Abstract
Cytotoxic T-lymphocyte antigen-4 (CTLA-4) polymorphisms have been associated with susceptibility to lymphoid malignancies. However, results from the published single studies are inconsistent. Therefore, the present meta-analysis was conducted to get a more accurate estimation of the relationship between CTLA-4 gene polymorphisms and the lymphoid malignancy risk. We identified nine independent studies accounting for 3090 subjects up to January 30, 2016. Summary odds ratios (OR) and 95 % confidence intervals (CI) were used to evaluate the risk of lymphoid malignancies. Overall, no significant association was found between +49A/G (rs231775), −318C/T (rs5742909), and +6230A/G (rs3087243) CTLA-4 gene polymorphisms and lymphoid malignancies. Furthermore, ethnicity (Asian and Caucasian) and histopathology subgroup analyses (non-Hodgkin’s lymphoma) also failed to detect an association between the studied polymorphisms and lymphoid malignancy risk. Our study shows that common CTLA-4 gene polymorphisms may not contribute to lymphoid malignancy susceptibility based on the current evidence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lymphoid malignancies, including lymphoma, lymphoid leukemia, and myeloma, are large and heterogeneous disease groups, which were classified by the World Health Organization (WHO) in 2001 [1] and updated in 2008 [2]. Lymphoid malignancies arise from the malignant transformation of normal B- and T-lymphoid cells at various stages of differentiation and comprise the seventh most common cancer worldwide among men and women [3]. However, the etiological factors contributing to most lymphoid malignancies remain poorly understood [4]. The potential molecular mechanism of lymphomagenesis is very complex. However, several studies support that immune-related genetic factors are associated with the etiology of lymphoid malignancies [5]. An increased incidence of lymphoid malignancies has been observed among individuals with autoimmune disease and congenital and acquired immunodeficiencies, suggesting that immune dysfunction might represent a risk factor for lymphoid malignancies [6].
Cytotoxic T-lymphocyte antigen-4 (CTLA-4), also known as CD152, is an immune regulatory molecule expressed exclusively in T cells. It has been known that CTLA-4 acts as an immune checkpoint and regulates immune function through pleiotropic mechanism suppression, the activity of effector T cells, and establishment of peripheral T cell tolerance [7]. Mouse model studies have demonstrated that deletion of the CTLA-4 gene results in a lethal systemic immune activation by profound lymphoproliferation [8]. Furthermore, several studies have verified that blocking the CTLA-4 function can enhance antitumor immunity [9, 10].
The human CTLA-4 gene, located on chromosome 2q33 in an immune regulatory gene region, is composed of four exons. Several relevant polymorphisms have been described in this region, including −318C/T (rs5742909) located in the promoter region [11], +49A/G (rs231775) located in exon 1 [12], and +6230A/G (rs3087243) located in the 3′-untranslated region (3′ UTR) [13]. It has been shown that these single-nucleotide polymorphisms (SNPs) in the CTLA-4 gene might influence the host immune response by affecting gene transcription, protein expression, and interaction with the CD80/CD86 ligand [14–16].
Numerous studies have shown that CTLA-4 polymorphisms play an important immunoregulatory role in lymphoid malignancies; however, the results were inconsistent and even contradictory [17–25]. Considering that a single study is unable to elucidate the overall effects, we conducted a comprehensive meta-analysis of all eligible case-control studies to evaluate the association between CTLA-4 SNPs (rs231775, rs5742909, and rs3087243) and the susceptibility to lymphoid malignancies.
Materials and methods
Publication search
We searched the PubMed, Embase, and Chinese National Knowledge Infrastructure (CNKI) databases for studies published prior to January 2016 (last search: January 30, 2016). The keywords searching were performed with and without Medical Subject Headings (MeSH) terms for “Cytotoxic T-lymphocyte antigen 4/CTLA-4/CD152,” “polymorphism,” and “lymphoid malignancies/lymphoid neoplasms.” To identify relevant publications, reference lists of research papers were also reviewed by manual search.
Inclusion and exclusion criteria
All studies included in the meta-analysis met the following criteria: (1) evaluation of the association of CTLA-4 polymorphisms and lymphoid malignancy risk, (2) case-control design, (3) available information on genotype frequency, and (4) the controls had no malignant disease. Additionally, the following exclusion criteria were also used: (1) the study was not regarding the polymorphisms of interest; (2) it was a repeated study, a review, or an abstract; and (3) the study did not have a control group.
