Abstract
Recent studies in adult lymphoma patients have indicated a correlation between polymorphisms of Fc gamma-receptors (FcγRs, encoded by the respective FCGR genes) and the response to rituximab treatment. In vitro, cells expressing FcγRIIIa-158V mediate antibody-dependent cellular cytotoxicity (ADCC) more efficiently than cells expressing FcγRIIIa-158F. The impact of the FCGR2A-131HR polymorphism is unclear. In this study, the FCGR polymorphisms FCGR3A-158VF and FCGR2A-131HR were analyzed in pediatric patients with mature aggressive B cell non-Hodgkin lymphoma/leukemia (B-NHL). Pediatric patients received a single dose of rituximab monotherapy. Response was evaluated on day 5 followed by standard chemotherapy for B-NHL. Among 105 evaluable patients, a response to rituximab was observed in 21 % of those homozygous for FcγRIIa-131RR (5/24) compared to 48 % of patients who were HH and HR FcγRIIa-131 allele carriers (18/34 and 21/47, respectively; p = 0.044). Among patients with the FCGR3A-158 polymorphism, those homozygous for the FF genotype had a significantly favorable rituximab response rate of 59 % (22/37) compared to 32 % in patients who were FcγRIIIa-158VV and FcγRIIIa-VF allele carriers (2/9 and 20/59, respectively; p = 0.022). A stringent phase II response evaluation of children and adolescents with B-NHL after one dose of rituximab monotherapy showed a significant association between the rituximab response rate and FCGR polymorphisms. These findings support the hypothesis that FCGR polymorphisms represent patient-specific parameters that influence the response to rituximab.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Current chemotherapy regimens result in event-free survival (EFS) rates of 65–100 % for pediatric and adolescent patients with mature B cell non-Hodgkin lymphoma (B-NHL) or leukemia (B-AL) [1–8]. Due to the significant acute toxicities of these treatment regimens and the poor rescue chances at the time of relapse, new treatment strategies with personalized and targeted therapies are needed. Rituximab, a chimeric monoclonal anti-CD20 immunoglobulin G1 (IgG1) antibody widely used in adult lymphoma patients, currently represents one of the most promising candidates to accomplish these goals in pediatric patients.
The efficacy of rituximab treatment in adults is influenced by the characteristics of the lymphoma cells and by those of the host [9]. According to recent studies, genetic polymorphisms of Fc gamma-receptors (FcγRs), which recognize IgGs, are a host parameter with potential prognostic value. FcγRs play an essential role in antibody-dependent cellular cytotoxicity (ADCC), by linking the rituximab-targeted CD20-positive cells to cytotoxic cells. FcγRIIIa is expressed on natural killer (NK) cells and macrophages, the most important ADCC effector cells. A genetic polymorphism (rs396991) of the encoding gene (FCGR3A) results in either a valine (V) or a phenylalanine (F) at amino acid position 158 of FcγRIIIa. Human IgG1 binds more strongly to homozygous FcγRIIIa-158VV NK cells than to homozygous FcγRIIIa-158FF or heterozygous FcγRIIIa-158VF NK cells in vitro [10, 11]. Accordingly, an association between FcγRIIIa-158V and favorable rituximab response compared to FcγRIIIa-158F has been identified in some clinical trials [12–18], although not in others [19–27]. Moreover, the polymorphism (rs1801274) of FCGR2A, expressed on neutrophils and mononuclear phagocytes, in which either a histidine (H) or arginine (R) at amino acid position 131 of FcγRIIa has been analyzed for its prognostic impact on rituximab response. Association of rituximab response with FCGR2A-131 could be shown in two trials, while other clinical trials failed to show an association with response to treatment [16, 20–22, 24, 26, 28, 29].
Besides the FCGR genotype, biology of the disease, administration of rituximab as monotherapy or combined with chemotherapy, pre-treatment of patients, time and method of response, and outcome evaluation, impact the evaluated rituximab response. Rituximab serum levels and thus the efficacy of this drug are also influenced by tumor burden and antigenic mass [9].
Taken together, in vitro and in vivo data indicate that FCGR2A-158 and FCGR3A-131 polymorphisms influence the therapeutic activity of rituximab [30, 31]. The objective of this study was to evaluate the prognostic implications of FCGR2A-158 and FCGR3A-131 polymorphisms in pediatric and adolescent patients with newly diagnosed mature B-NHL treated in a phase II clinical trial in which a single dose of rituximab was administered 5 days prior to standardized response evaluation and the start of standard chemotherapy. The advantage of the study was its well-standardized clinical setting, which allowed systematic investigations with potential implications for future treatment. The confirmation of FCGR polymorphisms as a relevant patient-specific prognostic parameter for rituximab response could allow patient-specific dosing schedules of rituximab in a personalized medicine approach to treatment.
