Abstract
Our understanding of MM genomics has expanded rapidly in the past 5–10 years as a consequence of cytogenetic analyses obtained in routine clinical practice as well as the ability to perform whole-exome/genome sequencing and gene expression profiling on large patient data sets. This knowledge has offered new insights into disease biology and is increasingly defining high-risk genomic patterns. In this manuscript, we present a thorough review of our current knowledge of MM genomics. The epidemiology and biology of chromosomal abnormalities including both copy number abnormalities and chromosomal translocation are described in full with a focus on those most clinically impactful such as 1q amplification and del(17p) as well as certain chromosome 14 translocations. A review of our ever-expanding knowledge of genetic mutations derived from recent whole-genome/exome data sets is then reviewed including those that drive disease pathogenesis from precursor states as well as those that may impact clinical outcomes. We then transition and attempt to elucidate how both chromosomal abnormalities and gene mutations are evolving our understanding of disease risk. We conclude by offering our perspectives moving forward as to how we might apply whole-genome/exome-level data in addition to routine cytogenetic analyses to improve patient outcomes as well as further knowledge gaps that must be addressed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple myeloma (MM) is an acquired malignant plasma cell disorder that typically develops late in life with a median age at diagnosis of 69 years. Although it is a rare disorder accounting for just 1.8% of all new cancers in the USA and a lifetime risk of just 0.76%, it is the second commonest hematological malignancy [1]. With the advent of new therapeutics and the increasing utilization of high-dose melphalan and autologous stem cell transplantation (ASCT) over the last 20 years [2], 5- and 10-year overall survivals (OS) have improved across all age, race, and ethnic groups [3]. These benefits are more tempered in those with high-risk disease with revised international staging system (R-ISS) stage III [4] patients achieving only a 24% 5-year progression-free survival (PFS) and 40% 5-year overall survival (OS). MM remains incurable with only 10–15% of MM patients achieving or exceeding expected survival as compared with the matched general population.
Our understanding of MM genomics has expanded rapidly in the past 5–10 years. This is a consequence of cytogenetic analyses obtained in routine clinical practice guided by the International Myeloma Working Group (IMWG) [5] and R-ISS [4] staging systems as well as the ability to perform whole-exome/genome sequencing and gene expression profiling on large patient data sets. Consequently, the driving genomic events that lead to the development of MM from its precursor states as well as the cytogenetic and mutational changes that drive disease progression and relapse are being elucidated. Given the clonal heterogeneity between and within MM patients, this knowledge has not directly translated into targeted therapeutics but has greatly increased our understanding of drug resistance mechanisms and the biology of risk.
Below, we first present a thorough review of our current knowledge of MM genomics including chromosomal abnormalities and genetic mutations. We subsequently explore how they drive MM disease pathogenesis and have translated into better risk stratification in newly diagnosed multiple myeloma (NDMM) patients. We conclude with our perspective moving forward as to how we might apply whole-genome/exome-level data in addition to routine cytogenetic analyses to improve patient outcomes as well as define and prioritize further knowledge gaps moving forward.
Chromosomal abnormalities in multiple myeloma pathogenesis: copy number abnormalities and translocations
MM is known to evolve from the precursor diseases monoclonal gammopathy of undetermined significance (MGUS) and smoldering myeloma (SMM). Genetic events detected at the MGUS stage are likely to be primary events involved in tumor development whereas events present at the MM stage and absent in MGUS are likely secondary events leading to tumor progression. The genetic abnormalities in MM are complex and heterogenic but molecular subgroups have been clearly defined. Primary events in MM can be generally grouped into two categories: cases with primary immunoglobulin translocations; and those that are hyperdiploid (HD) with trisomies of the odd-number chromosomes also referred to as cases with copy number abnormalities (CNAs known as aneuploidies, abnormal in whole chromosome or arm level) [6].
Of note, the methodology of detecting cytogenetic abnormalities has evolved in MM over the past several decades. During early risk stratification and genomic assessment of MM patients, specific abnormalities on metaphase cytogenetics were found to be associated with inferior survival. However, this assay relies on the presence of actively dividing cells, and as terminally differentiated B cells, plasma cells have limited proliferative capacity [7, 8]. For example, the t(4;14) translocation is karyotypically silent with conventional cytogenetics but detected by FISH probes for breakpoints on chromosome 4 in the FGFR3 gene. Consequently, only one-third of MM patients have metaphase cytogenetic abnormalities at diagnosis. Interphase FISH (iFISH) is a more sensitive modality for identifying specific cytogenetic abnormalities of pathological significance. Subsequent to the International Staging System (ISS) development [9], chromosomal abnormalities (CA) detected by iFISH have become a standard of care in risk stratifying MM patients [10, 11]. MM may have chromosomal aberrations carried by only a subset of tumor cells, and the cytogenetic heterogeneity of individual cases reflects the coexistence of cytogenetically defined aberrant plasma cell clones [12].
Copy number abnormalities
Karyotyping and iFISH studies have demonstrated that almost all MM are aneuploid. Based on the types of CNAs, MM is divided into two main groups: HD and non-hyperdiploid (NHD) which compose 60% and 40% of MM respectively. HD (with a number of chromosomes between 48 and 74) is characterized by trisomies of odd-numbered chromosomes 3, 5, 7, 9, 11, 15, 19, and/or 21 [13,14,15]; while NHD is characterized by hypodiploid (up to 44/45 chromosomes), pseudodiploid (44/45 to 46/47), near-tetraploid (more than 74), hyperhaploid (loss of nearly a haploid set of chromosomes with 24–34 chromosomes), and tetraploid [14]. The HD state may be a consequence of simultaneous gain of all additional chromosomes in a single abnormal mitosis. Only a limited percentage of HD tumors (<10%) have a concurrent primary translocation affecting IGH [16]. Single-cell sequencing has shown that HD can precede primary translocations affecting the Ig heavy chain (IgH) locus in some patients [17].
