Abstract
Background
In the past few years, immunotherapy has changed the way we treat solid tumors. People pay more and more attention to the immune microenvironment of laryngeal squamous cell carcinoma (LSCC). In this study, our immunotherapy research took advantage of the clinical database and focused our in-depth analysis on the tumor microenvironment (TME).
Methods
This study evaluated the relationship between the clinical outcome and the local tissue and overall immune status in 412 patients with primary LSCC. We constructed and validated a risk model that could predict prognosis, assess immune status, identify high-risk patients, and develop personalized treatment plans through bioinformatics. In addition, through immunohistochemical analysis, we verified the differential expression of CTSL and KDM5D genes with the largest weight coefficients in the model in LSCC tissues and their influence on the prognosis and tumor-infiltrating lymphocytes (TILs).
Results
We found that interstitial tumor-infiltrating lymphocytes, tumor parenchymal-infiltrating lymphocyte volume, tumor infiltrates lymphocytes of frontier invasion, and the platelet-to-lymphocyte ratio (PLR) were independent factors affecting the prognosis of patients with LSCC. A novel risk model can guide clinicians to accurately predict prognosis, identify high-risk patients, and formulate personalized treatment plans. The differential expression of genes such as CTSL and KDM5D has a significant correlation with the TILs of LSCC and the prognosis of patients.
Conclusion
Local and systemic inflammatory markers in patients with laryngeal squamous cell carcinoma are reliable prognostic factors. The risk model and CTSL, KDM5D gene have important potential research value.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Laryngeal squamous cell carcinoma (LSCC) is currently one of the most common head and neck malignancies and the second most common cancer in respiratory tumors after lung cancer [1]. There are approximately 151,000 new cases of LSCC worldwide, accounting for approximately 5.7–7.6% of systemic tumors [2]. Despite the development of improved strategies and more accurate treatments, the 5-year survival rate for LSCC has unfortunately decreased from 66 to 63% in the past 40 years [3]. To improve the living conditions of patients with LSCC, precision medicine is imperative.
In addition to surgical removal of the tumor, patients with LSCC with distant metastasis and focal recurrence accompanied by risk factors are usually assisted with cisplatin-based chemotherapy. Various immunotherapies are currently being studied as alternative options to the treatment of the disease [4, 5]. The emergence of checkpoint inhibitors has brought revolutionary changes to the outlook of the head and neck squamous cell carcinoma therapy, shifting it from cancer-centered to immune-centered research. The latest encouraging progress in cancer immunotherapy includes activating the host’s natural immune defense capabilities to effectively identify, target, and destroy cancer cells, which gives people some hope in the fight against LSCC [6,7,8,9].
Researchers have made great progress in understanding the immune microenvironment of LSCC [10]. The deciphering of tumor immunology and molecular structure may help determine the subset of immunogenic LSCC and identify potential predictive markers to select patients for immunotherapy [11]. Besides malignant cells, the LSCC solid tissue also contains various other innate immune cells (granulocytes, mast cells, and monocytes/macrophages), adaptive immune cells (T cells and B cells), fibroblasts, and endothelium cell. These cell interactions promote the inflammation and/or immunological state of tumor tissues through cell-to-cell contacts and/or the production of cytokines and chemokines. The tumor-infiltrating lymphocytes are a mixture of T cells, B cells, natural killer (NK) cells, macrophages, and other innate cells in different proportions, with the highest content of T cells [12]. Innate and adaptive immune responses can detect the presence of cancer cells. Dendritic cells swallow dying cancer cells and present antigens to T cells, which then migrate to tumors to fight tumor growth (Fig. S1). Thus, there is a clear, rational underpinning of the hypothesis that a high-grade in situ immune response may be associated with better tumor control [13]. TILs mainly appear in the human body to indicate the existence of the host, thus reflecting the dynamic process of cancer immunity [14, 15].
The International Immuno-Oncology Biomarker Working Group introduced guidelines for the assessment of TILs in solid tumors in 2017 and developed a standardized methodology for visual assessment of TILs on H&E sections [16]. Further studies showed that these criteria were reproducible and applicable to daily practice [17]. Although the histological evaluation of TILs in our H&E-stained samples did not reveal different subgroups of lymphocytes, it may still be a useful biomarker for evaluating tumor behavior [18]. However, this methodology is a single-factor assessment of the local immune status of the tumor and does not involve the tumor–stroma ratio and the percentage of TILs at the invasion front. Recognizing the multifaceted involvement of the TILs in LSCC, we propose the concepts of tumor parenchymal-infiltrating lymphocyte volume (TILv) and tumor infiltrates lymphocytes of frontier invasion (TILf). TILv was calculated using the formula TILv = % stroma in tumor × % stromal iTILs. In this study, we conducted a detailed assessment of the immune response in the three-dimensional structure of the tumor and analyzed the relationship between iTILs and TILv and TILf in LSCC overall survival (OS) and recurrence-free survival (RFS).
Tumor-infiltrating lymphocytes refer to lymphocytes that leave the blood and migrate into the tumor and are an important part of the TME. High TILs in local tumor tissues benefited from the enrichment of peripheral circulating immune cells [19, 20]. Therefore, host inflammation is also a key factor in tumor progression, the status of which can be indicated by peripheral blood cell counts [21]. We constructed a validation model based on the preoperative peripheral blood cell count and evaluated the prognostic value of the host inflammatory response to LSCC patients.
In order to further explore the specific molecular mechanisms that lead to differences in TILs, we conducted a bioinformatics analysis of LSCC data in the Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC). We defined a molecular prediction model based on the co-expression module of TIL-related genes and identified key genes that are significantly associated with TILs and prognosis. Figure S2 is our specific analysis flowchart. These findings are of great significance for predicting the prognosis of LSCC and the development of stratified immunotherapy.
Methods
Patients
This study retrospectively analyzed 412 cases. These patients were pathologically diagnosed with LSCC from December 2011 to December 2014 and underwent laryngectomy at the Head and Neck Nasopharyngeal Surgery Department of the Tumor Hospital of Harbin Medical University or the Head and Neck Department of Tumor Hospital of the Chinese Academy of Medical Sciences. It was reviewed and approved by the ethics committees of the two institutions and was carried out in accordance with the principles of the Declaration of Helsinki and its amendments. All participants provided informed consent to participate in the study. The clinical data (sex, age, BMI, history of drinking and smoking, tumor location, differentiation, TNM, T-stage, and N-stage) and follow-up information (clinical outcome and survival time) were collected from the electronic medical records. The prescribed inclusion criteria were as follows: (1) LSCC confirmed by histopathology; (2) no history of anticancer treatment; (3) complete clinical, laboratory, imaging, and follow-up data; (4) the remaining paraffin-fixed tissue was sufficient, and the structure is clear; (5) had at least one slice to assess the edge of tumor invasion; (6) no history of other malignant tumors and no distant metastasis. In this study, a total of 412 patients with LSCC were enrolled. There were 2–3 pathological tissue slices in each case, and a total of 1112 pathological tissue slices were reviewed. The samples were reviewed by two pathologists in a double-blind manner, and the patients were staged according to the eighth edition of the American Joint Committee on Cancer (AJCC) staging system. (7) Blood routine tests, which were based upon a single blood sample of each patient, were measured by an autoanalyzer (Sysmex XE-2100, Kobe, Japan). Data on peripheral blood cell counts, including platelets, neutrophils, lymphocytes, monocytes, eosinophils, and basophils, were extracted from the results of the first blood routine tests (limit to 30 days prior to surgery).