Methods
Patients, treatment, and response evaluation
All analyzed patients were enrolled after written informed consent for participation in the B-NHL BFM (Berlin-Frankfurt-Muenster) rituximab trial [32, 33]. This was obtained from the parents of the patients or from their legal guardians. Eligibility criteria were newly diagnosed CD20-positive mature B-NHL or B-AL, age below 19 years, and at least one index lesion available for response evaluation within 24 h prior to rituximab administration. Material for FCGR genotyping was available for 128 patients. B-NHL subtypes were classified according to the classification system of the World Health Organization and reviewed centrally. Initial staging was performed according to the St. Jude staging system [34]. Studies associated with the B-NHL BFM rituximab trial were approved by the Ethical Board of the Justus Liebig University, Giessen, Germany. The research was conducted in accordance with the Declaration of Helsinki.
Patients received one dose of rituximab, either 375 mg/m2 (88 patients) or 700 mg/m2 (40 patients), intravenously on day 1 of treatment. The dose of rituximab was escalated from 375 to 700 mg/m2 after an interim analysis of the phase II trial [33, 32]. For initial staging, the most appropriate imaging method was used, either ultrasound, magnetic resonance imaging, or computed tomography. Within 24 h before starting rituximab administration, at least one index lesion was identified and then measured by determining the product of the two largest perpendicular diameters. In addition, the ratio of blasts in bone marrow (BM) to those in peripheral blood (PB) was determined. Re-staging after rituximab treatment was performed with identical imaging methods and instrument settings and by the same investigator on day 5 and prior to the beginning of chemotherapy. In patients with initial BM/PB blasts, this ratio was re-examined as well. Evaluation records were reviewed centrally to assess the rituximab response. Patients with stable or progressive disease were classified as non-responders, defined as a <25 % reduction of the tumor or disease progression. For the early detection of tumor lysis syndrome (TLS), serum lactate dehydrogenase (LDH) levels were evaluated prior to and 4, 8, 12, 16, and 20 h after the start of rituximab infusion and on days 2, 3, 4, 5, and 10. Following this window, the patients were treated according to the B-NHL BFM 04 trial. Patients did not receive any form of chemotherapy and/or corticosteroids during the window, except for patients with central nervous system (CNS) disease, who received intrathecal therapy on days 1 and 3. Patients requiring corticosteroid therapy during the rituximab window (due to anaphylactic reactions to rituximab) were excluded from the response analyses.
Determination of FCGR2A-131 and FCGR3A-158 genotypes
PB and BM samples with sufficient quality and quantity were available for 128 patients. Mononuclear cells were isolated from PB or BM with Lymphoprep (Axis Shield, Oslo, Norway) using standard protocols. Genomic DNA was isolated from mononuclear cells using the QIAmp DNA blood mini-kit (Qiagen, Hilden, Germany). FCGR3A-158VF (rs396991) was genotyped using a Taqman-based assay for rs396991 (Life Technologies, Darmstadt, Germany). The probe specific for the V allele was labeled with VIC, and the probe for the F allele with FAM. Genotyping was carried out using an Applied Biosystems 7500 real-time PCR system. Allelic discrimination was performed using SDS software v1.4 (Life Technologies).
The FCGR2A-131HR polymorphism (rs1801274) was analyzed according to a PCR-based approach. PCR amplification was carried out using the following oligonucleotides: 5′-GGAAAATCCCAGAAATTCTCGC-3′ (forward) and 5′-CAACAGCCTGACTACCTATTACGCGGG-3′ (reverse) [35]. The forward primer introduced a BstUI restriction site if the nucleotide at the polymorphic site was G. The reverse primer introduced a BstUI site in all PCR products. PCR was performed with 200 ng of DNA, 1 unit (U) of AccuPrime Taq high fidelity DNA polymerase (Life Technologies), 10 pmol of each primer, and 1× AccuPrime PCR buffer II in a volume of 50 μl and standard amplification conditions. The PCR products were purified using the Illustra GFX PCR DNA and gel band purification kit (GE Healthcare, Freiburg, Germany) and digested with 20 U BstUI (New England Biolabs, Frankfurt, Germany) overnight at 60 °C. Samples were analyzed on 3 % agarose gels and visualized by ethidium bromide staining. Subgroups of FCGR2A-131 and FCGR3A-158 genotypes were evaluated according to tumor histology and risk group. B-NHL histology was classified as Burkitt’s lymphoma or leukemia (BL, B-AL) or diffuse large B cell lymphoma (DLBCL). Risk group was dichotomized as low-risk (R2 patients) or high-risk (R3/R4 patients). The R2 risk group was defined by the absence of CNS and BM involvement and an initial LDH serum level < 500 U/l. The higher risk groups, R3 and R4, consisted of patients with stage III or IV disease and an initial LDH level > 500 U/l.