In addition to aneuploidy changes, copy number gains or losses of chromosomes at an entire arm level are frequent including del13q (45–59%), +1q (35–40%), del14q (39%), del6q (33%), del1p (30%), and del17p (8%) [18]. Below, the biology and pathogenesis of several CNAs of note are discussed.
1q amplification (+1q)
The gain/amplification of CKS1B gene at chromosome region 1q21 (1q+) is seen in approximately one-third of NDMM patients making it one of the commonest copy number abnormality [19]. CKS1B is a member of the cyclin kinase subunit 1 protein family and is essential for cell growth and division [20]. It is widely expressed in various tissues but universally in the bone marrow where it associates with p27kip1-Cdk/cyclin complex and acts as a cofactor for Skp2-dependent ubiquitination of p27 [19,20,21]. When amplified, it activates the Cdk/cyclin complex and leads to greater degradation of p27, and ultimately cell cycle upregulation by promoting the G1/S transition this leading to MM cell survival and growth.
1p deletion
CKS1B amplification often co-occurs with CDKN2C gene deletion at chromosome 1p32.3 (1p-) locus [19]. Approximately 10% of NDMM will have a 1p deletion [22]. CDKN2C is a tumor suppressor gene that leads to deregulation of the G1/S transition resulting in proliferation of plasma cells in patients with MM [22].
Del(17p)/TP53
Loss of the short arm of chromosome 17 [17p13 [del(17p)] is seen in 5–10% of ND-MM. The methodology for identifying deletions varies but the techniques employed include metaphase cytogenetics, iFISH, Seq-FISH, and whole-genome/exome sequencing. Importantly, cytogenetic analysis of chromosome 17p deletions which spans the TP53 gene is typically performed by FISH probes against 17p and does not probe TP53 in isolation. Although the clinical relevance of del17p is well established in MM, the exact mechanism by which del17p promotes aggressive disease biology remains unclear [23]. The length of the deleted region can vary from a few megabases (MBs) to deletion of the entire short arm of chromosome 17. The TP53 gene is located in the minimally deleted region (0.25 MB) suggesting that it is a critical gene in the 17p13 region [24]. TP53 mutation has been identified as a driver mutation in MM [25]. However, a deletion event usually involves several genes, and co-deletion of TP53 along with Eif5a and Alox15b has resulted in more aggressive disease [26] and it remains unclear how genes other than TP53 contribute to tumorigenesis.
From the myeloma genome project (MGP), Walker et al. [ 25] demonstrated that TP53 deletion is the most common abnormality at 8%, followed by mutation (~6%) and bi-allelic inactivation (~4%). Deletions in TP53 induce clonal immortalization and survival of tumor cells as well as drug resistance which is thought to drive poor prognosis [27, 28]. Missense mutations of TP53 might associate with even worse outcomes in some cases. As in other tumor types, TP53 mutations in MM are spread across the entire gene, with many mutations occurring within the DNA-binding domain. Missense mutations in TP53 produce mutant TP53 proteins that not only result in loss of normal TP53 function (LOF), but also gain of oncogenic functions (GOF) [29, 30].
Early studies from analyses of small numbers of patients suggested an association between deletion on one allele and mutation on the second allele of chromosome 17p, putatively resulting in complete inactivation of P53 function [31]. In a larger NDMM dataset (n = 779) where both TP53 mutation and FISH data were available (n = 72), a significant correlation between mutation and deletion was observed [32]. The MGP data also showed a significant association between the presence of del(17p) cancer clone fraction (CCF) >0.55 and mutation on the second allele of TP53, where 27 of 28 patients with a TP53 mutation had CCF >0.55 [25]. Finally, the relationship between mono and bi-allelic del(17p) and TP53 mutational status remains to be clarified. Additional research is needed to improve our understanding of drivers of high-risk biology in MM patients with del17p and or TP53 deletions.
Chromosome translocations
NHD MM includes tumors that harbor chromosomal translocations and often involve the IgH locus, light chain kappa (IgK) locus at chromosome 2, light chain lambda (IgL) locus at chromosome 22, and MYC locus in the chromosome 8q24 region. The largest majority (>90%) of chromosomal translocations in MM affect chromosome 14, specifically the IGH locus at 14q32.33, which is one of the most heavily transcribed genes in plasma cells [33]. The landscape of translocations with particular attention to those involving the immunoglobulin (Ig) heavy chains (IGH) is described below.
Translocations into the immunoglobulin heavy chain locus located at 14q23
The translocation of oncogenes to the IgH locus at 14q32 is thought to be the seminal event in the development of MM [33, 34]. Translocations into the immunoglobulin heavy chain locus located at 14q23 occur in early stages of disease development suggesting this may be a primer driver event.
There are well-established translocation partners with varying frequencies:
-
Common; seen in more than 10% of patients: t(4;14) and t(11;14) translocations [34]
-
Less common; ≤5% of patients: t(14;16), t(6;14), t(8;14), and t(14;20) translocations [34].
Deregulation of a D group cyclin is consistent across translocation subgroups. This can occur either directly as seen in t(11;14) (cyclin D1) and t(6;14) (cyclin D3); or indirectly as seen in t(4;14) or in the MAF translocation (t(14;20) and t(14;16)) [33,34,35,36,37,38]. Translocations into 14q32 influence prognosis due to an upregulation of oncogenes. Depending on the partner, this varies but can include both the MAF family members (such as MafA, MafB, and c-Maf) as well as D-type cyclins (such as cyclin D1, D2, and D3) [33,34,35,36].
t(4;14) was initially identified based on breakpoints on chromosome 4 in the FGFR3 gene and subsequently involved the MMSET gene (MMSET: multiple myeloma SET domain; also known as Wolf-Hirschhorn syndrome candidate 1 (WHSC1) or nuclear receptor-binding SET domain 2 (NSD2)). The t(4:14) translocation was the first example of an IgH translocation that simultaneously dysregulated two genes with oncogenic potential: FGFR3 on der(14) and MMSET on der(4) [36]. Importantly, FGFR3 shows only weak transforming activity and is eventually lost in 30% of patients suggesting that it is not the main oncogenic factor [37], whereas MMSET is known to have histone methyl transferase activity and is deregulated early on in the genesis of developing MM [38].
