylori cells
Helicobacter pylori cells were transformed with plasmids (pSJHK-1069, pSJHC-0169, pSJHK-0788, or pSJHC-0788) by electroporation with the procedure similar to that described by Clayton et al. [17]. Briefly, H. pylori cultured on plates was scraped and suspended in 20 ml ice-cold double-distilled water. After harvested by centrifugation at 4360×g for 5 min, the pellet was washed using 20 ml ice-cold 10 % glycerol twice and resuspended in 1 ml 10 % glycerol. Plasmid DNA (1 μg in 5 μl TE buffer) was mixed with 0.2-ml cell suspension. The mixture was added to a cooled 0.2-cm electroporation cuvette (Bio-Rad), and subjected to single-pulse electroporation (2.5 kV, 25 mF, 200 Ω). The sample was then transferred onto a serum plate and incubated for 24 h at 37 °C. Finally, the cells were inoculated onto selective media with 15 mg/ml kanamycin or 10 mg/ml chloramphenicol for 5 days to allow the growth of transformants.
Identification of Gene Deletion Mutant
Total DNA of the transformant was isolated using a bacterial DNA kit (Omega), which was used as the template to do diagnostic PCR. For kanamycin-resistant transformants in the disruption of hp0169, primers located upstream of 0169-1 (0169test-1) and inside of aphA (km-test-2) were designed to analyze if homologous recombination had occurred in the upstream homologous arm. Primers located inside of aphA (km-test-3) and downstream of 0169-4 (0169test-4) were designed to analyze if homologous recombination had occurred in downstream homologous arm. If homologous recombination had occurred both in upstream and downstream homologous arms, gene deletion of hp0169 could be confirmed. Wild-type strain was used as the negative control.
To analyze the gene deletion of hp0788, primers design and diagnostic PCR were carried out with the same method. For chloramphenicol-resistant transformants, primers cm-test-2 and cm-test-3 were designed inside of catGC. All the primers are shown in Table S1.
Results
Construction of the Template Plasmids pSJHK and pSJHC for Gene Deletion in H. pylori
Plasmid pLYL03 could not be directly used in knockout studies in H. pylori because of the improper antibiotic marker. The derivative plasmids pSJHK and pSJHC suitable for gene knockout in H. pylori were generated based on the backbone of pLYL03. The modification was made as previously described. First, genes on pLYL03 plasmid were rearranged, and the redundant sequences were reduced to generate pSJHE, which includes erythromycin-resistant gene and multiple cloning sites on the flanks of antibiotic-resistant gene. Then, erythromycin-resistant gene on pSJHE was replaced by aphA or catGC, yielding template plasmids pSJHK (kanamycin-resistant) and pSJHC (chloramphenicol-resistant), respectively. The sizes of pSJHK and pSJHC are 4371 and 3949 bp, respectively. The construction process of the plasmids is illustrated in Fig. 1. The upstream and downstream homologous arms would be inserted into multiple cloning sites when generating the double-crossover gene-targeting plasmid.
Gene Targeting in H. pylori
Genes hp0169 and hp0788 were selected as targeted genes to test the efficiency of the template plasmids pSJHK and pSJHC. Double-crossover recombination gene knockout plasmids (pSJHK-1069, pSJHC-0169, pSJHK-0788, pSJHC-0788) were constructed according to the procedure described in “Materials and Methods” section. Schematic figures of pSJHK-1069 and pSJHC-0169 are shown in Fig. 2. These plasmids containing the double-crossover cassettes were transformed into H. pylori cells by electroporation.
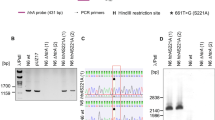
The positive antibiotic-resistant transformants of ∆hp0169km were subjected to PCR assay with the diagnostic primers 0169test-1 and km-test-2. As shown in Fig. 3, an amplicon of 1.68 kbp for the mutant strain was absent from the wild-type strain, which suggested that homologous recombination had occurred in the upstream homologous arm. An amplicon of 1.53 kbp from diagnostic PCR with primers km-test-3 and 0169test-4 was also only for the mutant indicated that homologous recombination had occurred in the downstream homologous arm. These results suggested that homologous recombination replacement had occurred at the hp0169 locus. Finally, PCR products from the amplification with primers 0169-1 and 0169-4 were sequenced for further verification of the disruption.