Statistical analyses
Associations of FCGR genotypes with rituximab response, B-NHL subtypes, involved compartments and LDH level were analyzed using the χ 2 test or Fisher’s exact test. Cumulative incidences of relapse were calculated according to Kalbfleisch and Prentice [36]. The association of FCGR2A-131 and FCGR3A-158 genotypes with rituximab response was calculated using the Cochran-Armitage trend test for analyzing the association of two variables, the second of them with more than two categories. This modified Pearson χ 2 test is able to incorporate a suspected ordering in the effects of the categories of the second variable [37]. The areas under the curves (AUCs) of the logarithmic values of the individual changes in LDH serum level compared to the initial LDH level were also determined. The values were calculated with the rectangle method. Missing values were replaced by the previous value (last value carried forward) or, in case only one value was missing, by the mean of the neighboring values. Data were updated as of July 2015.
Results
Material for genotyping was evaluable in 128 pediatric B-NHL patients with the FCGR2A-131 polymorphism and in 127 pediatric patients with the FCGR3A-158 polymorphism. The patients characteristics are summarized in Table 1. Their comparison according to FCGR status revealed no significant associations except for a higher proportion of female patients in the FCGR2A-131RR subgroup (Table 1). The distribution of FCGR genotypes was similar to that described for other populations of patients with lymphoma [38].
Clinical response
After the exclusion of 23 patients whose response evaluation was not according to protocol guidelines or who received steroids during the 5-day window, a total of 105 patients with evaluable response data and known FCGR status were available for analyses of the prognostic impact of FCGR polymorphisms on rituximab response. The response rate was better in patients with the low-affinity FCGR3A-158FF genotype than in patients carrying the high-affinity valine allele. Patients with the former genotype had a response rate of 59 % (22/37) compared to 22 % for patients homozygous for the valine isotypes (2/9) and 33.9 % for heterozygous patients (20/59 patients; p = 0.02) (Table 1). Additional parameters with potential impact on rituximab response were analyzed within each patient subgroup defined by their FCGR3A genotype. FCGR3A-158VF patients had a significantly better response rate when the fluid compartment rather than solid lymphoma was used for response evaluation (p = 0.0005). Neither histological B-NHL subtype nor rituximab dose was significantly associated with rituximab response in these subgroups (Table 2).
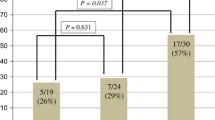
In addition to FCGR3A, we analyzed the prognostic impact of FCGR2A genotype status. A response to rituximab was observed in 21 % of patients with homozygous for FCGR2A-131RR compared to 53 % and 45 % of the FCGR2A-131 histidine allele carriers (HH and HR respectively; p = 0.04). Superior rituximab response rates were significantly associated with the R3/R4 risk group of FCGR2A-131HR patients [65 % (13/20) vs. 31 % (8/26) in the R2 risk group; p = 0.04] and fluid lymphoma compartment of FCGR2A-131HH patients. A response rates of 100 % (8/8) within the BM compartment contrasted with the 38 % (10/26) response rate of nodal lesions (p = 0.003) (Table 3).
Association of rituximab response and LDH serum level changes
For the early detection of TLS, serum LDH levels were closely monitored, with control carried out at 4 h intervals on the day of rituximab infusion followed by daily controls until day 5. Follow-up data on serum LDH levels were available for 119 patients, with 104 also evaluable for rituximab response. In 34 out of 119 patients (29 %), follow-up serum LDH levels did not exceed the individual upper normal limit until day 5. In the remaining 85 patients, the maximum LDH level was reached between 4 h and 5 days following rituximab infusion. Since the majority of LDH changes occurred within 48 h, this 48 h interval was analyzed in greater detail. Changes in the LDH serum level compared to the initial LDH level were quantified by calculating the AUC of all 96 patients for whom initial LDH levels and follow-up levels on day 2 were available, together with a sufficient rituximab response evaluation on day 5 and FCGR genotyping. Serum LDH changes were more prominent in patients who responded to rituximab than in rituximab non-responders (Table 4). This difference was seen in all genetic subgroups and reached statistical significance in FCGR3A-158VF, FCGR2A-131HR, and FCGR2A-131-HH patients.