In 15–20% MM cases whose IgH spilt can be detected by FISH, no specific partner chromosomes can be identified. Despite its remarkable prevalence with t(14; unknown) being as common as t(4;14) or t(11;14), its impact on risk and prognosis as well as biology is not well described albeit it is thought to be neutral [39]. Finally, the partners in ~15% of IgH translocations are defined most recently by using next-generation sequence techniques. Partner genes in this group are mainly MYC, and less frequently WWOX, B2M, ERF, RND3, JUN PAX5, DPF3, and MTMR1 [40, 41].
IgK and IgL translocations
IgK and IgL translocations are detected in 4.5% and 10% of ND-MM cases [42, 43]. Approximately 41% of IgL translocations and most IgK translocations involve MYC locus. Other recurrent partners include MAP3K14, CD40, MAFB, TXNDC5, CCND1, CCND2, and CCND3 but with lower frequencies (1 to 7%).
Myc translocations
Myc translocations and aberrations are common and frequently lead to an upregulation of the oncogene MYC which like in other B-cell malignancies has been shown to be unfavorable. In the CoMMpass study, a comprehensive natural history myeloma genomic project spearheaded by the Multiple Myeloma Research Foundation (MMRF) that has currently enrolled over 1150 patients from 76 sites in North America and Europe, MYC translocations occurred in 182 (23%) of 795 patients evaluated on trial and were juxtaposed to a large number of regions [43]. Among MYC structural alterations, translocations involving the immunoglobulin lambda (IgL) locus are present in 9.8% of patients and of which IgL-MYC translocations accounted for 41% of IgL translocations overall. t(8;14) is uncommon in myeloma with a prevalence of only 1–2%. Of note, there are significant associations between Myc and other abnormalities highlighting oncogenic dependencies in MM. For example, Myc rearrangements can lead to deregulation of FAM46C which has been associated with hyperdiploid MM [25]. 8q24 breakpoints have been found to partner with the immunoglobulin enhancers IGH, IGK, and IGL, important B-cell maturation loci including XBP1, FAM46C, CCND1, KRAS, and other superenhancers such as NSMCE2, TXNDC5, FOXO3, IGJ, and PRDM1 [40].
Genetic mutations in MM pathogenesis
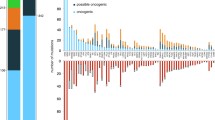
The development of MM from precursor diseases such as MGUS and SMM is driven by secondary events which provide a fitness advantage to a particular subclone and are required for tumor progression. For example, most copy number variations (CNV), translocations involving MYC, and somatic mutations affecting MAPK, NF-κB, and DNA repair pathways are observed during MM and less frequently in pre-malignant stages. Next-generation sequence techniques have demonstrated recurrent gene mutations detected in almost all MM cases. There is significant molecular heterogeneity and a well-defined hierarchy of frequencies but most mutations are found in less than 10% of patients. Genes that are involved in MM mutations include KRAS (23–36%), NRAS (20–21%), BRAF (6–8%), FAM46C (11–12%), DIS3 (11–16%), TP53 (8–16%), TRAF3 (5%), CYLD (2%), RB1 (3%), PRDM1/BLIMP1 (5%), ACTG1 (2%), RYR2 (5%), SVIL (5%), TTN (8%), MUC16 (8%), IGKC (2%), FAT3 (4–7%), SP140 (7–12%), CCND1 (3–10%), ROBO1 (2–5%), EGR1 (4–6%), P140 (5–7%), IgHJ6, and SDNAH5 [25, 44,45,46,47,48]. Through utilization of whole-genome sequencing in NDMM patients, Bolli et al. [ 48] showed the pervasiveness of driver mutations. When considering recurrent translocations and aneuploidies, deletions of tumor suppressor genes, amplification of oncogenes, and mutations pertaining to “oncogenic” or “possible oncogenic” classes, at least one such driver event was present in >99% of patients and overall a median of 6 events were present in each patient.
Affected signaling pathways by genetic abnormalities in MM
Among all the genetic abnormalities in MM pathogenesis, the majority are involved in two main pathways: RAS/MAPK--43% (KRAS, NRAS, BRAF, FGFR3, RASA2, PTPN11); and NF-κB--17% (TRAF2, TRAF3, CYLD, BIRC2, BIRC3, SP140, TWEAK, MAP3K14, COBLL1, and PRKD2). Such mutations lead to abnormal activation of RAS/MAPK and NF-κB signaling, promoting the proliferation and survival of the mutant cells. NF-κB gene signature analysis suggests that NF-κB signaling is inversely associated with RAS-RAF mutations, suggesting 2 distinct subsets of diseases [25, 46, 49, 50]. The remaining affected genes are involved in the following pathways [51,52,53,54] (Fig. 1):
-
Cell cycle: LEMD2, CCND1/2/3, RB1, CDKN1B, c-MAF, and MAF-B
-
p53-DNA repair: TP53, ATM, ATR, BAX, ZNFHX4, SAMHD1
-
Plasma differentiation: IRF4, PRDM1/BLIMP1, MYC-MAX, ZNF208, HOXB3
-
RNA process: DIS3, FAM46C, FCF1, RPL10, RP53A, and PABPC1
-
Protein translation: FBXO4, RPN1, PTH2, and ST6GAL1
-
Epigenomic modification: MMSET, PHF19, EZH2, HIST1H1E, KMT2B, KMT2C, CREBBP, ARID1A, ATRX, EP300, SETD2, TET2, KDM5C, ARID2, DNMT3A, KDM6A, NCOR1, IDH1, HistH1E, HistH14H
During this disease development process, multiple genetic abnormalities are dynamically accumulated and can be detected in the same patient suggesting the collaboration of these abnormalities in disease development and progression. Primary genetic events including hyperdiploid and IgH translocations can be detected in all disease stages from MGUS to SMM to MM [6]. On the contrary, genetic mutations are thought to be secondary genetic events that are acquired during disease progression to smoldering myeloma and eventually to symptomatic MM [55]. These secondary genetic events cause activation of oncogenic signaling and inactivation of tumor suppression signaling. Del17p/TP53 double-hit and Myc mutations (a rare event) are associated with drug resistance which are expanded in RR-MM cases [56, 57]. In addition, mutations that involve the proteasome, ER-stress, and CRBN as well as those that lead to an increased exportin 1 (XPO1) activation are also expanded in RR-MM cases and related to drug resistance (Fig. 2) [56, 57].