Data extraction
According to the inclusion and exclusion criteria listed above, two investigators independently revised all potentially relevant studies, and disagreements were resolved through discussion with a third researcher. For each study, the following information was collected: first author, year of publication, country of origin of the study, subjects’ ethnicity, source of control, genotyping method, total number of cases and controls, number of different genotypes in cases and controls, and Hardy-Weinberg value of controls. All data were obtained from published articles. All patients were confirmed by histology or pathology. Histopathological subtypes of lymphoid malignancies were classified based on the WHO classification guidelines [2, 26].
Statistical analysis
We calculated the odds ratios (OR) and 95 % confidence intervals (CI) to evaluate the lymphoid malignancy risk associated with CTLA-4 polymorphisms, based on the allele frequencies in cases and controls for each eligible study. The following five different ORs were computed: (i) B vs. A (allele comparison), (ii) BB vs. AA (homozygous carriers), (iii) BB vs. AA + AB (recessive model), (iv) BB + AB vs. AA (dominant model), and (v) AB vs. AA (heterozygous carriers). Statistical heterogeneity between studies was evaluated by chi-square-based Q test and I 2 test. Higher I 2 values indicated higher levels of heterogeneity (I 2 = 75–100 %: extreme heterogeneity; I 2 = 50–75 %: large heterogeneity; I 2 = 25–50 %: moderate heterogeneity; I 2 < 25 %: no heterogeneity). If the p value of the Q test was higher than 0.1, the pooled OR estimate of the study was calculated by the fixed-effects model. Otherwise, the random-effects model was used. Subgroup analysis was performed to assess the ethnicity-specific effects. Egger’s and Begg’s tests were adopted to assess publication bias. A p < 0.05 was considered statistically significant. Sensitivity analysis was performed to estimate the stability of the results by removing the studies, one at a time. All statistical analyses were performed with STATA 12.0 software (Stata Corp, College Station, TX).
Results
Study characteristics
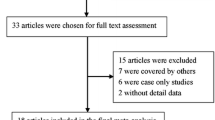
As shown in Fig. 1, 102 publications were retrieved from the initial screening. After skimming the titles and reviewing the abstracts, 72 studies were excluded by their irrelevance to the current analysis. Furthermore, 21 studies were excluded because of lack of detailed data or lack of data regarding the target polymorphisms. Thus, nine case-control studies involving 3090 subjects were selected for the meta-analysis. The main characteristics of the included studies are summarized in Table 1. Eligible studies presented data for several different lymphoid malignancies including chronic lymphocytic leukemia (CLL), mucosa-associated lymphoid tissue (MALT) lymphoma, non-Hodgkin’s lymphoma (NHL), and multiple myeloma. There were seven studies based on Caucasian and two on Chinese population. Additionally, six studies were hospital-based, and three were population-based case-control studies. Detailed data on genotype distribution in each eligible study and minor allele frequency (MAF) as well as Hardy-Weinberg equilibrium (HWE) of controls are shown in Table 2. Among the nine case-control studies, in one study regarding CTLA-4 rs231775 polymorphism, a significant departure from HWE was observed [24].
Meta-analysis results
Nine studies assessed the association between CTLA-4 rs231775 polymorphism and lymphoid malignancy susceptibility. As shown in Fig. 2, the heterogeneity of GG vs. AA + AG was assessed for all case-control studies. The I 2 value was 0 %, indicating no heterogeneity. Meta-analysis results were as follows: χ 2 = 7.66, degrees of freedom = 8, and p = 0.41 in a fixed-effects model. We observed a lack of association between the rs231775 polymorphism and lymphoid malignancy risk under a recessive model. Further studies confirmed that neither the subgroup analyses by histologic type nor those by ethnicity revealed a noteworthy association. All comparison results are listed in Table 3.
Forest plots of CTLA-4 rs231775 polymorphism and NHL risk based on recessive model (GG vs. AA + AG). The squares and horizontal lines correspond to the study-specific OR and 95 % CI, respectively. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95 % CI. OR odd ratio, CI confidence interval
Seven studies evaluated the association between rs5742909 polymorphism and lymphoid malignancy risk including 1037 cases and 1332 healthy controls. As shown in Table 3, the meta-analysis results indicate that the rs5742909 polymorphism is not associated with lymphoid malignancy predisposition in the overall population, neither in the NHL subgroup nor in Caucasian or Chinese populations. However, trends were observed in a heterozygous comparison model in Caucasians (CT vs. CC: 95 % CI = 1.00–1.86, P = 0.05).
Five studies including those on three Caucasian and two Chinese populations evaluated the association between CTLA-4 rs3087243 polymorphism and risk of lymphoid malignancies. A fixed-effects model was selected for all genetic comparisons showing no significant differences in terms of heterogeneity. The results of quantitative synthesis evidenced the lack of a significant relationship in the overall population and subgroup analyses by histological type or ethnicity (Table 3).