The changes in LDH serum level according to rituximab response within the subgroup of 36 patients with FCGR3A-158FF genotype is exemplarily shown in Fig. 1, which compares the LDH changes to both the initial LDH levels, prior to the start of rituximab infusion (Fig. 1 a, b), and the individual upper normal limit of each patient (Fig. 1 c, d).
Changes in serum LDH levels exemplarily shown for patients with the FCGR3A-158FF genotype, available initial and follow-up LDH levels, and sufficient rituximab response evaluation on day 5. Shown are the changes in the LDH serum levels compared to the initial LDH level in rituximab responders (a) and non-responders (b) and according to the individual upper normal limit (UNL) in rituximab responders (c) and non-responders (d). The hours (h) or days (d) after the start of rituximab infusion are indicated
Multivariate analysis was carried out with the co-variables rituximab dose, evaluated compartment, disease extent, FCGR2A-131 and FCGR3A-158 genotypes, and quantitative changes in serum LDH levels. FCGR2A-131 and FCGR3A-158 genotypes and changes in serum LDH levels remained statistically significant prognostic parameters for rituximab response on day 5 (Table 5).
Outcome
Among the evaluated patient cohort, nine patients suffered disease relapse (7 % of evaluated patients). The incidences of the FCGR3A-158 variants VV, VF, and FF for these nine patients were 0 (0 %), 8 (89 %), and 1 (11 %) compared to 13 (11 %), 61 (52 %), and 44 (37 %) in the 118 patients without relapse, respectively as illustrated in Fig. 2. The VF polymorphism was not evaluable in one patient without relapse. The RR, HR, and HH FCGR2A-131 genotypes were detected in 1 (11 %), 5 (56 %), and 3 (33 %) of patients with disease relapse compared to 25 (21 %), 56 (47 %), and 38 (32 %) of the 119 patients without relapse. The cumulative incidences of relapse were similar for all genetic subgroups.
Outcome of the evaluable patients according to FCGR3A-158 genotypes illustrated in absolute numbers of patients. Shown is the distribution of relapsed and non-relapsed patients according to FCGR3A-158-genotype. The number of patients with relapse is indicated in dark gray, the number of patients without relapse is shown in light gray
Discussion
With the goal of individualized treatment of patients with B-NHL, relevant efforts have been undertaken to elucidate the mechanisms responsible for the interindividual variability in the response to rituximab, with most evidence pointing to FCGR polymorphisms. However, according to the published data in the literature, the impact of these polymorphisms on the effectiveness of rituximab treatment is unclear [12–24]. An important question is whether data obtained in adult patients can be transferred to pediatric patients because of relevant differences in the treatment, response evaluation, outcome, and biology of the disease between the two age groups. In fact, there are as yet no data on the role of FCGR polymorphisms in the rituximab response of children and adolescents with B-NHL.
The rituximab window-trial, described herein, was conducted by the NHL-BFM group with the aim of systematically investigating the rituximab response in pediatric mature B-NHL patients. The major strengths of the study were the large number of patients with uniform treatment and the stringent response evaluation after the rituximab window. All patients in the study were simultaneously enrolled in a phase II trial with the endpoint rituximab response, the response evaluation of our study was highly standardized concerning timelines and procedure. All cases were centrally reviewed, with the strict exclusion of patients in case of protocol violations. Almost 130 of the patients enrolled in the trial were genotyped for FCGR2A and FCGR3A status, which was then correlated with clinical characteristics, rituximab response, and treatment outcome.
Patient characteristics were similar among the subgroups defined by FCGR2A and FCGR3A genotypes, indicating no association of specific genotypes with specific patient characteristics. However, the rituximab response rates of the genetic subgroups significantly differed, with a much higher response rate in patients with the FCGR2A-131HH and FCGR2A-131HR genotypes than in those with the RR genotype. This finding is in line with the clinical observations of two other trials [13, 28], although seven clinical trials failed to show an association with response to treatment [16, 18–22, 24]. The homozygous low-affinity FCGR3A-158FF genotype was significantly associated with a favorable rituximab response compared to patients carrying the high-affinity valine allele. This is in contrast to the clinical observations reported for adult lymphoma patients, in whom either no prognostic impact or a lack of a favorable response was determined in carriers of the valine allele [12, 13, 15–22, 24, 25, 27, 38]. An association of the FCGR3A-158FF genotype with unanticipated clinical findings was noted in three other studies: Two of those studies, on indolent and aggressive lymphoma or chronic lymphocytic leukemia (CLL), reported a trend towards a favorable rituximab response and/or outcome in FCGR3A-158FF patients [20, 23]. The third study analyzed a series of 24 lymphoma patients and found that the FCGR3A-158FF genotype correlated significantly with lower immunoglobulin levels after autologous stem cell transplantation and adjuvant rituximab [39]. The precise mechanisms underlying these lower immunoglobulin levels are unknown.