“Double-hit” and myeloma pathogenesis
In HD-MM and NHD-MM, the increase or decrease in the numbers of whole chromosomes results in expansion or reduction of both oncogenes and tumor repressor genes which may partially neutralize the oncogenic effects. This might explain why such dramatic change fails to induce malignant transformation when it happens alone. Such level change of oncogenes or tumor suppressor genes seems insufficient to induce malignant transformation. Clinical studies suggest that “double-hit” in the same gene or functional pathway are commonly observed in MM suggesting a critical role of threshold activity of the functional pathway in disease development. For example:
-
1.
Several tumor repressor genes (RB1, DIS3, DLEU2/miR-15a/16-1) have been identified in the deletion region of del13q. DIS3 inactive mutations commonly coincide with del13q which results in bi-allelic inactivation of DIS3 [58].
-
2.
Putative oncogenes in +1q include CKS1B (a negative regulator of CDKN1B), MCL1, BCL9, and ADAR1 [59,60,61]. The common coincidence of +1q and del13q results in double hits on the CDKN1B-RB1 pathway.
-
3.
The t(11;14) leads to cyclin D1 overexpression. An increase in CCND1 mutations is detected in MM with t(11;14) [34,35,36,37,38].
-
4.
The t(14;16) and t(14;20) lead to overexpression of c-MAF and MAF-B respectively. MAF is only significantly mutated in t(14;16) MM, while MAFB mutations are only detected in t(14;20) MM [62, 63].
-
5.
The t(4;14) results in MMSET overexpression in 100% of cases and FGFR3 overexpression in 70% of cases depending on the breakpoint site. FGFR3 mutations are detected in 17% of patients harboring t(4;14), and probably result from somatic hypermutations on der14 [11, 64, 65].
-
6.
MM with del1p often co-occurs with mutations of tumor repressors CDKN2C or FAM46C leading to double hits on the CDKN2C or FAM46C gene [66].
-
7.
MYC region implication or duplication is commonly detected in MYC translocations, leading to double hit on the MYC gene [40, 41].
-
8.
TP53 mutation or deletion often detected in patients with del17p, especially RR-MM cases, results in bi-allele deletion (or mutation) in the TP53 gene [19, 31].
MM pathogenesis is a result of multiple genetic abnormalities which collaboratively stimulate malignant transformation and uncontrolled expansion of malignant cells by inducing differentiation blockage and enhanced survival and uncontrolled proliferation. This explains the preferential coexistence of certain genetic abnormalities in the same patient. Several well-described and notable oncogenic co-occurrences and or dependencies of note include:
-
1.
Patients harboring t(4;14) co-segregate with mutations in FGFR3, DIS3, and PRKD2, or with del12p, del13q, and gain of 1q9 [36, 67].
-
2.
Patients with t(14;16) preferentially have co-occurring mutations in MAF, BRAF, DIS3, or ATM genes [25, 67].
-
3.
Mutations in CCND1, KRAS, IRF4, LTB, and HUWE1 genes are commonly detected in patients with t(11;14) [36, 48, 49, 67].
-
4.
CDKN1B, FUBP1, NFKB2, PRDM1, PTPN11, RASA2, RFTN1, and SP140 are significantly mutated only in HD-MM samples [68, 69].
- 5.
-
6.
Monosomy 17 or del17p is identified in patients with hyperhaploid [68, 69].
Several recently published large clinical databases have elucidated true genomic clustering resulting from oncogenic dependencies. In an enlightening analysis of both clinical and genomic data from the MGP, Walker and colleagues [25] evaluated whole-exome and in many cases whole-genome sequencing of 1273 NDMM patients. They identified 9 total clusters, two of which were hyperdiploid (cluster 1: gains in chromosomes 3, 5, 9, 15, 19, and 21; cluster 2: gains in the same chromosomes plus chromosome 11 and mutation of FAM46C) while 7 were non-hyperdiploid (cluster 3: del1p; cluster 4 by del12p and 13q; cluster 5: del13q and 14q and mutations in MAX, TRAF3, and NFKBIA; cluster 6: del16q; cluster 7: t(14;16), del11q, 1q gain, and mutation of DIS3; cluster 8: t(11;14); cluster 9: t(11;14) with gain 11q). Other groups have shown similar clustering and co-segregating genomic patterns [46, 49].
The clinical significance of genetic abnormalities in multiple myeloma
As with other hematological malignancies, the accumulation of genomic aberrations is not only a hallmark of MM but also is associated with patient outcomes. Whole-genome/exome sequencing as well as the review of large patient data sets in the modern era has improved our knowledge of disease biology and pathogenesis from mutations to chromosomal. Although direct correlations with mutations have been linked to risk and patient outcomes, the implementation of this granulation genomic data into routine clinical practice remains an enormous barrier to improving risk stratification in NDMM. Thus, defining uniform characteristics of risk in NDMM remains elusive despite several validated risk stratification systemic in routine clinical use. The accurate assessment of risk at diagnosis is critical for many reasons including but not limited to:
-
1.
The longest remission period being achieved by initial therapy and thus the duration of the first remission is one of the most important factors impacting patient prognosis and long-term outcomes in general
-
2.
Accurate definition of risk for clinical trial design and enrollment
-
3.
Establishing which clinical data should be obtained routinely in practice given certain tools such as gene expression profiling are challenging to obtain while others such as serum lactate dehydrogenase (LDH) can easily be incorporated into routine clinical care.