Experimental methods [suggestion: use past tense or imperative mood]
Tumor tissue sampling and laryngeal cancer tissue wax block preparation
(1) Surgically excised laryngeal tissue specimens were cut and fixed with 10% formalin solution; (2) 3*3*0.5 cm tissue blocks were cut from the laryngeal cancer tissue and placed in a tissue embedding box and placed in 10% formalin solution; (3) laryngeal cancer tissue blocks were dehydrated by gradient alcohol of low concentration to high concentration; (4) laryngeal cancer tissue blocks were soaked in xylene to remove alcohol transparent tissue; (5) laryngeal cancer tissue blocks were embedded in paraffin to make laryngeal cancer tissue wax blocks.
Preparation of white slices of laryngeal cancer tissue
(1) Section the laryngeal cancer tissue wax blocks in 4-μm-thick slices with a microtome; (2) place the slices in 30 °C water and flatten with a glass slide; (3) bake the slides at 72 °C for 1–2 h.
Hematoxylin–Eosin staining (H&E staining)
(1) White slices of laryngeal cancer tissue are deparaffinized in xylene solution; (2) white slices are hydrated with high- to low-concentration gradient alcohol; (3) white slices are stained with hematoxylin; (4) after washing, the sections are placed in hydrochloric acid alcohol to return to blue and differentiated in the differentiation solution; (5) after the sections are rinsed, they are dehydrated in low- to high-concentration gradient alcohol; (6) the sections are stained in alcohol and eosin; (7) the sections are placed dehydrate in pure alcohol; (8) place the slices in xylene to be transparent; (9) use neutral resin to seal the slices after drying.
TILs histological scoring in laryngeal cancer tissues
We evaluated TIL-related parameters according to the scoring method introduced by the International Immuno-Tumor Biomarker Working Group recently [22]. The evaluation of iTILs does not include any stromal areas that are not directly related to the tumor. In addition, areas of fibrosis or central necrosis are not included in the iTILs assessment. The percentage of iTILs was evaluated in two areas of each sample (the front and the center of the tumor invasion). The TILs working group guidelines recommend “Don't focus on hot spots” [22]. Therefore, the average value of TILs in the region should be used when reporting iTILs, TILv, and TILf. We evaluated at least five regions to assess the average value of TILs. As recommended, we used the whole untrimmed tumor sections. Each case in our study had at least one representative section (4–5 µm). Low-quality tumor sections, such as tumor sections without tumor–stroma interface, were excluded.
Follow-up methods
For each patient, extensive demographic, clinicopathological, and treatment information were extracted from the electronic medical record system. The demographic data and clinicopathological characteristics of the patients were collected from the database of two agencies. All patients who met the inclusion criteria were followed up by a combination of inpatient case review and telephone through January 2020. The median follow-up time was 59.9 months (range 1.9–83.2 months). The median OS time was 68.1 months (95% CI: 65.6–70.5). The primary outcome was OS from diagnosis to death, and the second outcome was RFS from cancer diagnosis to disease recurrence or metastasis or cancer-specific death, whichever came first.
Bioinformatics analysis of TILs
(1)Clinical sample and data collection
Gene expression quantification data (FPKM and counts format) for LSCC were downloaded from TCGA (https://portal.gdc.cancer.gov/). Then, 111 LSCC samples were obtained. The RNA expression matrix was extracted separately by annotations using the Gencode (GENCODE v 26) GTF file and normalized. Genes whose expression was “0” in 90% of LSCC patients were removed. Clinical data were downloaded from the UCSC Xena website (https://xena.ucsc.edu/). To analyze the correlation of TIL-related gene expression signatures with the prognosis of LSCC patients, we filtered the samples without survival information, and finally, all samples were included in the prognostic analysis.
Gene expression quantification and single nucleotide polymorphism data for LSCC were downloaded from ICGC (https://dcc.icgc.org/). Then, 85 LSCC samples were obtained. The RNA expression matrix was extracted separately by annotations using the Gencode (GENCODE v 26) GTF file. Genes whose expression was “0” in 90% of LSCC patients were removed. Clinical data were downloaded from ICGC (https://dcc.icgc.org/).
(2)Single-sample Gene Set Enrichment Analysis
An immune gene chip was downloaded from GSEA (http://software.broad institute.org/gsea/index.jsp), and ssGSEA was performed for LSCC data by GSVA and the GSEABase package of R.
(3) Construction of WGCNA Co-Expression Modules related to TILs
Weighted gene co-expression network analysis (WGCNA) is an analytical method to identify gene co-expression networks based on the topological overlap. The clusters of highly interconnected genes were determined by hierarchical clustering based on the connectivity and covariation coefficients of the genes. Eigengene expression patterns within each module are condensed into a “Module eigengene (ME).” MEs in the same cluster are considered to have a high correlation, consistent expression patterns, and similar biological functions, which help further explore the functions of different clusters [23]. We used the “goodSamplesGenes” function in the “WGCNA” R package to check the missing values of gene expression. After excluding genes with an average expression amount less than 1, genes with top 25% variance were performed WGCNA analysis to construct co-expression modules [24]. The power value is an important soft threshold parameter for defining highly positive correlations among genes in the same module. The “WGCNA” R package was employed to test the independence and average connectivity of different modules under different power values, and the power value corresponding to an independence index of R2 = 0.9 was selected. The minimum numbers of genes in the Gene module were set to 30 [25]. The “WGCNA” R package was used to cluster thousands of genes into different modules (identified by arbitrary colors). Based on TILs, the correlation between modules and TILs was calculated, and modules related to TILs were screened to obtain TIL-related genes (WGCNA hub genes) with P < 0.05 as the statistical significance threshold.
(4)mRNAs Position and Function Annotation
The Gencode (GENCODE v 26) GTF file is used to annotate the position of genes on chromosomes. Further, we visualized the results using the “karyoplote” R package. Gene ontology (GO) analysis was performed on the WGCNA hub genes to annotate the biological processes (BP), the cellular component (CC), and the molecular function (MF) in which they are involved. Meanwhile, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was used to annotate the signaling pathways associated with these WGCNA-genes. These analyses were performed by the R software package “clusterProfiler,” with P < 0.05 as the threshold of statistical significance. The “ggplot2” R package was used to visualize the GO and KEGG enrichment results [26].