Capuano and colleagues showed that NK cell interaction with Rituximab opsonized target cells resulted in a downmodulation of FcγRIIIa. Furthermore the ability of NK cells to kill target cells was impaired after the interaction with the rituximab opsonized target cells [40]. In this context the low-affinity homozygous FcγRIIIa-F receptor may contribute to a more favorable NK cell responsiveness compared to the FcγRIIIa-158VF or VV receptors.
In terms of the prognostic impact of FCGR polymorphisms in the context of lymphoma biology and disease extent, the response rates of patients homozygous for the FCGR3A-158FF genotype were favorable throughout all analyzed subgroups with no significant differences according to lymphoma histology, risk group stratification, and analyzed compartment (Table 2).
Our analyses also indicated a potential prognostic impact of FCGR2A-131 polymorphisms on rituximab response. In the group of favorably responding patients with FCGR2A-131HH genotypes, a good rituximab response was achieved in those with a BM response. This is in line with earlier reports of a faster and superior response in fluid compartments compared to solid lymphoma [33, 41]. Increasing the dose of rituximab from 375 mg/m2 to 700 mg/m2 did not enhance the rituximab response rates, except in FCGR2A-131RR patients. Larger series will be necessary to evaluate the role of the rituximab dose in the context of FCGR2A-131 and FCGR3A-158 polymorphisms. The small number of patients with disease relapse (n = 9) precluded detailed outcome analyses according to genetic subgroups; thus, larger patient series are required here as well. Therefore, the current analyses will be continued in the upcoming clinical trial of the NHL-BFM group.
Analysis of early changes in LDH serum levels revealed larger increases in patients with a clinical response to rituximab than in non-responders. Thus, a sufficient rituximab response might correlate with cell lysis and the liberation of cytoplasmic LDH.
Other relevant mechanisms of action of monoclonal antibodies besides ADCC are complement-dependent cytotoxicity (CDC), phagocytosis, and direct apoptosis induction [42]. As a type I anti-CD20 antibody, rituximab demonstrates efficient phagocytosis, ADCC, and CDC but it does not directly induce apoptosis. This contrasts with type II antibodies, which strongly induce apoptosis but only weakly activate CDC [42]. Thus, the most important mechanisms of action of rituximab seem to be ADCC and CDC, while a direct induction of apoptosis is probably negligible. None of the available parameters in the current study allowed the observed response to be attributed to CDC vs. ADCC. But in line with the hypothesis that CDC occurs immediately after the start of rituximab therapy while ADCC requires a certain latency period, the observed kinetics of the LDH levels, as an indirect parameter of tumor cell lysis, support ADCC as the relevant mechanism of rituximab response in patients with newly diagnosed B-NHL. However, the experimental setup of this study might also have measured mechanisms other than ADCC.
Our results show that a detailed evaluation of host genotype characteristics in combination with disease parameters, such as histology, disease extent, and/or involved compartments, can identify subgroups likely to exhibit a specific response to rituximab. Analyses of larger patient series are necessary to allow correlation with disease outcome and to achieve the long-term goal of personalized treatment, by modifying rituximab administration according to the individual response profile.