Clinical staging systems
The international staging system (or ISS) is an early risk stratification for NDMM patients [9]. It incorporated a combination of serum beta2 microglobulin (B2M) and albumin into a simple powerful three-stage classification predictive of OS. Subsequent to the ISS development, chromosomal abnormalities detected by iFISH have become a standard of care in risk stratifying MM patients and thus integrated genetic abnormalities into routine clinical practice. In 2014, the IMWG published and updated risk stratification focusing on differentiating high-risk patients, those with an expected OS of less than 2 years despite the use of novel agents, from low-risk patients, those with expected survival greater than 10 years. The staging system combines the ISS with certain high-risk iFISH changes including t(4;14), 17p13, and 1q21 [5]. The revised international staging system (R-ISS) was also developed which combines iFISH changes, serum (LDH), and ISS features and is the most widely recognized risk stratification tool for NDMM patients today [4]. Although it incorporates important genetic markers including t(4;14), t(14;16), and del17p, it does not include some important genomic data including 1q gain/amplification an increasingly important prognostic marker [19, 25] nor mutational data from TP53 as the data were not available.
High-risk genetic lesions including copy number abnormalities, chromosomal translocations, and mutations
In addition to well established clinical staging system incorporating iFISH as outlined above, certain high-risk genomic lesions are well established. Table 1 outlines select high-risk iFISH changes detectable in routine clinical practice with supporting literature to summarize their impact on risk. In addition to having a single high-risk genomic lesion, what is frequently being referred to as double-hit myeloma (different from the double hit concept described above driving myeloma pathogenesis) includes patients who have more than 1 adverse cytogenetic lesions (such as del(17p), t(4;14), and gain of 1q) or an adverse cytogenetic lesion concurrently with a high-risk mutation. These patients display dismal survival outcomes. For example, in the Bolli et al. [ 49] experience, patients with both t(4;14) and PRDM1 deletion had a dismal median OS of 265 days whereas data from the MGP shows that patients with bi-allelic TP53 mutation have a PFS of only 15.4 months and OS of just 20.7 months [79].
Mutations
Although the duration of follow-up is limited on large datasets of NDMM with extensive genomic sequencing, it is apparent that few solitary mutations drive prognosis. TP53 mutation is the only consistent adverse finding in addition to a link between general mutational burden and clinical outcomes [25, 28, 49]. The impact of bi-allelic/double hit v monoallelic TP53 inactivation or del(17p) remains unclear. The MPG had only 33 patients with monoallelic deletion/mutation of TP53 [25]. These patients did not have an inferior outcome when compared to patients without a TP53 abnormality. This observation could be explained by the subclonal nature of TP53 deletions in these patients [28]. Table 1 outlines a higher cancer clone fraction (CCF) may be required to drive poor outcomes. TP53 mutations in isolation may have additional pathological significance yet to be determined such as driving and/or propagating MM clones leading to a selective clonal advantage [28] and the co-occurrence of TP53 mutations with other gene mutations may in the end be synergistic.
The MGP found 63 driver genes including novel previously unidentified oncogenes PTPN11 (activator of MEK/ERK signaling), PRKD2 (protein kinase D), IDH1 and IDH2 (DNA methylation), and SF3B1 (spliceosome factor). Novel tumor suppressor genes including UBR5 (a ubiquitin ligase) and HUWE1 (a ubiquitin ligase that can affect MUC expression via MIZ1) were also described. Of the 63 driver genes identified in their study, only TP53, TRAF3, and TGDS had an impact on outcome in univariate analyses. Driver gene mutational burden did lead to worse PFS and OS (P=0.001). Furthermore, two markers of genomic instability were associated with outcomes including an APOBEC mutational signature and loss of heterozygosity [25]. Interestingly, the extent of LOH was positively correlated with the APOBEC signature (P=0.039), loss of TP53 (P=0.001), and presence of mutation in at least 1 of 15 genes involved in homologous recombination deficiency (P < 0.001). Updated analyses focused on the key clinical problem of identifying high-risk patients destined for early relapse and death where a change in treatment strategy could result in improved outcome [79]. On multivariate modeling, only t(4;14) and bi-allelic TP53 inactivation were predictive of PFS while bi-allelic TP53 inactivation and amplification of CKS1B (1q21) were predictive of OS.
Bolli et al. [ 49] found similar results performing whole-genome sequencing of a total of 418 NDMM patients. Risk as defined by worsening survival leads to clustering of patients based on the number of mutations as well as the number/type of cytogenetic abnormalities. In multivariate analyses, they found that mutations in SP140 and NRAS, t(4;14), amp(1q), and del(17p13) and deletions of FAT1 and PRDM1 negatively impacted PFS. Conversely, t(4;14), amp(1q), del(17p13), and del(1p) negatively impacted OS. The only mutation that negatively impacted both PFS and OS was TP53; DNAH11 mutations lead to worse OS only [49]. Similar to Walker et al. [25], they discovered clusters of patients stratified based on the overall number of mutations and number/type of CNAs that lead to distinct effects on survival. Thus, individual mutations and performing an extended genotype at diagnosis likely lead to improved prognostication in NDMM. Finally, as seen from the MGP, they found distinct genomic clustering patterns leading to poor prognosis. For example, cluster 2 was enriched for IGH translocations and a high number of CNAs, enriched for amp(1q), del (13), del(17p), and deletions of BIRC2/3 and XBP1, and carried more TP53 mutations [49].
The CoMMpass study (as described above) found 55 genes that were significantly mutated and there was a 65% overlap with the MGP. After a median follow-up of 39 months, early progressive disease (PD) was detected in 191/926 (20.6%) patients. In a multivariate logistic regression model, independent factors increasing the early PD risk were TP53 mutation (OR, 3.78, P < 0.001), high LDH (OR, 3.15, P 0.006), IgL-chain translocation (OR, 2.25, P 0.033), and IGLL5 mutation (OR, 2.15, P 0.007) [67].