Heterogeneity assessment and sensitivity analysis
Because of significant heterogeneity in the literature regarding the CTLA-4 rs231775 variant (G vs. A: p < 0.001; GG vs. AA: p = 0.03; GG + AG vs. AA: p < 0.001; AG vs. AA: p < 0.001), sensitivity analysis was conducted to identify the source, by sequentially omitting one study at a time. The results indicate that the studies by Monne et al. [18] and Piras et al. [19] contributed to the major heterogeneity. The exclusion of these two studies significantly decreased the heterogeneity (G vs. A: p = 0.21; GG vs. AA: p = 0.18; GG + AG vs. AA: p = 0.09; AG vs. AA: p = 0.12). It was noted that the initial lack of association was quantitatively altered, when the two studies were excluded in the homozygous model (OR = 1.27, 95 % CI = 0.99–1.62). Sensitivity analyses were also conducted for the remaining two variants, and the pooled results were not statistically influenced when each study was sequentially omitted.
Publication bias
Begg’s funnel plot and Egger’s test were performed to evaluate the publication bias for rs231775, rs5742909, and rs3087243 polymorphisms. As shown in Fig. 3, the funnel plots do not evidence any obvious asymmetry (Begg: p = 0.10, p = 0.88, p = 0.62, respectively), and the results of Egger’s test suggest no evidence of publication bias (Egger: p = 0.36, p = 0.82, p = 0.84, respectively).
Discussion
Lymphoid tumors differ from other tumors in that the malignancy originates from the immune system itself. The vast majority of lymphoid neoplasms arise from mature B and T lymphocytes. Therefore, the role of immune pathways in lymphomagenesis is very complex. As an archetypal immune regulatory checkpoint, CTLA-4 plays an important role in T cell anergy and in T and B cell suppression responses [7]. Therefore, an abnormal expression of CTLA-4 may have an effect on the pathogenesis of lymphoid malignancies. Furthermore, SNPs in the CTLA-4 gene have been extensively considered to modify promoter activity and regulate the expression levels of CTLA-4 [27, 28].
In the present study, we summarized the association between three CTLA-4 gene SNPs (rs231775, rs5742909, and rs3087243) and the risk of lymphoid malignancies including lymphoma, multiple myeloma, and CLL. To our knowledge, this is the first meta-analysis focusing on CTLA-4 polymorphisms and lymphoid malignancy susceptibility. The results of our study suggest that the studied SNPs were not associated with an increased risk of lymphoid malignancies.
The rs231775 polymorphism of the CTLA-4 gene was investigated in several case-control studies on lymphoid malignancy risk; however, the results are controversial. Reports by Khorshied et al. [23] and Piras et al. [19] showed that rs231775 A alleles were associated with an increased risk of non-Hodgkin’s lymphoma; however, the sample sizes in the two studies were relatively small and included different histological types of lymphomas. Contrary results were presented by Liu et al. [22] and Bonzheim et al. [24], who showed that rs231775 variant was not associated with the risk of T cell non-Hodgkin’s lymphoma. Pavkovic et al. [17] found that rs231775 G allele was associated with increased risk of CLL. However, no significant association between this polymorphism and the risk of CLL development was observed by Suwalska et al. [21]. Our meta-analysis suggested that the rs231775 variant might not contribute to the risk of lymphoid malignancies in the overall population, in NHL population, or in Caucasian or Chinese populations.
The CTLA-4 rs5742909 polymorphism has been widely studied in several different cancers. However, reports on lymphoid malignancy patients remain scarce, and the association between this locus and lymphoid malignancy risk is inconclusive. A study carried out by Cheng et al. [20] examined the role of this SNP in MALT lymphoma development in Chinese population and evidenced that the rs5742909 CT genotype was associated with a lower risk of developing MALT lymphoma. Conversely, the lack of association between this variant and lymphoid malignancy susceptibility has been reported in Chinese populations by Liu et al. [22], in Egyptian populations by Khorshied et al. [23], and in Europe Caucasians by Suwalska et al. [21] and Bonzheim et al. [24]. The results of the present meta-analysis suggest that the rs5742909 polymorphism is not associated with lymphoid malignancy risk, although trends were observed in Caucasians based on the heterozygous model (CT vs. CC: 95 % CI = 1.00–1.86). Further multicenter clinical studies on the CTLA-4 rs5742909 polymorphism association with lymphoid malignancy risk in Caucasians are necessary.