References
Gerrard M, Cairo MS, Weston C, Auperin A, Pinkerton R, Lambilliote A, Sposto R, McCarthy K, Lacombe MJ, Perkins SL, Patte C, Committee FLIS (2008) Excellent survival following two courses of COPAD chemotherapy in children and adolescents with resected localized B-cell non-Hodgkin’s lymphoma: results of the FAB/LMB 96 international study. Br J Haematol 141(6):840–847. doi:10.1111/j.1365-2141.2008.07144.x
Cairo MS, Gerrard M, Sposto R, Auperin A, Pinkerton CR, Michon J, Weston C, Perkins SL, Raphael M, McCarthy K, Patte C (2007) Results of a randomized international study of high-risk central nervous system B non-Hodgkin lymphoma and B acute lymphoblastic leukemia in children and adolescents. Blood 109(7):2736–2743. doi:10.1182/blood-2006-07-036665
Patte C, Auperin A, Gerrard M, Michon J, Pinkerton R, Sposto R, Weston C, Raphael M, Perkins SL, McCarthy K, Cairo MS, Committee FLIS (2007) Results of the randomized international FAB/LMB96 trial for intermediate risk B-cell non-Hodgkin lymphoma in children and adolescents: it is possible to reduce treatment for the early responding patients. Blood 109(7):2773–2780. doi:10.1182/blood-2006-07-036673
Woessmann W, Seidemann K, Mann G, Zimmermann M, Burkhardt B, Oschlies I, Ludwig WD, Klingebiel T, Graf N, Gruhn B, Juergens H, Niggli F, Parwaresch R, Gadner H, Riehm H, Schrappe M, Reiter A, Group BFM (2005) The impact of the methotrexate administration schedule and dose in the treatment of children and adolescents with B-cell neoplasms: a report of the BFM Group Study NHL-BFM95. Blood 105(3):948–958. doi:10.1182/blood-2004-03-0973
Patte C, Auperin A, Michon J, Behrendt H, Leverger G, Frappaz D, Lutz P, Coze C, Perel Y, Raphael M, Terrier-Lacombe MJ (2001) The Societe Francaise d’Oncologie Pediatrique LMB89 protocol: highly effective multiagent chemotherapy tailored to the tumor burden and initial response in 561 unselected children with B-cell lymphomas and L3 leukemia. Blood 97(11):3370–3379. doi:10.1182/blood.V97.11.3370
Link MP, Shuster JJ, Donaldson SS, Berard CW, Murphy SB (1997) Treatment of children and young adults with early-stage non-Hodgkin’s lymphoma. N Engl J Med 337(18):1259–1266. doi:10.1056/NEJM199710303371802
Pillon M, Arico M, Mussolin L, Mainardi C, Giraldi E, Garaventa A, Lombardi A, Santoro N, Carraro E, d’Amore ES, Rosolen A, Italian Association of Pediatric Hematology OAN-HLWG (2014) Mediastinal Burkitt lymphoma in childhood. Pediatr Blood Cancer 61(11):2127–2128. doi:10.1002/pbc.25127
Tsurusawa M, Mori T, Kikuchi A, Mitsui T, Sunami S, Kobayashi R, Takimoto T, Saito A, Watanabe T, Fujimoto J, Nakazawa A, Ohshima K, Horibe K, lymphoma committee of Japanese Pediatric Leukemia/Lymphoma Study G (2014) Improved treatment results of children with B-cell non-Hodgkin lymphoma: a report from the Japanese pediatric leukemia/lymphoma study group B-NHL03 study. Pediatr Blood Cancer 61(7):1215–1221. doi:10.1002/pbc.24975
Cartron G, Trappe RU, Solal-Celigny P, Hallek M (2011) Interindividual variability of response to rituximab: from biological origins to individualized therapies. Clin Cancer Res 17(1):19–30. doi:10.1158/1078-0432.CCR-10-1292
Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, Salmon JE, Kimberly RP (1997) A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest 100(5):1059–1070. doi:10.1172/JCI119616
Mishima Y, Terui Y, Mishima Y, Kuniyoshi R, Matsusaka S, Mikuniya M, Kojima K, Hatake K (2012) High reproducible ADCC analysis revealed a competitive relation between ADCC and CDC and differences between FcgammaRllla polymorphism. Int Immunol 24(8):477–483. doi:10.1093/intimm/dxs048
Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H (2002) Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 99(3):754–758
Weng WK, Levy R (2003) Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol 21(21):3940–3947. doi:10.1200/JCO.2003.05.013
Persky DO, Dornan D, Goldman BH, Braziel RM, Fisher RI, Leblanc M, Maloney DG, Press OW, Miller TP, Rimsza LM (2012) Fc gamma receptor 3a genotype predicts overall survival in follicular lymphoma patients treated on SWOG trials with combined monoclonal antibody plus chemotherapy but not chemotherapy alone. Haematologica 97(6):937–942. doi:10.3324/haematol.2011.050419