Perspectives moving forward
Our ability to define risk in NDMM has vastly improved as our knowledge of the pathogenesis, biology, and genomics of MM has rapidly expanded. However, we continue to struggle to standardize risk as is evident by the routine use of several risk staging systems that do not completely align including the R-ISS, IMWG, and mSMART [77]. With the R-ISS being now nearly a decade old, it is likely time to formally revisit and develop an updated risk stratification system wielding the power of our increased knowledge of myeloma genomics. The biggest barrier to implementing our improved knowledge of mutations and other genomic factors impacting risk is the ability to perform such assays (advanced mutational profiling, whole-genome/exome sequencing, and even gene expression profiling (GEP) as outlined below) in routine clinical practice outside of clinical trials or major academic centers where the vast majority of NDMM patients are treated. Furthermore, payer reimbursement for such testing is likely to continue to be a problem.
This is particularly important given the rapid expansion and Food and Drug Administration (FDA) approval of new therapeutics since the R-ISS 2014 publication including but not limited to ixazomib (2015), carfilzomib (in combination with lenalidomide and dexamethasone; 2015), daratumumab (2015), elotuzumab (2015), selinexor (2019), belantamab mafodotin-blmf (2020), isatuximab (2020), and idecabtagene vicleucel (2021). These therapeutics have improved MM outcomes and may influence the impact of genomic risk in MM patients. It is critical to accurately identify high-risk patients at diagnosis and move toward the establishment of risk-adapted treatments. While many studies are targeting improved outcomes in high-risk myeloma [91], there has yet to be a randomized trial that has demonstrated a definitive preferred approach in this challenging patient population with the most recent negative study being the SWOG1211 (bortezomib, dexamethasone, and lenalidomide with or without elotuzumab in treating patients with newly diagnosed high-risk multiple myeloma) [92].
Is there a way to incorporate whole-genome/exome-level genomic data into routine clinical use? Gene expression profiling may very well be the answer. Gene expression profiling measures which genes are being expressed in a myeloma cell and specifically measures mRNA levels, showing the pattern of genes expressed by a cell at the transcription level. Given that DNA-based assays such as whole genomic sequencing are able to identify individual lesions and markers of global genomic instability and ultimately prognosis, it is not surprising that the development and now validation of several GEP score systems that have prognostic value have increased interest in incorporating GEP into routine clinical practice. Various reports, using distinct approaches, have identified gene expression signatures capable of predicting event-free survival and OS in multiple myeloma [93,94,95]. Although a variety of other GEP have been developed, only two have matured into validated clinical tests: MMprofiler (EMC92/SYK92) and MyPRS (UAMS GEP 70). Despite growing evidence of the prognostic value of GEP, its application to routine clinical care remains challenging. There is not a consensus on a universal adaptation of a particular GEP system. GEP remains an investigational tool and is not yet validated by the FDA.
In order to best define risk in NDMM patients, we would recommend the following cytogenetic and mutational assessments be obtained at diagnosis prior to treatment initiation whenever possible. We understand that currently the scope of advanced genomic sequencing is limited thus many of the mutations listed below. Most recommended cytogenetic studies below are readily available in MM FISH panels:
-
High risk:
-
Cytogenetics:
-
t(14;16)
-
t(4;14)
-
IgL-MYC translocation
-
+1q amplification (≥4 copies)
-
1p-
-
del(17p)
-
-
Mutations/advanced genomic sequencing
-
TP53 deletion
-
LOH and APOEBEC signature
-
CKS1B amplification
-
-
-
Potentially high risk:
-
Cytogenetics:
-
t(14;20)
-
t(8;14) and other MYC translocations
-
+1q gain (3 copies)
-
del 13q/-13
-
-
Mutations/advanced genomic sequencing
-
TRAF3
-
TGDS
-
PRDM1
-
DNAH11
-
FAT1
-
NRAS
-
SP140
-
IGLL5
-
Driver gene mutational burden
-
-
In conclusion, as we move forward, there is a need for improved longitudinal genomic data in addition to that already described above. This will require large-scale collaboration between researchers and institutions but will help further elucidate key genomics driving pathogenesis and risk. Importantly, this may translate into an improved understanding of poor outcomes, drug resistance, and potentially targetable pathways for new therapeutics.
Data Availability
Not applicable as this manuscript does not include human subjects or laboratory-derived data.