The CTLA-4 rs3087243 polymorphism has been poorly explored in lymphoid malignancy case-control studies. A study involving 200 multiple myeloma patients and 380 controls showed that the rs3087243 G allele was associated with an increased risk of developing multiple myelomas in the Polish population [25]. Other studies by Khorshied et al. [23], Liu et al. [22], and Bonzheim et al. [24] had not observed a correlation between the rs3087243 polymorphism and lymphoid malignancy risk. The results of our meta-analysis support the negative findings of the latter studies, as no significant correlation was determined between CTLA-4 rs3087243 polymorphism and the risk of lymphoid malignancies.
Meta-analysis can overcome a few issues caused by a single study, such as small sample size, selection bias, and low test power; therefore, it is considered a powerful tool for integrating conflicted results from different studies [29]. However, it is inevitable to note limitations in our meta-analysis. First, significant heterogeneity was observed among some comparisons, especially for CTLA-4 rs231775. The heterogeneity may come from the classification of histopathological differences of lymphoid malignancies, the source of eligible studies in different countries, and the differences in gene detection methods. Although we analyzed NHL as a subgroup, it was not possible to conduct a further subgroup analysis based on a single tumor type, because of the lack of detailed data on pathological classification of malignant lymphomas in most of the eligible studies. Second, owing to the absence of detailed original data for conducting appropriate haplotype analysis, we could not assess potential effects of combined genotypes of these three SNPs. Third, tumorigenesis in humans is a multistep process; other factors such as gene-gene, gene-microbe, and gene-environment interactions may also contribute to the development of lymphoid malignancies. Finally yet importantly, we have not found published studies on the association between CTLA-4 gene polymorphisms and Hodgkin’s lymphoma risk.
In conclusion, our study represents, for the first time, a comprehensive meta-analysis of the role of CTLA-4 polymorphisms on lymphoid malignancy risk. The results of the present study indicate that the three CTLA-4 gene SNPs studied were not significantly associated with lymphoid malignancy susceptibility. More large-scale epidemiological studies are necessary to confirm our findings and clarify the real genetic effect.
References
Jaffe ES (2001) Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Iarc.
Sabattini E, Bacci F, Sagramoso C, Pileri SA (2010) WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: an overview. Pathologica 102(3):83–87
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63(1):11–30
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES (2016) The 2016 revision of the World Health Organization (WHO) classification of lymphoid neoplasms. Blood. doi:10.1182/blood-2016-01-643569
Knoechel B, Lohr JG (2013) Genomics of lymphoid malignancies reveal major activation pathways in lymphocytes. J Autoimmun 45:15–23
Baecklund E, Smedby KE, Sutton LA, Askling J, Rosenquist R (2014) Lymphoma development in patients with autoimmune and inflammatory disorders—what are the driving forces? Semin Cancer Biol 24:61–70
Armand P (2015) Immune checkpoint blockade in hematologic malignancies. Blood 125(22):3393–3400
Chambers CA, Sullivan TJ, Allison JP (1997) Lymphoproliferation in CTLA-4-deficient mice is mediated by costimulation-dependent activation of CD4+ T cells. Immunity 7(6):885–895
Leach DR, Krummel MF, Allison JP (1996) Enhancement of antitumor immunity by CTLA-4 blockade. Science 271(5256):1734–1736
Ribas A, Glaspy JA, Lee Y, Dissette VB, Seja E, Vu HT, Tchekmedyian NS, Oseguera D, Comin-Anduix B, Wargo JA, Amarnani SN, McBride WH, Economou JS, Butterfield LH (2004) Role of dendritic cell phenotype, determinant spreading, and negative costimulatory blockade in dendritic cell-based melanoma immunotherapy. J Immunother 27(5):354–367
Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29(2):233–237
Donner H, Rau H, Walfish PG, Braun J, Siegmund T, Finke R, Herwig J, Usadel KH, Badenhoop K (1997) CTLA4 alanine-17 confers genetic susceptibility to Graves’ disease and to type 1 diabetes mellitus. J Clin Endocrinol Metab 82(1):143–146