(5) Construction and evaluation of signatures based on WGCNA genes
Univariate Cox regression analysis was performed on these WGCNA-Genes in the TCGA cohort, and WGCNA hub genes with p < 0.05 were considered prognostic-related WGCNA hub genes and used for subsequent analysis. Lasso Cox regression analysis was performed on the above-mentioned prognostic-related WGCNA hub genes. Eventually, an optimal model composed of 19 WGCNA hub genes was determined. Subsequently, the optimal model based on WGCNA-Genes signature of each patient was calculated. [Based on the optimal model, the WGCNA hub gene signature of each patient was calculated subsequently.] In the 3-year overall survival TCGA cohort, time-dependent receiver operating character curve (ROC) analysis was used to determine the optimal cutoff value for the WGCNA hub genes signature [27]. Patients were divided into high- and low-risk groups according to the cutoff value for further analyses. The log-rank test was used to evaluate the overall survival difference between the low-risk group and the high-risk group, and the KM survival curve was drawn. ROC curve analysis was used to evaluate the sensitivity and specificity of WGCNA hub genes. An ROC curve, including the clinical characteristics, was drawn, and the AUC was calculated. Finally, univariate and multivariate Cox regression analyses were used to investigate whether the prognostic value of the WGCNA hub genes was affected by other clinical characteristics.
(6) Construction and evaluation of nomograms
We combined the clinical characteristics of the TCGA data set with the WGCNA hub genes signature to construct a nomogram. We used the C index to evaluate the discriminative power of the nomogram and drew a calibration chart and decision curve analysis (DCA) to evaluate the accuracy of the nomogram.
(7) Estimation of immune infiltration
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) is a tool for predicting tumor purity and the presence of infiltrating stromal/immune cells in tumor tissues using gene expression data. ESTIMATE algorithm is based on single-sample gene set enrichment analysis (ssGSEA) and generates three scores.
Firstly, the immune infiltration assessment was performed using the “microenvironment cell population count (MCP-counter)” method [28]. Using the normalized FPKM expression matrix converted by log2 as input, the absolute abundance scores of ten immune cell and stromal cell populations are generated through the “MCP-counter” package. Research shows that immune cell infiltration assessed by the MCP-counter algorithm performs well when comparing between samples [29]. Subsequently, Cell type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) was used to infer the relative proportion of 22 infiltrating immune cells in each sample for supplementation.
(8) Analysis of the Immunosuppressive Molecules Expressing Related to immune checkpoint inhibitors (ICIs)
To study the relationship between the model and the expression level of genes related to ICIs, we performed ggstatsplot package and violin plot visualization.
(9) Exploration of the Significance of the Model in the Clinical Treatment
The difference in the ICIs between the high- and low-risk groups was compared by Wilcoxon signed-rank test. The results are shown as violin plots using gglpot2 in R.
(10) Gene set enrichment analyses
Gene set enrichment analyses (GSEA) software (version 4.0.1) was used to perform gene set enrichment analysis between the high-risk and low-risk groups. KEGG and Hallmark were used to identify enriched terms between the high-risk group and the low-risk group. P < 0.05 and false discovery rate (FDR) < 0.05 were considered statistically significant.
Immunohistochemistry
The expressions of cathepsin L (CTSL) and lysine (K)-specific demethylase 5D (KDM5D) on the laryngeal cancer tissue were performed by immunohistochemistry (IHC) staining. Tissue sections were incubated with a primary antibody against CTSL (1:100, ProteintechTM) or KDM5D (1:200, Affinity) at 4 ℃ overnight and then incubated with horseradish peroxidase combined with goat anti-rabbit antibody (PV-6001, ZSGB) at room temperature for 30 min. Tissue sections were stained using DAB and counterstained with hematoxylin.
The results of the experiment were analyzed by two doctors and two pathologists. The rules are as follows: 7, about 50% of tumor cells are strongly stained; 6, about 50% of tumor cells are weakly stained; 5, about 25% of tumor cells are strongly stained; and 4, about 25% of tumor cells showed weak staining; 3,5–25% of tumor cells showed strong staining; 2, about 5–25% of tumor cells showed weak staining; 1, < 1% of tumor cells showed low or no staining; 0, no staining was detected in the tumor cells (0%). Samples with a staining score of 0–2 were considered low expression, while samples with a score of 3–7 were considered high expression.
Data analysis
We first divide the patients into two groups according to the best cutoff point for iTIL, TILv, TILf level, or the hematological index. The cutoff points were determined by the surv_cutpoint function in the survival R package; overall survival status was used as the dependent variable (0, alive;1, death). We report the mean and standard deviation or count and frequency of continuous and categorical variables, respectively. The differences of continuous covariates and categorical covariates between groups were compared using Student’s t test and Chi-square test, respectively.
We then conducted univariate and multivariate Cox regression analyses and reported hazard ratios (HRs) and 95% confidence intervals to assess the association between iTILs, TILv, TILf, the hematological index, and the prognosis of LSCC patients. The likelihood ratio backward stepwise method was used for the multivariate Cox regression analysis. Kaplan–Meier curves and log-rank tests were then conducted to compare the OS rates between groups. Two-sided statistical significance was defined as P < 0.05. All other statistical analyses were performed using R software 4.0.3 (Institute for Statistics and Mathematics, Vienna, Austria).
Results
Patient characteristics
A total of 412 patients are eligible for this study. There were 336 males (81.6%) and 76 females (18.4%). Most subjects had a current or history of smoking (72.1%). As shown in Table 1, 52.2% of patients had supraglottic squamous cell carcinoma, and 47.8% of patients had glottal laryngeal squamous cell carcinoma. Most patients have localized early tumors (T1 or T2) (65.3%), but most of them are moderately or poorly differentiated (59.0%) (Table 1).