Ghielmini M, Rufibach K, Salles G, Leoncini-Franscini L, Leger-Falandry C, Cogliatti S, Fey M, Martinelli G, Stahel R, Lohri A, Ketterer N, Wernli M, Cerny T, Schmitz SF (2005) Single agent rituximab in patients with follicular or mantle cell lymphoma: clinical and biological factors that are predictive of response and event-free survival as well as the effect of rituximab on the immune system: a study of the Swiss Group for Clinical Cancer Research (SAKK). Ann Oncol 16(10):1675–1682. doi:10.1093/annonc/mdi320
Kim DH, Jung HD, Kim JG, Lee JJ, Yang DH, Park YH, Do YR, Shin HJ, Kim MK, Hyun MS, Sohn SK (2006) FCGR3A gene polymorphisms may correlate with response to frontline R-CHOP therapy for diffuse large B-cell lymphoma. Blood 108(8):2720–2725. doi:10.1182/blood-2006-01-009480
Zhang W, Wang X, Li J, Duan MH, Zhou DB (2010) Fcgamma receptor IIIA polymorphisms and efficacy of rituximab therapy on Chinese diffuse large B-cell lymphoma. Chin Med J 123(2):198–202
Cornec D, Tempescul A, Querellou S, Hutin P, Pers JO, Jamin C, Bendaoud B, Berthou C, Renaudineau Y, Youinou P (2012) Identification of patients with indolent B cell lymphoma sensitive to rituximab monotherapy. Ann Hematol 91(5):715–721. doi:10.1007/s00277-011-1369-y
Carlotti E, Palumbo GA, Oldani E, Tibullo D, Salmoiraghi S, Rossi A, Golay J, Pulsoni A, Foa R, Rambaldi A (2007) FcgammaRIIIA and FcgammaRIIA polymorphisms do not predict clinical outcome of follicular non-Hodgkin’s lymphoma patients treated with sequential CHOP and rituximab. Haematologica 92(8):1127–1130
Farag SS, Flinn IW, Modali R, Lehman TA, Young D, Byrd JC (2004) Fc gamma RIIIa and Fc gamma RIIa polymorphisms do not predict response to rituximab in B-cell chronic lymphocytic leukemia. Blood 103(4):1472–1474. doi:10.1182/blood-2003-07-2548
Dornan D, Spleiss O, Yeh RF, Duchateau-Nguyen G, Dufour A, Zhi J, Robak T, Moiseev SI, Dmoszynska A, Solal-Celigny P, Warzocha K, Loscertales J, Catalano J, Afanasiev BV, Larratt L, Rossiev VA, Bence-Bruckler I, Geisler CH, Montillo M, Wenger MK, Weisser M (2010) Effect of FCGR2A and FCGR3A variants on CLL outcome. Blood 116(20):4212–4222. doi:10.1182/blood-2010-03-272765
Galimberti S, Palumbo GA, Caracciolo F, Benedetti E, Pelosini M, Brizzi S, Ciabatti E, Fazzi R, Stelitano C, Quintana G, Conte E, Tibullo D, Di Raimondo F, Petrini M (2007) The efficacy of rituximab plus hyper-CVAD regimen in mantle cell lymphoma is independent of FCgammaRIIIa and FCgammaRIIa polymorphisms. J Chemother 19(3):315–321
Pennell NM, Bhanji T, Zhang L, Seth A, Sawka CA, Berinstein NL (2008) Lack of prognostic value of FCGR3A-V158F polymorphism in non-Hodgkin’s lymphoma. Haematologica 93(8):1265–1267. doi:10.3324/haematol.12638
Mitrovic Z, Aurer I, Radman I, Ajdukovic R, Sertic J, Labar B (2007) FCgammaRIIIA and FCgammaRIIA polymorphisms are not associated with response to rituximab and CHOP in patients with diffuse large B-cell lymphoma. Haematologica 92(7):998–999
Varoczy L, Zilahi E, Gyetvai A, Kajtar B, Gergely L, Sipka S, Illes A (2012) Fc-gamma-receptor IIIa polymorphism and gene expression profile do not predict the prognosis in diffuse large B-cell lymphoma treated with R-CHOP protocol. Pathol Oncol Res 18(1):43–48. doi:10.1007/s12253-011-9414-7
Ahlgrimm M, Pfreundschuh M, Kreuz M, Regitz E, Preuss KD, Bittenbring J (2011) The impact of Fc-gamma receptor polymorphisms in elderly patients with diffuse large B-cell lymphoma treated with CHOP with or without rituximab. Blood 118(17):4657–4662. doi:10.1182/blood-2011-04-346411
Martinelli G, Schmitz SF, Utiger U, Cerny T, Hess U, Bassi S, Okkinga E, Stupp R, Stahel R, Heizmann M, Vorobiof D, Lohri A, Dietrich PY, Zucca E, Ghielmini M (2010) Long-term follow-up of patients with follicular lymphoma receiving single-agent rituximab at two different schedules in trial SAKK 35/98. J Clin Oncol 28(29):4480–4484. doi:10.1200/JCO.2010.28.4786
Paiva M, Marques H, Martins A, Ferreira P, Catarino R, Medeiros R (2008) FcgammaRIIa polymorphism and clinical response to rituximab in non-Hodgkin lymphoma patients. Cancer Genet Cytogenet 183(1):35–40. doi:10.1016/j.cancergencyto.2008.02.001