Hughes TA (2006) Regulation of gene expression by alternative untranslated regions. Trends Genet 22(3):119–122
Antczak A, Pastuszak-Lewandoska D, Gorski P, Domanska D, Migdalska-Sek M, Czarnecka K, Nawrot E, Kordiak J, Brzezianska E (2013) Ctla-4 expression and polymorphisms in lung tissue of patients with diagnosed non-small-cell lung cancer. Biomed Res Int 2013:576486
Perez-Garcia A, Osca G, Bosch-Vizcaya A, Kelleher N, Santos NY, Rodriguez R, Gonzalez Y, Roncero JM, Coll R, Serrando M, Lloveras N, Tuset E, Gallardo D (2013) Kinetics of the CTLA-4 isoforms expression after T-lymphocyte activation and role of the promoter polymorphisms on CTLA-4 gene transcription. Hum Immunol 74(9):1219–1224
Linsley PS, Greene JL, Brady W, Bajorath J, Ledbetter JA, Peach R (1994) Human B7-1 (CD80) and B7-2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity 1(9):793–801
Pavkovic M, Georgievski B, Cevreska L, Spiroski M, Efremov DG (2003) CTLA-4 exon 1 polymorphism in patients with autoimmune blood disorders. Am J Hematol 72(2):147–149
Monne M, Piras G, Palmas A, Arru L, Murineddu M, Latte G, Noli A, Gabbas A (2004) Cytotoxic T-lymphocyte antigen-4 (CTLA-4) gene polymorphism and susceptibility to non-Hodgkin’s lymphoma. Am J Hematol 76(1):14–18
Piras G, Monne M, Uras A, Palmas A, Murineddu M, Arru L, Bianchi A, Calvisi A, Curreli L, Gaviano E, Lai P, Murgia A, Latte GC, Noli A, Gabbas A (2005) Genetic analysis of the 2q33 region containing CD28-CTLA4-ICOS genes: association with non-Hodgkin’s lymphoma. Br J Haematol 129(6):784–790
Cheng TY, Lin JT, Chen LT, Shun CT, Wang HP, Lin MT, Wang TE, Cheng AL, Wu MS (2006) Association of T-cell regulatory gene polymorphisms with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. J Clin Oncol 24(21):3483–3489
Suwalska K, Pawlak E, Karabon L, Tomkiewicz A, Dobosz T, Urbaniak-Kujda D, Kuliczkowski K, Wolowiec D, Jedynak A, Frydecka I (2008) Association studies of CTLA-4, CD28, and ICOS gene polymorphisms with B-cell chronic lymphocytic leukemia in the Polish population. Hum Immunol 69(3):193–201
Liu J, Liu J, Song B, Wang T, Liu YH, Hao J, Yu JM (2013) Genetic variations in CTLA-4, TNF-alpha, and LTA and susceptibility to T-cell lymphoma in a Chinese population. Cancer Epidemiol 37(6):930–934
Khorshied MM, Gouda HM, Khorshid OM (2014) Association of cytotoxic T-lymphocyte antigen 4 genetic polymorphism, hepatitis C viral infection and B-cell non-Hodgkin lymphoma: an Egyptian study. Leuk Lymphoma 55(5):1061–1066
Bonzheim I, Geissinger E, Chuang WY, Roth S, Strobel P, Marx A, Reimer P, Wilhelm M, Puppe B, Rosenwald A, Muller-Hermelink HK, Rudiger T (2008) Analysis of single nucleotide polymorphisms in the FAS and CTLA-4 genes of peripheral T-cell lymphomas. J Hematop 1(1):11–21
Karabon L, Pawlak-Adamska E, Tomkiewicz A, Jedynak A, Kielbinski M, Woszczyk D, Potoczek S, Jonkisz A, Kuliczkowski K, Frydecka I (2012) Variations in suppressor molecule ctla-4 gene are related to susceptibility to multiple myeloma in a polish population. Pathol Oncol Res 18(2):219–226
Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES (2016) The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127(20):2375–2390
Ghaderi A (2011) CTLA4 gene variants in autoimmunity and cancer: a comparative review. Iran J Immunol 8(3):127–149
Banelli B, Morabito A, Laurent S, Piccioli P, Dozin B, Ghio M, Ascierto PA, Monteghirfo S, Marasco A, Ottaviano V, Queirolo P, Romani M, Pistillo MP (2014) A novel multiplex pyrosequencing assay for genotyping functionally relevant CTLA-4 polymorphisms: potential applications in autoimmunity and cancer. Hum Immunol 75(8):730–739
Munafo MR, Flint J (2004) Meta-analysis of genetic association studies. Trends Genet 20(9):439–444
Acknowledgments
This study was supported by the Science and Technology Foundation of Shaanxi Province, China (No. 2014K11020107).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Zhiming Dai and Chuanjie Feng contributed equally to this work.
Rights and permissions
About this article
Cite this article
Dai, Z., Feng, C., Zhang, W. et al. Lack of association between cytotoxic T-lymphocyte antigen-4 gene polymorphisms and lymphoid malignancy risk: evidence from a meta-analysis. Ann Hematol 95, 1685–1694 (2016). https://doi.org/10.1007/s00277-016-2753-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-016-2753-4