Schematic representation of the deletion of hp0169 and verification of genetic recombination by PCR. a Schematic representation of the deletion of hp0169. Two homologous arms of hp0169 (H1 and H2) were amplified from the genome of H. pylori with two sets of primers 0169-1/0169-2 and 0169-3/0169-4, digested, and ligated into the template plasmid pSJHK. The gene-targeting plasmid was transformed into H. pylori by electroporation, and transformants were selected by kanamycin resistance. Black arrows show approximate locations and orientations of primers; black-filled boxes indicate homologous arms; open arrowheads show arrangements and orientations of genes; open boxes indicate residual genes. b Verification of genetic recombination by PCR. Lane M DNA molecular weight standard (DL10000), Lane 1 PCR products from WT using primers 0169test-1 and km-test-2, Lane 2 PCR products from mutant using primers 0169test-1 and km-test-2, Lane 3 PCR products from WT using primers km-test-3 and 0169test-4, Lane 4 PCR products from mutant using primers km-test-3 and 0169test-4
The results of diagnostic PCR assay for other mutants were not shown.
The Efficiency Detection of Plasmids pSJHK and pSJHC for Gene Deletion in H. pylori
Usually, electroporation of the gene-targeting plasmid would lead to both single-crossover and double-crossover gene deletions. In this study, we tested 160 positive antibiotic-resistant transformants in the disruption of genes hp0169 and hp0788. For the disruption of gene hp0169, there were 84 double-crossover transformants and 6 single-crossover transformants in 90 tested transformants, and for the disruption of gene hp0788, there were 65 double-crossover transformants and 5 single-crossover transformants in 70 tested transformants. Almost all the transformants were double-crossover gene deletions. Therefore, in calculation of the efficiency of plasmids pSJHK and pSJHC for gene deletion in H. pylori, the amount of positive antibiotic-resistant transformants was regarded as double-crossover gene deletions.
For the same gene with the same homologous arms, the efficiency to generate the transformants was not same between pSJHK and pSJHC. For plasmids pSJHK-0169 and pSJHK-0788, the efficiency is 0.6 × 102 and 0.9 × 102 transformants per microgram plasmid DNA, respectively. For the plasmids pSJHC-0169 and pSJHC-0788, the efficiency is 1.3 × 102 and 1.5 × 102 transformants per microgram plasmid DNA, respectively. Therefore, the efficiency of plasmids pSJHC is a little higher than pSJHK in gene deletion in H. pylori (Fig. 4).
We found that the transformation of 1 μg plasmid with 800-bp homologous sequence for each arm to approximately 109 electrocompetent cells was sufficient for obtaining transformants. In fact, 400 bp of homologous sequence for each arm was adequate for recombination after trying different fragment sizes of homologous arms (data not shown).
Discussion
Although many virulence genes of H. pylori have been deeply studied, what determines the different outcomes of an infection was still an unsolved mystery. Studies on other bacterial pathogens suggested that H. pylori-specific factors may exist and influence the pathogenicity of H. pylori. Several novel pathogenic properties associated with increased risks of special clinical outcomes were reported recently that confirmed this speculation. The jhp0947 gene was reported to be associated with an increased risk of both duodenal ulcers and gastric cancers [11], while the dupA gene was reported to be associated with an increased risk of duodenal ulcers and protection against gastric cancers [12]. Therefore, more potential pathogenic genes need to be discovered for better understanding of the pathogenic mechanism and illustrating the different outcomes of the infection by H. pylori.