The prognostic value of tumor local inflammation infiltration indicators peripheral blood inflammatory markers
In our study, 226 patients had lower iTILs (Fig. S3a), and 186 patients had higher iTILs (Fig. S3b). The 5-year OS rate of the high iTILs group (50.5%) was significantly higher than that of the low iTILs group (42.5%, P < 0.001, Fig. 1a). According to the TILv stratification of patients, 126 patients had lower TILv (Fig. S3c), and 286 patients had higher TILv (Fig. S3d). The 5-year OS rate of the high TILv group (48.3%) was significantly higher than the low TILv group (41.3%, P < 0.001, Fig. 1b). Lastly, we stratified the patients again according to TILf, 142 patients having lower TILf (Fig. S3e) and 270 patients having higher TILf (Fig. S3f). The 5-year OS rate of the high TILf group (47.8%) was significantly higher than the low TILv group (42.9%, P < 0.001, Fig. 1c). In conclusion, the levels of iTILs, TILv, and TILf are closely related to the improvement of patient survival outcomes. In addition, we also evaluated the prediction accuracy of the three indicators for OS, and the ROC curve shows the good prediction performance of our three indicators (Fig. 1d). The statistical analysis of the relationship between iTILs, TILv, and TILf levels and the disease recurrence did not find any significant correlation. In the univariate cox analysis, BMI ≥ 24, poorly differentiated, supraglottic carcinoma, high T, N stage or TNM, low iTILs, TILv, and TILf levels were identified as predictors of poor prognosis. Next, we included statistically significant indicators into the multivariate analysis, and the results showed that BMI (P = 0.041, HR: 0.642, 95% CI: 0.420–0.983), iTILs (P < 0.001, HR: 0.502, 95% CI: 0.338–0.746), TILv (P < 0.001, HR: 0.462, 95% CI: 0.318–0.672), and TILf (P = 0.004, HR: 0.585, 95% CI: 0.405–0.845) levels showed significant statistical significance (Table 1). Therefore, we can consider that high iTILs, TILv, and TILf levels are independent predictors of a good prognosis.
Correlation analysis of inflammatory markers and prognosis a, b, c, Kaplan–Meier curves showed that high iTILs(b), fTILs(c), and TILV(d) density correlated with superior OS of patients with LSCC, P-values calculated by log-rank test. d A receiver operating characteristic (ROC) curve analysis for iTILs, TILv and TILf. e, f, g, h Kaplan–Meier curves showed that high LMR (e), low PLR (f),NLR (g), and SII (h)correlated with superior OS of patients with LSCC. P-values calculated by log-rank test
The Kaplan–Meier curve is shown in Fig. 1e–h. The 5-year OS rate of the high lymphocyte-to-monocyte ratio (LMR), platelet-to-lymphocyte ratio (PLR), neutrophil-to-lymphocyte ratio (NLR) and systemic immune-inflammation index (SII, platelets × neutrophils/lymphocytes) group was significantly higher than that of the low group (all P < 0.05). In the univariate Cox analysis, high PLR, NLR, and SII were significantly correlated with better OS (Table 1). In the multivariate Cox analysis, PLR is still statistically associated with better OS in LSCC patients (P < 0.05, HR: 0.585, 95% CI: 1.000–1.006), which indicates that PLR is LSCC independent prognostic indicators (Table 2).
In the TCGA database, the TILs of LSCC patients are related to the prognosis
We first retrieved the transcriptome data and clinical characteristics of LSCC from the TCGA database. Excluding samples with repeated sequencing and zero survival time, a total of 111 tumor samples were included in the study. We used the GTF file to annotate the probes and the “mean” method to delete duplicate gene names. According to immune gene sets, we analyze tumor samples by immune-related single-sample gene set enrichment analysis (ssGSEA) and use heat maps for visualization (Fig. S4a). Next, the samples were grouped according to the enrichment of TILs, and the KM curve showed that LSCC patients with high TILs had longer survival times than those with low TILs (P < 0.001) (Fig. 2a). We continued to use the Pearson correlation test to evaluate the correlation between TILs and survival time. As shown in Fig. 2b, the survival time of patients gradually increased with the increase in TILs score (R = 0.23, P = 0.013).
Analysis of the prognostic value of TILs and enrichment analysis of TIL-related genes a Patients in the high TILs’ group experienced a longer survival time, tested by the Kaplan–Meier test. b The survival time of patients with LSCC longer as the TILs increased (R = 0.23, P = 0.013). c Module–trait associations. Each row corresponds to a module, and each column corresponds to a trait. Each cell contains the corresponding correlation and P value. The table is color-coded by correlation according to the color legend. d The Venn diagram was used to extract the intersection of ICP and TIL-related genes, and 802 common genes were obtained. e The genes involved in the blue module were analyzed by KEGG. The node size reflects the gene count, and the node color reflects the P value. f Gene Ontology analysis of the genes involved in the blue module regarding biological process, cellular component, and molecular function
Screening of gene co-expression modules related to TILs and functional analysis of module characteristic genes
We eliminated genes with low expression levels and selected the top 25% of the genes with variance variation rates. We cluster the samples to identify whether there are any obvious outliers. The height cutoff value was set to 120, and 102 samples were included in our analysis (Fig. S5a). After preliminary sorting, we constructed the WGCNA co-expression module of 6322 genes from 102 samples. To construct a WGCNA network, we first calculated the soft thresholding power β, to which the co-expression similarity is raised to calculate adjacency. We used the pick soft threshold function in WGCNA for the network topology analysis. The soft thresholding power β was set at 5 in the subsequent analysis because the scale independence reached 0.9 and had relatively high-average connectivity (Fig. S5b). We constructed the gene network and identified modules using the one-step network construction function of the WGCNA R package. To cluster splitting, the soft thresholding power was set at 5, the minimum module size was set at 30, and the deep Split was set at 2 (which implies a medium sensitivity). Finally, 17 genes’ co-expression modules were constructed (Fig. S5c). We mapped the relationships between the identified modules (Fig. S5d). The heatmap depicts the topological overlap matrix (TOM) among all genes included in the analysis. The light color represents a high overlap, and the progressively darker red color represents a decreasing overlap. The results of this analysis indicated that the gene expression was relatively independent between modules. We correlated modules with the TILs and immune checkpoint (ICP) scores of LSCC samples and searched for the most important associations. The analysis results show that the blue module is significantly associated with TILs and ICPs (P < 0.0001) (Fig. 2c and Fig. S4b, S4c).
The Venn diagram shows the intersection of the ICP-related gene set and the TIL-related gene set (Fig. 2d), and finally, 802 genes are obtained. We found that these genes are mainly concentrated in chr1 and chr19 (Fig. S5e). In order to understand the composition of these genes in the cell and the biological functions they perform, we performed GO and KEGG enrichment analysis on 802 WGCNA hub genes. KEGG results showed that they are mainly involved in the regulation of pathways including cytokine–cytokine receptor interaction, chemokine signaling pathway, T cell, and B cell receptor signaling pathway, PD-L1 expression, and PD-1 checkpoint pathway in cancer (Fig. 2e). GO results show that 802 WGCNA hub genes are mainly enriched in biological processes, such as T cell activation, regulation of immune effector process and lymphocyte proliferation; cellular component, such as MHC protein complex, immunological synapse, and MHC class II protein complex; and molecular functions such as immune receptor activity, cytokine receptor binding, and cytokine receptor activity (Fig. 2f). The P values of the above enrichment analysis results were all P < 0.05.