Abbreviations
- amp:
-
Amplification, ≥4 copies
- ASCT:
-
Autologous hematopoietic cell transplant
- B2M:
-
Beta2 microglobulin
- CA:
-
Chromosomal abnormalities
- CCF:
-
Cancer clone fraction
- CNAs:
-
Copy number alterations
- CNV:
-
Copy number variations
- EMN:
-
European myeloma network
- FDA:
-
Food and Drug Administration
- HD:
-
Hyperdiploid
- HR:
-
Hazard ratio
- IFM:
-
Institute Francophone du Mye´lome
- IGH:
-
Ig heavy chain locus
- IMWG:
-
International Myeloma Working Group
- ISS:
-
International Staging System
- LDH:
-
Lactate dehydrogenase
- IgK:
-
Light chain kappa
- IgL:
-
Light chain lambda
- MGP:
-
Myeloma genome project
- MGUS:
-
Monoclonal gammopathy of undetermined significance
- MM:
-
Multiple myeloma
- MMRF:
-
Multiple Myeloma Research Foundation
- MMSET:
-
Multiple myeloma SET domain
- MV:
-
Multivariate
- NDMM:
-
Newly diagnosed multiple myeloma
- NHD:
-
Non-hyperdiploid
- OS:
-
Overall survival
- PCL:
-
Plasma cell leukemia
- PFS:
-
Progression-free survival
- PI:
-
Prognostic index
- R-ISS:
-
Revised international staging system
- RRMM:
-
Relapsed refractory multiple myeloma
- SMM:
-
Smoldering myeloma
- XPO1:
-
Exportin 1
References
SEER Cancer Stat Facts: Myeloma. National Cancer Institute. Bethesda. https://seer.cancer.gov/statfacts/html/mulmy.html
Cowan AJ et al (2020) The global state of hematopoietic cell transplantation for multiple myeloma: an analysis of the Worldwide Network of Blood and Marrow Transplantation Database and the Global Burden of Disease Study. Biol Blood Marrow Transplant 26:2372–2377
Costa LJ et al (2018) Recent trends in multiple myeloma incidence and survival by age, race, and ethnicity in the United States. Blood advances 1:282–287
Palumbo A et al (2015) Revised international staging system for multiple myeloma: a report from International Myeloma Working Group. J Clin Oncol 33:2863–2869
Chng WJ et al (2014) IMWG consensus on risk stratification in multiple myeloma. Leukemia 28:269–277
Smadja NV et al (1998) Chromosomal analysis in multiple myeloma: cytogenetic evidence of two different diseases. Leukemia 12:960–969. https://doi.org/10.1038/sj.leu.2401041
Rajkumar S et al (1999) Abnormal cytogenetics predict poor survival after high-dose therapy and autologous blood cell transplantation in multiple myeloma. Bone Marrow Transplant 24:497–503
Dewald GW et al (2005) Relationship of patient survival and chromosome anomalies detected in metaphase and/or interphase cells at diagnosis of myeloma. Blood 106:3553–3558
Greipp PR et al (2005) International staging system for multiple myeloma. J Clin Oncol 23:3412–3420
Neben K et al (2010) Combining information regarding chromosomal aberrations t(4;14) and del(17p13) with the International Staging System classification allows stratification of myeloma patients undergoing autologous stem cell transplantation. Haematologica 95:1150–1157
Boyd KD et al (2012) A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia 26:349–355
Jones J et al (2019) Clonal evolution in myeloma: the impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica 104(7):1440–1450
Kumar SK, Rajkumar SV (2018) The multiple myelomas - current concepts in cytogenetic classification and therapy. Nat Rev Clin Oncol 15:409–421
Chretien ML et al (2015) Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood 126:2713–2719
Chng W et al (2006) Analysis of genetic abnormalities provides insights into genetic evolution of hyperdiploid myeloma. Genes Chromosomes Cancer 45:1111–1120
Manier, S. et al. (2017) Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol
Pawlyn C et al (2015) Coexistent hyperdiploidy does not abrogate poor prognosis in myeloma with adverse cytogenetics and may precede IGH translocations. Blood 125:831–840
Walker BA et al (2010) A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 116:e56–e65
Varma A et al (2020) Outcome of multiple myeloma with chromosome 1q gain and 1p deletion after autologous hematopoietic stem cell transplantation: propensity score matched analysis. Biol Blood Marrow Transplant 26:665–671
Spruck C et al (2001) A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol Cell 7:639–650
Ganoth, D. et al. (2001) The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol
Leone P et al (2008) Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res 14(19):6033–6041
Lakshman A et al (2019) Impact of acquired del(17p) in multiple myeloma. Blood advances 3:1930–1938
Boyd KD et al (2011) The clinical impact and molecular biology of del(17p) in multiple myeloma treated with conventional or thalidomide-based therapy. Genes Chromosomes Cancer 50:765–774
Walker BA et al (2018) Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 132:587–597
Liu Y et al (2016) Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531:471–475
Lee MK et al (2012) Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell 22:751–764
Flynt E et al (2020) Prognosis, biology, and targeting of TP53 dysregulation in multiple myeloma. Cells 9(2):287
de Vries A et al (2002) Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc Natl Acad Sci 99:2948–2953
Boettcher S et al (2019) A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365:599–604
Lode L et al (2010) Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 95:1973–1976
Besse A et al (2018) Carfilzomib resistance due to ABCB1/MDR1 overexpression is overcome by nelfinavir and lopinavir in multiple myeloma. Leukemia 32:391–401
Gonzalez D et al (2007) Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood 110:3112–3121
Kalff A, Spencer A (2012) The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: prognostic implications and current clinical strategies. Blood Cancer J 2:e89
Ross FM et al (2010) The t(14;20) is a poor prognostic factor in myeloma but is associated with long-term stable disease in monoclonal gammopathies of undetermined significance. Haematologica 95:1221–1225
Chesi M et al (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92:3025–3034
Keats JJ et al (2003) In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood 101:1520–1529
Mirabella F et al (2013) MMSET is the key molecular target in t(4;14) myeloma. Blood Cancer J 3:e114
Kaufman GP et al (2016) Impact of cytogenetic classification on outcomes following early high-dose therapy in multiple myeloma. Leukemia 30:633–639
Affer M et al (2014) Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 28:1725–1735
Walker BA et al (2014) Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J 4:e191
Kuehl WM, Bergsagel PL (2002) Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer 2:175–187
Barwick BG et al (2019) Multiple myeloma immunoglobulin lambda translocations portend poor prognosis. Nat Commun 10:1911
Leich E et al (2013) Multiple myeloma is affected by multiple and heterogeneous somatic mutations in adhesion- and receptor tyrosine kinase signaling molecules. Blood cancer journal. 3:e102
Walker BA et al (2015) APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun 6:6997
Lohr JG et al (2014) Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25:91–101
Chapman MA et al (2011) Initial genome sequencing and analysis of multiple myeloma. Nature 471:467–472
Bolli N et al (2014) Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 5:2997