Plasmid pLYL03 could not be directly used for knockout in H. pylori because of the improper antibiotic marker. The derivative plasmids pSJHK and pSJHC in this study were generated based on the backbone of pLYL03 and carrying kanamycin-resistant gene (aphA) and chloramphenicol-resistant gene (catGC), respectively, which are suitable for gene knockout in H. pylori. Compared to pLYL03, the sizes of pSJHK and pSJHC are 4371 and 3949 bp, respectively (smaller than pLYL03, 5998 bp). Compared with other gene knockout plasmids for H. pylori, like pBluescript II SK, pSJHK and pSJHC provide two optional antibiotic markers, which would improve selectivity in generation of gene knockout cassette. Moreover, multiple cloning sites were designed on the flanks of antibiotic-resistant gene in pSJHK and pSJHC, which was more beneficial to insert the upstream and downstream homologous arms when generating the double-crossover gene-targeting plasmid.
Based on these plasmids, we have disrupted genes hp0169, hp0788, and several other genes (data not shown), indicating that these plasmids were efficient tools for gene deletion in H. pylori. The efficiency of plasmids pSJHC is a little higher than pSJHK in gene deletion in H. pylori with unknown reason. We speculated that it may be related to the different efficiency of promoters before the antibiotic-resistant genes. Anyway, plasmids pSJHK and pSJHC provide convenient genetic tools for further research of novel pathogenic factors in H. pylori.
By replacement of the antibiotic-resistant genes, several plasmids with different antibiotic resistance could be obtained, such as pSJHE (erythromycin resistance, mentioned in the methods and results), pSJHT (tetracycline resistance), and pSJHCF (cefoxitin resistance). Plasmids pSJHT and pSJHCF were not shown in this paper. These derivative plasmids will also provide effective tools in gene deletion in other strains and be valuable for the study of their functional genes.
Both hp0169 and hp0788 genes were reported to be essential for H. pylori to colonize in the gerbil stomach by Kavermann [18]. Gene hp0169 encodes a putative collagenase (HP0169, PrtC), which is noted as a protease in U32 protein family by sequence homology. Most proteases in this family are identified as pathogenic factors in destroying host tissues. Therefore, HP0169 is speculated to be associated with gastric or duodena tissue injury and delay in the healing process. Gene hp0788 encodes an outer-membrane protein (HP0788), which might have a function of adhesion, just as AlpAB, BabA, HopZ, and other outer-membrane proteins described before. Otherwise, it might play an essential role in stabilizing the integrity of the outer membrane to resist the harsh conditions in the gastric mucus. However, the detail functions of hp0169 and hp0788 are not clear, and gene deletion strains developed in this study would be used in the further study to clarify the pathogenic mechanisms of these genes.
References
Atherton JC, Cao P, Peek RM Jr, Tummuru MK, Blaser MJ, Cover TL (1995) Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem 270(30):17771–17777
Backert S, Clyne M, Tegtmeyer N (2011) Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori. Cell Commun Signal 9:28
Backert S, Ziska E, Brinkmann V, Zimny-Arndt U, Fauconnier A, Jungblut PR, Naumann M, Meyer TF (2000) Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell Microbiol 2(2):155–164
Baldwin DN, Shepherd B, Kraemer P, Hall MK, Sycuro LK, Pinto-Santini DM, Salama NR (2007) Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun 75(2):1005–1016
Bauerfeind P, Garner RM, Mobley LT (1996) Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun 64(7):2877–2880
Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A (1995) Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res 55(10):2111–2115
Braun TF, Khubbar MK, Saffarini DA, McBride MJ (2005) Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187(20):6943–6952