Predictive model construction and evaluation
We included 111 TCGA samples in the survival correlation analysis. Univariate Cox analysis was performed on these WGCNA hub genes (P < 0.05), and 69 WGCNA hub genes related to prognosis were obtained (Fig. S6a). In order to prevent overfitting, Lasso Cox regression analysis was performed on these prognostic WGCNA hub genes, and 19 WGCNA hub genes were obtained (Fig. S6b, c). We selected 19 WGCNA hub genes to construct features and obtained a well-balanced prognostic model. (The list of immunization pathways and coefficients of WGCNA hub genes is shown in Table S1.) We separately constructed the ROC curves of WGCNA hub genes signature, TNM stage, age, and gender. The area under the curve (AUC) of the WGCNA hub genes signature is 0.790 (Fig. 3a), which shows that our signature has excellent predictive power. Then, we plotted the ROC curve of WGCNA hub genes’ signature over time. We found that the areas under the WGCNA hub genes signature curve are: 1 year: 0.811; 3 years: 0.953; 5 years: 0.939. This shows that our model has a good predictive ability for patients with survival (Fig. 3b). We used the time-dependent ROC to determine the cutoff value for the best WGCNA hub genes signature. The best cutoff value for WGCNA hub genes signature is 3.510 (Fig. S6d). According to the cutoff value, patients were divided into high-risk groups and low-risk groups. We found that compared to patients in the low-risk group, the overall survival rate of patients in the high-risk group was significantly lower (Fig. 3c). Subsequently, we drew a survival status graph that included survival time and risk score regression curves. The results showed that as the risk score increased, the mortality rate of patients gradually increased, and the survival time gradually shortened (Fig. 3d). Moreover, as the risk score increases, the expression values of genes such as NUPR2, MT-ATP8, and AQP9 increase. On the contrary, the expression value of genes such as KDM5D, MAP3K14, and TNFRSF4 decreased with the increase in risk score (Fig. 3e).
Analysis of TIL-related genes risk scores of patients with LSCC in TCGA. a A comparison of ROC curves with other common clinical characteristics showed the superiority of the WCGNA-Genes signature. b The 1-, 3-, and 5-year ROC curve of the optimal model suggested that all AUC values were over 0.80. c Patients were sorted by increasing risk score in the LSCC set. d The Kaplan–Meier survival curve with log-rank test was drawn to demonstrate the relationship between risk model and OS. Compared with the high-risk group, patients in the low-risk group experienced a longer survival time. The survival time and survival status of patients worsened as the risk score increased (y = -1.58x + 8.21, R2 = 0.25). e Heat map of the expression of 19 key genes in LSCC. Colors from blue to red indicate a trend of expression from low to high
ICGC database verification model
In the ICGC-GENEs data set, the risk scores of 85 LSCC samples were calculated according to the same method. We drew the ROC curve of the signature over time. We found that the areas under the ICGC-GENEs signature curve are: 1 year: 0.628; 3 years: 0.815; 5 years: 0.821. This shows that our model still has excellent predictive capabilities in the validation set (Fig. S7a). The patients were divided into high-risk groups and low-risk groups. As shown in Fig. S7b, patients in the low-risk group have a longer overall survival. Next, we used the same method to draw a survival status graph with regression curves. The results showed that as the risk score increased, the patient’s mortality rate gradually increased, and the survival time gradually shortened (Fig. S7c). This is consistent with the results we obtained in the TCGA database. Unfortunately, we only found a significant correlation between gender differences and risk scores in the correlation analysis between clinical features and risk models. Other clinical features may need to be verified in a larger clinical sample size (Fig. S7d and S7e).
Correlation between predictive models and clinical characteristics
We evaluated the prognostic value of the LSCC risk score. In the univariate analysis, we found that OS and risk score (HR = 5.658, 95% CI = 3.430–9.331, P < 0.001), M staging (HR = 8.355, 95% CI = 1.912–36.513, P < 0.01), N staging (HR = 1.437, 95% CI = 1.042–1.980, P < 0.05), gender (HR = 3.142, 95% CI = 1.511–6.533, P < 0.05) were significantly correlated (Fig. 4a). Multivariate analysis showed that risk score (HR = 6.156, 95% CI = 3.644–10.401, P < 0.001) and M stage (HR = 12.416, 95% CI = 2.714–56.799, P = 0.01) were independent predictors of OS (Fig. 4b). In order to further improve the prediction accuracy, we constructed a new nomogram prediction map based on the WGCNA hub genes (Fig. 4c). The nomogram C-index is 0.835. By calculating the total score, oncologists can easily obtain the probability of OS predicted by the nomogram of a single patient. Besides, we used the calibration curve to evaluate the model’s prediction accuracy. The results showed that the prediction calibration curve of the three calibration points in 1, 3, and 5 years was close to the standard curve, which indicated that the model had good predictive performance (Fig. 4d). We also used the DCA to evaluate the reliability of the model (Fig. 4e). It can be seen that the profit of this model was significantly higher than the limit curve and clinical characteristic; that is to say, it had good reliability.
Establishment and evaluation of clinical prognostic model a Forest plot of univariate Cox regression results shows that risk score (P < 0.001), metastasis (P < 0.05), and sex (P < 0.05) are prognostic-related factors. b Forest plot of multivariate Cox regression results show that risk score (P < 0.001) and metastasis (P = 0.001) are independent influencing factors for prognosis. c Nomogram predicting the probability of TCGA patient’s mortality based on risk score and clinical variables. d Calibration curves of the nomogram for 1, 3 and 5 years. e Decision curve analysis of the nomograms based on the WCGNA-Genes signature. *P < 0.05, **P < 0.01, ***P < 0.001
The relationship between immune cell infiltration and risk model
We explore the difference in immune cell infiltration between the two groups. Based on the ESTIMATE algorithm, we first calculated the immune score for each LSCC sample. As shown in Fig. 5a, b, compared to the high-risk group, the immune score of the low-risk group (P = 0.029) is higher, and it is negatively correlated with the risk score (the correlation coefficient is −0.36, P < 0.001). Next, we used the MCP counting method to calculate the abundance of 10 immune-related cells. Compared to the high-risk group, the low-risk group had more abundant four types of cell populations (CD8 T cells, myeloid dendritic cells, neutrophils, T cells) (Fig. 5c). We further explored the relationship between immune cell infiltration and risk score. The results showed that the degree of immune cell infiltration was significantly negatively correlated with the risk score (Fig. 5d). We use CIBERSORT to further supplement the relative proportion of 22 immune infiltrating cells in each sample (Fig. 5e). The relative proportions of T cell CD8 and T cell CD4 memory resting in the low-risk group were higher. The relative proportions of macrophages M0 and eosinophils in the high-risk group were higher. There is a big difference in immune cell infiltration between high-risk and low-risk groups. It is worth noting that the radar chart shows that T cell CD4 memory cessation and M0 macrophage infiltration rate are higher in all patients.