Bolli N et al (2018) Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia 32:2604–2616
Stein CK et al (2017) The varied distribution and impact of RAS codon and other key DNA alterations across the translocation cyclin D subgroups in multiple myeloma. Oncotarget 8:27854–27867
Misiewicz-Krzeminska I et al (2016) Post-transcriptional modifications contribute to the upregulation of cyclin D2 in multiple myeloma. Clin Cancer Res 22:207–217
Bergsagel PL et al (2005) Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106:296–303
Dimopoulos K et al (2014) The role of epigenetics in the biology of multiple myeloma. Blood Cancer J 4(5):e207
Chourhury S et al (2002) Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer 2(3):175–187
Landgren O et al (2009) Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood 113:5412–5417
Davis L, Sherbenou D (2021) Emerging therapeutic strategeis to overcome drug resistance. Cancers 13(7):1686
Paradzik T et al (2021) The landscape of signaling pathways and proteasome inhibitors combinations in multiple myeloma. Cancers 13(6):1235
Boyle E et al (2020) BRAF and DIS3 mutations associate with adverse outcome in a long-term follow-up of patients with multiple myeloma. AACR 26(10):2422–2432
Shaughnessy J et al (2005) Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27Kip1 and an aggressive clinical course in multiple myeloma. Hematology 10(Suppl 1):117–126
Samo AA et al (2018) MCL1 gene co-expression module stratifies multiple myeloma and predicts response to proteasome inhibitor-based therapy. Genes Chromosomes Cancer 57:420–429
Teoh PJ et al (2018) Aberrant hyperediting of the myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis. Blood 132:1304–1317
Hurt EM et al (2004) Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 5:191–199
Hanamura I et al (2001) Ectopic expression of MAFB gene in human myeloma cells carrying (14;20)(q32;q11) chromosomal translocations. Japanese J Cancer Res 92:638–644
Keats JJ et al (2005) Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood 105:4060–4069
Walker BA et al (2013) Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 121:3413–3419
Zhu YX et al (2017) Loss of FAM46C promotes cell survival in myeloma. Cancer Res 77:4317–4327
D'Agostino M et al (2020) Early relapse risk in patients with newly diagnosed multiple myeloma characterized by next-generation sequencing. Clin Cancer Res 26(18):4833–4841
Ashby C et al (2019) Poor overall survival in hyperhaploid multiple myeloma is defined by double-hit bi-allelic inactivation of TP53. Oncotarget 10:732–737
Peterson JF et al (2019) Hyperhaploid plasma cell myeloma characterized by poor outcome and monosomy 17 with frequently co-occurring TP53 mutations. Blood Cancer J 9:20
Fonseca R et al (2001) Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia 15:981–986
Avet-Loiseau H et al (1999) Monosomy 13 is associated with the transition of monoclonal gammopathy of undetermined significance to multiple myeloma. Intergroupe Francophone du Myelome. Blood 94:2583–2589
Chiecchio L et al (2006) Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia 20:1610–1617
Jurczyszyn A et al (2018) The prognostic impact of t(14;16) in multiple myeloma: a multicenter retrospective study of 213 patients. Is it time to revise the revised ISS? Blood 132(suppl 1):4452
Shah V et al (2017) Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia 32:102–110
Avet-Loiseau H et al (2011) Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood 117:2009–2011
Chan H et al (2018) Single-center experience in treating patients with t(4;14) multiple myeloma with and without planned frontline autologous stem cell transplantation. CLML 18(3):225–234
Mikhael JR et al (2013) Mayo Clinic. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013 [published correction appears in Mayo Clin Proc. 2013;88(7):777]. Mayo Clin Proc 88(4):360–376
Giri S et al (2019) Chromosome 1 abnormalities and clinical outcomes in multiple myeloma in the era of novel agents. ASCO 37(15):suppl 8044
Walker BA et al (2019) A high-risk, double-hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia 22:159–170
D’Agostino M et al (2020) Impact of gain and amplification of 1q in newly diagnosed multiple myeloma patients receiving carfilzomib-based treatment in the forte trial. Blood 136:38–40
An G et al (2015) The impact of clone size on the prognostic value of chromosome aberrations by fluorescence in situ hybridization in multiple myeloma. Clin Cancer Res 21:2148–2156
Boyd KD et al (2011) Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin Cancer Res 17(24):7776–7784
Hebraud B et al (2014) Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: the IFM experience on 1195 patients. Leukemia 28(3):675–679
Zojer N et al (2000) Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood 95(6):1925–1930
Binder M et al (2017) Prognostic implications of abnormalities of chromosome 13 and the presence of multiple cytogenetic high-risk abnormalities in newly diagnosed multiple myeloma. Blood Cancer J 7:e600
Tricot G et al (1995) Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood 86:4250–4256
Perrot A et al (2019) Development and validation of a cytogenetic prognostic index predicting survival in multiple myeloma. JCO 37(19):1657–1665
Corre J et al (2021) del(17p) without TP53 mutation confers a poor prognosis in intensively treated newly diagnosed patients with multiple myeloma. Blood 137(9):1192–1195
Thakurta A et al (2019) High subclonal fraction of 17p deletion is associated with poor prognosis in multiple myeloma. Blood 133(11):1217–1221
Kuiper R, et al. (2015) Prediction of high- and low-risk multiple myeloma based on gene expression and the International Staging System. Blood; 126(17): 1996-2004
Pawlyn C, Davies F (2019) Toward personalized treatment in multiple myeloma based on molecular characteristics. Blood 133(7):660–675
Usmani S et al (2021) Bortezomib, lenalidomide, and dexamethasone with or without elotuzumab in patients with untreated, high-risk multiple myeloma (SWOG-1211): primary analysis of a randomised, phase 2 trial. Lancet Haematol 8:e45–e54
Shaughnessy JD Jr et al (2007) A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109:2276–2284
Szalat R et al (2016) Gene expression profiles in myeloma: ready for the real word? CCR 22(22):5434–5442
Kuiper R et al (2012) A gene expression signature for high-risk multiple myeloma. Leukemia 26:2406–2413
Funding
This work was supported by NIH grants R01 HL133560 and R01 CA223194 through Loyola University Chicago, as well as Loyola program development funds to Jiwang Zhang.
Author information
Authors and Affiliations
Contributions
Patrick Hagen was the primary author of this manuscript with significant and equal contributions from the remaining authors.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable as this manuscript does not include human subjects.
Consent for publication
Not applicable as this manuscript does not include human subjects.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hagen, P., Sellin, M., Berg, S. et al. Increasing genomic discovery in newly diagnosed multiple myeloma: defining disease biology and its correlation to risk. Ann Hematol 101, 1407–1420 (2022). https://doi.org/10.1007/s00277-022-04856-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-022-04856-1