Colbeck JC, Hansen LM, Fong JM, Solnick JV (2006) Genotypic profile of the outer membrane proteins BabA and BabB in clinical isolates of Helicobacter pylori. Infect Immun 74(7):4375–4378
Copass M, Grandi G, Rappuoli R (1997) Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect Immun 65(5):1949–1952
de Bernard M, Josenhans C (2014) Pathogenesis of Helicobacter pylori infection. Helicobacter 19(Suppl 1):11–18
de Jonge R, Kuipers EJ, Langeveld SC, Loffeld RJ, Stoof J, van Vliet AH, Kusters JG (2004) The Helicobacter pylori plasticity region locus jhp0947-jhp0949 is associated with duodenal ulcer disease and interleukin-12 production in monocyte cells. FEMS Immunol Med Microbiol 41(2):161–167
Douraghi M, Mohammadi M, Oghalaie A, Abdirad A, Mohagheghi MA, Hosseini ME, Zeraati H, Ghasemi A, Esmaieli M, Mohajerani N (2008) dupA as a risk determinant in Helicobacter pylori infection. J Med Microbiol 57(Pt 5):554–562
Ferrero RL, Cussac V, Courcoux P, Labigne A (1992) Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol 174(13):4212–4217
Forman D (1998) Helicobacter pylori infection and cancer. Br Med Bull 54(1):71–78
Haas R, Meyer TF, van Putten JP (1993) A flagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol Microbiol 8(4):753–760
Kahrs AF, Odenbreit S, Schmitt W, Heuermann D, Meyer TF, Haas R (1995) An improved TnMax mini-transposon system suitable for sequencing, shuttle mutagenesis and gene fusions. Gene 167(1–2):53–57
Kato S, Onda M, Matsukura N, Tokunaga A, Matsuda N, Yamashita K, Shields PG (1997) Helicobacter pylori infection and genetic polymorphisms for cancer-related genes in gastric carcinogenesis. Biomed Pharmacother 51(4):145–149
Kavermann H, Burns BP, Angermuller K, Odenbreit S, Fischer W, Melchers K, Haas R (2003) Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Exp Med 197(7):813–822
Li LY, Shoemaker NB, Salyers AA (1995) Location and characteristics of the transfer region of a bacteroides conjugative transposon and regulation of transfer genes. J Bacteriol 177(17):4992–4999
JAMA (1994) NIH Consenus Conference. Helicobacter pylori in peptic ulcer disease. NIH consenus development panel on Helicobacter pylori in peptic ulcer disease. JAMA 272(1):65–69
Oleastro M, Menard A (2013) The role of Helicobacter pylori outer membrane proteins in adherence and pathogenesis. Biology 2(3):1110–1134
Peek RM Jr, Blaser MJ (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2(1):28–37
Posselt G, Backert S, Wessler S (2013) The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Commun Signal 11:77
Pyndiah S, Menard A, Zerbib F, Megraud F (2005) Evaluation of the homologous recombination in Helicobacter pylori. Helicobacter 10(3):185–192
Schmitt W, Odenbreit S, Heuermann D, Haas R (1995) Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol Gen Genet 248(5):563–572
Yamaoka Y (2008) Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. J Med Microbiol 57(Pt 5):545–553
Yuan JP, Li T, Shi XD, Hu BY, Yang GZ, Tong SQ, Guo XK (2003) Deletion of Helicobacter pylori vacuolating cytotoxin gene by introduction of directed mutagenesis. World J Gastroenterol 9(10):2251–2257
Zhang J, Qian J, Zhang X, Zou Q (2014) Outer membrane inflammatory protein A, a new virulence factor involved in the pathogenesis of Helicobacter pylori. Mol Biol Rep 41(12):7807–7814
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81471561 and 81501718), the Natural Science Foundation of Shandong Province (ZR2015PC021 and ZR2014CP020), and the Scientific Research Starting Foundation of Binzhou Medical University (BY2014KYQD10 and BY2014iKYQD11). We sincerely thank Professor Lu (Shandong University, China) for providing plasmids pLYL03 and pHimarEm1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors have no conflict of interests.
Additional information
Xiaofei Ji and Huilin Zhao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ji, X., Zhao, H., Zhang, Y. et al. Construction of Novel Plasmid Vectors for Gene Knockout in Helicobacter pylori . Curr Microbiol 73, 897–903 (2016). https://doi.org/10.1007/s00284-016-1140-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1140-7