Correlation between WGCNA-Genes signature and immune cell infiltration and immune score. a The Wilcoxon rank-sum test was used to compare differences in immune scores between low- and high-risk groups (P < 0.05). b Spearman's correlation coefficients were computed to investigate the potential relationship between risk score and immune scores. c The Wilcoxon rank-sum test compared the absolute abundance scores of eight immune cells and two stromal cells populations in two groups of patients. d Spearman's correlation coefficients were computed to investigate the potential relationship between absolute abundance scores of immune cells and stromal cells and risk score. The area of fan represents the degree of correlation (red represents a positive correlation and blue represents a negative correlation). e Radar chart of the relationship between 22 immune cell infiltration and risk group. (Wilcoxon test). *P < 0.05, **P < 0.01, ***P < 0.001
Mutational situation in the ICGC database and assessment of response to immunotherapy
The association of immune subtypes with mutational status higher tumor mutation burden (TMB) and somatic mutation rates is correlated to stronger anticancer immunity30. Therefore, we calculated the TMB and mutations in each patient using the mutect2-processed mutation dataset of ICGC and analyzed the difference between the mutation and TMB in the high- and low-risk groups. The waterfall chart shows the overall mutation status of the top 20 genes in the high- and low-risk groups (Fig. 6a, b). Compared to the high-risk group, the low-risk group showed a higher mutation rate. Next, we plotted the TMB situation in the high- and low-risk groups. Compared to the high-risk group, the low-risk group showed higher TMB (Fig. 6c). These findings suggest that our prediction model can predict the TMB and somatic mutation rates of LSCC patients and that patients in the low-risk group may have a positive response to immunotherapy.
The relationship between WGCNA-Genes signature and immunotherapy a, b Gene mutations in high- and low-risk groups in LSCC samples. c The Wilcoxon rank-sum test was used to compare differences in TMB between low- and high-risk groups. P < 0.05. d Spearman's correlation coefficients were computed to investigate the potential relationship between immune checkpoints and risk score. (Red represents a positive correlation and blue represents a negative correlation). e Different expressions of ICIs between risk groups defined by the WGCNA-genes signature. LAG3 and PDCD1 were overexpressed in the low-risk group (all P < 0.05)
The application of ICIs for tumor immunotherapy has become a promising treatment for recurrent and metastatic LSCC. In order to further study the relationship between risk models and immunotherapy, we explored the correlation between risk models and ICI-related biomarkers. The results showed that the expression levels of LAG3 and PDCD1 were upregulated in the low-risk group (both P < 0.05), and the risk score was similar to LAG3 (r = -0.40, P < 0.05), PDCD1 (r = -0.40, P < 0.05), and CD274 (r = -0.20, P < 0.05) which was negatively correlated, indicating that the low-risk group benefited more from immunotherapy (Fig. 6d, e).
Gene set enrichment analysis of the risk model
Since the risk model obtained is highly correlated with the prognosis, immune microenvironment, and treatment effect of LSCC, we tried to explore their functional meaning and internal connection through subsequent enrichment analysis. The results of KEGG functional enrichment analysis showed that the differentially expressed genes of high-risk and low-risk populations were significantly enriched in nine KEGG pathways (P < 0.05). Among them, the high-risk group had the most abundant TGF beta signaling pathway and ECM receptor interaction, and the low-risk group had the most significant way to produce primary immunodeficiency and intestinal immune network for IgA production (Fig. 7a). To investigate whether the signature was associated with tumor biological processes, gene set enrichment analysis (GSEA) was performed. As shown in Fig. 7b, a total of nine hallmark gene sets were enriched (P < 0.05). Four hallmarks (including HYPOXIA, KRAS_SIGNALING_UP AND MTORC1_SIGNALING) were associated with high-risk scores, suggesting that the activation of these biological processes may participate in LSCC progression. In contrast, the other five hallmarks (INTERFERON_ALPHA_RESPONSE, INTERFERON_GAMMA_RESPONSE, PI3K_AKT_MTOR_ SIGNALING, IL6_JAK_STAT3_SIGNALING, DNA_REPAIR and G2M_CHECKPOINT) were associated with low-risk scores, suggesting that their activation inhibits tumor progression and improves survival in LSCC patients.
Immunohistochemistry
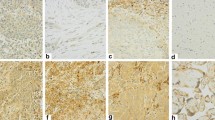
In our study, 112 patients had a high CTSL expression by employing IHC experiments (27.2%) and another 300 patients had a low CTSL expression (72.8%, Fig. 8a). The OS rate of patients with high CTSL expression was significantly lower than the low CTSL expression group (P < 0.001, Fig. 8b). According to the KDM5D stratification of patients, 270 patients had high KDM5D expression (65.5%), and 275 patients had low KDM5D expression (34.5%, Fig. 8a). The OS rate of patients with high KDM5D expression was significantly higher than the low KDM5D group (P < 0.001, Fig. 8c). In the correlation analysis of TILs, we found that CTSL was negatively correlated with TILs (P < 0.05), and KDM5D was significantly positively correlated with TILs (P < 0.001) (Fig. 8d, e). In addition, we provided more representative pictures and corresponding positive controls in Figure S8.
CTSL and KDM5D gene expression levels were associated with clinical outcomes a We selected eight representative immunohistochemical pictures to show the expression of CTSL and KDM5D in tissues. b,c The Kaplan–Meier curve showed that low CTSL expression and high KDM5D expression were associated with higher OS in LSCC patients, and the p value was calculated by log-rank test (all P < 0.001). d Box plot shows that low CTSL expression is significantly correlated with higher TILs (P < 0.05). e High KDM5D expression is significantly correlated with higher TILs(P < 0.001)
Discussion
In this two-center, large-sample retrospective cohort study, we evaluated the relationship between the TILs and the prognosis and recurrence in multiple dimensions. A systematic evaluation was also conducted on the effects of preoperative peripheral blood NLR, PLR, MLR, and SII on the prognosis of LSCC patients. Through local and overall joint analysis, we highlighted that inflammation indicators were an independent factor influencing the prognosis of LSCC patients. As a double-validation cohort, we obtained LSCC samples from the TCGA and ICGC databases to analyze the prognosis and benefit of adjuvant therapy. Based on the TCGA database, we identified 19 genes that could reflect TILs status. Furthermore, we evaluated the immune microenvironment, immunotherapy response, chemotherapy response, and the effect of immune combined chemotherapy in patients with LSCC in the high and low TIL group. The relationship between TIL and the prognosis and recurrence suggested by the laboratory data was further corroborated by the data derived from the ICGC database. In addition, we evaluated the expression of proto-oncogenes and tumor-suppressor genes with the largest weight coefficients in the tissues and identified two genes that are significantly associated with patients’ TIL and prognosis. This was of great significance for clinicians to identify high-risk patients simply and accurately and to further choose personalized treatment methods.