Levy D, Bellesso M, Oliveira-Souza P, Maciel FV, Pereira J, Bydlowski SP (2011) The H/R FcgammaRIIA-131 polymorphism and survival in patients with diffuse large B-cell lymphoma (DLBCL) treated with R-CHOP: a study in a genetically mixed population. Clinics (Sao Paulo) 66(5):919–922
Clynes R (2007) IVIG therapy: interfering with interferon-gamma. Immunity 26(1):4–6. doi:10.1016/j.immuni.2007.01.006
Manches O, Lui G, Chaperot L, Gressin R, Molens JP, Jacob MC, Sotto JJ, Leroux D, Bensa JC, Plumas J (2003) In vitro mechanisms of action of rituximab on primary non-Hodgkin lymphomas. Blood 101(3):949–954. doi:10.1182/blood-2002-02-0469
Lisfeld J, Burkhardt B, Meinhardt A, Zimmermann M, Kabıckova E, Bielack S, Kontny U, Gnekow A, Sauerbrey A, Reiter A (2012) Phase II window study on rituximab in newly diagnosed pediatric mature B-cell non-Hodgkin’s lymphoma and Burkitt leukemia: dose-escalation does not increase the response rate. Br J Haematol 112(Suppl. 2012):Abstract 6
Meinhardt A, Burkhardt B, Zimmermann M, Borkhardt A, Kontny U, Klingebiel T, Berthold F, Janka-Schaub G, Klein C, Kabickova E, Klapper W, Attarbaschi A, Schrappe M, Reiter A, Berlin-Frankfurt-Munster g (2010) Phase II window study on rituximab in newly diagnosed pediatric mature B-cell non-Hodgkin’s lymphoma and Burkitt leukemia. J Clin Oncol 28(19):3115–3121. doi:10.1200/JCO.2009.26.6791
Murphy SB (1980) Classification, staging and end results of treatment of childhood non-Hodgkin’s lymphomas: dissimilarities from lymphomas in adults. Semin Oncol 7(3):332–339
Jiang XM, Arepally G, Poncz M, McKenzie SE (1996) Rapid detection of the Fc gamma RIIA-H/R 131 ligand-binding polymorphism using an allele-specific restriction enzyme digestion (ASRED). J Immunol Methods 199(1):55–59
Kalbfleisch JD, Prentice RL (1980) The statistical analysis of failure time data. John Wiley, New York, pp. 163–188
Armitage P (1955) Test for linear trends in proportions and frequencies. Biometrics 11:375–386
Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A (2013) A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol 6:1. doi:10.1186/1756-8722-6-1
Nishio M, Endo T, Fujimoto K, Yamamoto S, Obara M, Yamaguchi K, Takeda Y, Goto H, Kasahara I, Sato N, Koike T (2009) FCGR3A-158V/F polymorphism may correlate with the levels of immunoglobulin in patients with non-Hodgkin’s lymphoma after rituximab treatment as an adjuvant to autologous stem cell transplantation. Eur J Haematol 82(2):143–147. doi:10.1111/j.1600-0609.2008.01174.x
Capuano C, Romanelli M, Pighi C, Cimino G, Rago A, Molfetta R, Paolini R, Santoni A, Galandrini R (2015) Anti-CD20 therapy acts via FcgammaRIIIA to diminish responsiveness of human natural killer cells. Cancer Res 75(19):4097–4108. doi:10.1158/0008-5472.CAN-15-0781
Gong Q, Ou Q, Ye S, Lee WP, Cornelius J, Diehl L, Lin WY, Hu Z, Lu Y, Chen Y, Wu Y, Meng YG, Gribling P, Lin Z, Nguyen K, Tran T, Zhang Y, Rosen H, Martin F, Chan AC (2005) Importance of cellular microenvironment and circulatory dynamics in B cell immunotherapy. J Immunol 174(2):817–826
Okroj M, Osterborg A, Blom AM (2013) Effector mechanisms of anti-CD20 monoclonal antibodies in B cell malignancies. Cancer Treat Rev 39(6):632–639. doi:10.1016/j.ctrv.2012.10.008
Acknowledgments
The authors thank Gabriele Buck (cytomorphology), Bettina Paul, and Ulrike Meyer (data management) for their expert work. We also thank the physicians, radiologists, nurses, and data managers at the participating hospitals who cared for the children and supplied the data. This work was supported by “Forschungshilfe Peiper”, Giessen, Germany. EK was supported by a project of the Czech Ministry of Health: project for conceptual development of research organization 00064203.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Rights and permissions
About this article
Cite this article
Burkhardt, B., Yavuz, D., Zimmermann, M. et al. Impact of Fc gamma-receptor polymorphisms on the response to rituximab treatment in children and adolescents with mature B cell lymphoma/leukemia. Ann Hematol 95, 1503–1512 (2016). https://doi.org/10.1007/s00277-016-2731-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-016-2731-x