In terms of sample size, this is the largest study assessing the prognostic value of TILs in LSCC thus far. Significantly, we recognized the prognostic value of the volume of TILs and the TILs at the frontier invasion and proposed the concepts of TILv and TILf, which are necessary for a detailed understanding of the immune status of the tumor microenvironment. The infiltration of tumors by chronic inflammatory cells includes lymphocytes, plasma cells, and macrophages [31]. TILs are a manifestation of the host's immune response to tumor cells, represented by T cells, B cells, and natural killer cells [32]. Formalin-fixed paraffin-embedded sections can be used to assess tumor immunity from multiple perspectives such as TIL morphology, T cell subsets (e.g., CD3( +) or CD8( +)) immune score, and immunophenotype reaction [33,34,35]. In our study, three aspects of TILs were evaluated morphologically. The results show that high levels of iTILs, TILv, and TILf are reliable and independent prognostic indicators for LSCC. Our study confirmed and expanded the correlation between quantified TILs levels in tissues and outcomes reported by other researchers. [36,37,38,39]. Unlike the 120 LSCC cohort study by Wang et al., neither Kaplan–Meier curve nor single-factor multivariate analysis found a significant association between TILs, TILv, TILf and tumor recurrence in this study cohort. This result may require further examination [40].
The morphological evaluation of TILs is simple, can be carried out in routine clinical practice, and can provide better histopathological predictions based on immune response without additional costs. This prediction may prove to be valuable in clinical decision making. The immune response is one of the main factors affecting the clinical outcome of tumors. However, in the current staging system, LSCC patients with very different immune responses may be categorized into the same clinical stage and/or the same histopathological grade. We strongly believe that the evaluation of iTILs, TILv, and TILf should be included in the clinicopathological prognosis model of LSCC patients to identify high-risk patients more accurately. A similar proposal has been made in breast cancer. Loi’s comprehensive analysis of 2148 patients with the early triple-negative disease showed that TILs increase the prognostic value of known clinical variables [41]. In addition, the International Immuno-oncology Biomarker Breast Cancer Working Group has proposed a standardized method for pathologists to evaluate the role of iTILs in the residual disease environment after neoadjuvant. In this study, we assessed the local tumor immune microenvironment status and introduced two indicators, TILv, and TILf for the following reasons: First, iTILs only reflect the content of tumor–stroma, which is incomplete information. On the other hand, TILv considers the percentage of stroma to the overall tumor when calculating iTILs; hence, TILv may be more representativeness than iTILs. Secondly, many studies have shown that the frontier of tumor invasion best represents the real-time immune status of tumors [42,43,44]. Finally, when iTILs alone are not enough to evaluate the local immune status of tumors, TILv and TILf may provide additional information.
As an important sign of cancer, inflammation plays a key role in various stages of tumorigenesis [45]. It is well established that the circulating immune cells originate from bone marrow [46]. Mature immune cells migrate to the site of inflammation under the action of various driving factors, forming tumor-infiltrating lymphocytes [47]. Lymphocytes are an important part of the tumor microenvironment in inducing apoptosis, inhibiting cancer cell proliferation, and inhibiting cancer cell migration and invasion [48]. Neutrophils, as the first responders of inflammation, can metastasize to tumors through CXC chemokines [49, 50] to promote the spread of cancer cells to distant organs [51]. The monocytes in the tumor microenvironment are the potential cells with dendritic differentiation or tumor-associated macrophages [52, 53]. The role of platelets in the development and progression of cancer is controversial [54, 55]. We quantitatively evaluated the local real-time immune status of patients with LSCC based on the assessment of tumor-infiltrating lymphocytes. Peripheral blood cell count can reflect the degree of total inflammation and is easily obtained in clinical practice. In order to comprehensively evaluate the immune status of tumor patients, we further constructed an inflammation marker based on the ratio of the counts of immune cells in the peripheral blood. We found that NLR, PLR, MLR, and SII were all influencing factors of OS in LSCC patients and that PLR was an independent prognostic factor.
Clinicopathological data and peripheral blood indicators have verified the influence of inflammation indicators on the prognosis of cancer patients. In order to further study the specific molecular mechanisms and the sensitivity of high- and low-risk groups to adjuvant therapy, we identified 19 genes that could reflect TILs in the TCGA data set. Based on 19 genes, we constructed a good prognostic signature. The results showed that the 5-year survival rate of patients in the high-risk group was significantly worse than that of the low-risk group. As the risk score increased, the survival time of LSCC patients was gradually shortened, and vice versa. Furthermore, the risk score is also an independent factor for the prognosis of patients, verified by the analysis of the ICGC database. The differential analysis of immune cell infiltration showed that T cells, neutrophils and myeloid dendritic cells had higher infiltration in the low-risk group, whereas the macrophages had higher infiltration in the high-risk group of the LSCC patients. Many studies have shown that dense infiltration of T cells, especially T cells CD8, indicates a good prognosis [56,57,58]. In a complex microenvironment, macrophages, as a plastic and pluripotent cell group, play an important role in the occurrence and development of malignant tumors. In most tumors, macrophages have been shown to participate in tumor growth and development, with chronic inflammation and aggressive phenotypes. The macrophages are present in hepatocellular carcinoma, gastric cancer, pancreatic duct adenocarcinoma, and melanoma, and have a poor prognosis59–62. Our research results support these observations.
Chemotherapy is the main treatment for recurrent and metastatic LSCC. We attempted to explore the effectiveness of this risk model in LSCC chemotherapy patients. We found that this model was effective in identifying subgroups that might benefit from the gemcitabine treatment. Compared to chemotherapy, immunotherapy has the characteristics of less toxic side effects and lasting curative effects [63]. Tumor mutation burden (TMB) is the number of somatic mutations in the tumor genome without the germline mutations. Higher TMB indicates that the tumor produces more new antigens, and the tumor is more easily recognized by immune cells. Therefore, here we first explored the relationship between the risk model and TMB and then assessed the correlation between the risk model and immune checkpoints. We found that LSCC patients in the low-risk group had higher expression levels of TMB, LAG3, and PDCD1, and the expression levels of LAG3 and PDCD1 gradually decreased as the risk score increased. This observation provides strong evidence that patients in the low-risk group might benefit more from immunotherapy. In the last few years, immunotherapy has transformed the way we treat solid tumors, including head neck, melanoma, lung, renal, breast, and bladder cancers. Sustainable responses and long-term survival benefits have been observed in many cancer patients, with favorable toxicity profiles of immunotherapeutic agents relative to chemotherapy. Cures have become possible in some patients with metastatic disease.
In order to further explore the role of the internal gene composition of the model, we selected the potential proto-oncogene (CTSL) and tumor-suppressor gene (KDM5D) with the largest weight coefficient. Several studies have shown that overexpression of CTSL can promote EMT [64,65,66]. However, the down-regulation of KDM5D induced the progression of malignant tumors [67,68,69]. We also confirmed that the two differentially expressed genes are significantly associated with the TILs and prognosis of LSCC patients through immunohistochemical analysis. Moreover, our laboratory has embarked the study on the mechanism of action of the CTSL and KDM5D genes in the occurrence and development of head and neck squamous cell carcinoma.
Although our result showed excellent predictive power in predicting patient outcomes, identifying high-risk LSCC patients and individualizing treatment, there are several limitations. First, although this study is based on the largest cohort of 412 eligible patients, these analyses still need to be validated in larger patient cohorts. Second, there is an inherent gender bias in our patient cohort, due to the fact that LSCC is a male-dominated disease. Finally, as a retrospective study, our findings need to be verified in a prospective analysis.
Conclusion
To sum up, our research confirms that local tumor immune status (including iTILs, TILv, and TILf) and overall immune status (PLR) are independent factors influencing patient survival. A novel risk model can guide clinicians to accurately predict prognosis, identify high-risk patients, and develop personalized treatment plans. Studies have found that the differential expression of genes such as CTSL and KDM5D is significantly related to the TILs of LSCC and the prognosis of patients.
Data availability
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Abbreviations
- AJCC:
-
American Joint Committee on Cancer
- BP:
-
Biological processes
- CC:
-
Cellular component
- CCLE:
-
Cancer Cell Line Encyclopedia
- CIBERSORT:
-
Cell type Identification by Estimating Relative Subsets of RNA Transcripts
- CTSL:
-
Cathepsin L
- DCA:
-
Decision curve analysis
- ESTIMATE:
-
Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- H&E staining:
-
Hematoxylin–eosin staining
- HRs:
-
Hazard ratios
- ICIs:
-
Immune checkpoint inhibitors
- ICGC:
-
International Cancer Genome Consortium
- iTILs:
-
Interstitial tumor-infiltrating lymphocytes
- KDM5D:
-
Lysine (K)-specific demethylase 5D
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LSCC:
-
Laryngeal squamous cell carcinoma
- MCP-counter:
-
Microenvironment cell population count
- ME:
-
Module eigengene
- MF:
-
Molecular function
- OS:
-
Overall survival
- PLR:
-
Platelet-to-lymphocyte ratio
- ROC:
-
Receiver operating character curve
- RFS:
-
Recurrence-free survival
- TME:
-
Tumor microenvironment
- TILs:
-
Tumor-infiltrating lymphocytes
- TILv:
-
Tumor parenchymal-infiltrating lymphocyte volume
- TILf:
-
Tumor infiltrates lymphocytes of frontier invasion
- TCGA:
-
The Cancer Genome Atlas
- TMB:
-
Tumor mutation burden
- WGCNA:
-
Weighted gene co-expression network analysis
References
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115–132
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127(12):2893–2917
Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF (2017) An update on larynx cancer. CA Cancer J Clin 67(1):31–50
Wei FQ, Sun W, Wong TS, Gao W, Wen YH, Wei JW et al (2016) Eliciting cytotoxic T lymphocytes against human laryngeal cancer-derived antigens: evaluation of dendritic cells pulsed with a heat-treated tumor lysate and other antigen-loading strategies for dendritic-cell-based vaccination. J Exp Clin Cancer Res 35:18
Chen Y, Li ZY, Zhou GQ, Sun Y (2021) An Immune-Related Gene Prognostic Index for Head and Neck Squamous Cell Carcinoma. Clin Cancer Res 27(1):330–341
Kaderbhai C, Tharin Z, Ghiringhelli F. The Role of Molecular Profiling to Predict the Response to Immune Checkpoint Inhibitors in Lung Cancer. Cancers (Basel). 2019;11(2)
Havel JJ, Chowell D, Chan TA (2019) The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 19(3):133–150
Galluzzi L, Chan TA, Kroemer G, Wolchok JD, Lopez-Soto A. The hallmarks of successful anticancer immunotherapy. Sci Transl Med. 2018;10(459)
Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP et al (2020) Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 38(1):1–10
Xiang H, Ramil CP, Hai J, Zhang C, Wang H, Watkins AA et al (2020) Cancer-associated fibroblasts promote immunosuppression by inducing ROS-generating monocytic MDSCs in lung squamous cell carcinoma. Cancer Immunol Res 8(4):436–450
Mele L, Del Vecchio V, Marampon F, Regad T, Wagner S, Mosca L et al (2020) beta2-AR blockade potentiates MEK1/2 inhibitor effect on HNSCC by regulating the Nrf2-mediated defense mechanism. Cell Death Dis 11(10):850
Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J et al (2016) Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387(10030):1837–1846
Egen JG, Ouyang W, Wu LC (2020) Human anti-tumor immunity: insights from immunotherapy clinical trials. Immunity 52(1):36–54
Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411)
Fumet JD, Richard C, Ledys F, Klopfenstein Q, Joubert P, Routy B et al (2018) Prognostic and predictive role of CD8 and PD-L1 determination in lung tumor tissue of patients under anti-PD-1 therapy. Br J Cancer 119(8):950–960
Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B et al (2017) Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the international immuno-oncology biomarkers working group: part 2: TILs in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv Anat Pathol 24(6):311–335
Kojima YA, Wang X, Sun H, Compton F, Covinsky M, Zhang S (2018) Reproducible evaluation of tumor-infiltrating lymphocytes (TILs) using the recommendations of International TILs working group 2014. Ann Diagn Pathol 35:77–79
Alessandrini L, Franz L, Ottaviano G, Ghi MG, Lanza C, Blandamura S et al (2020) Prognostic role of programmed death ligand 1 (PD-L1) and the immune microenvironment in laryngeal carcinoma. Oral Oncol 108:104836
Coussens LM, Zitvogel L, Palucka AK (2013) Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science 339(6117):286–291
Yi M, Jiao D, Xu H, Liu Q, Zhao W, Han X et al (2018) Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer 17(1):129
Fu J, Zhu J, Du F, Zhang L, Li D, Huang H, et al. Prognostic Inflammatory Index Based on Preoperative Peripheral Blood for Predicting the Prognosis of Colorectal Cancer Patients. Cancers (Basel). 2020;13(1)
Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G et al (2015) The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an international TILs working group 2014. Ann Oncol 26(2):259–271
Kozela E, Juknat A, Gao F, Kaushansky N, Coppola G, Vogel Z (2016) Pathways and gene networks mediating the regulatory effects of cannabidiol, a nonpsychoactive cannabinoid, in autoimmune T cells. J Neuroinflammation 13(1):136
Ni Y, Zhang Z, Chen G, Long W, Tong L, Zeng J (2019) Integrated analyses identify potential prognostic markers for uveal melanoma. Exp Eye Res. 187:107780
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287
Heagerty PJ, Lumley T, Pepe MS (2000) Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56(2):337–344
Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F et al (2016) Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 17(1):218
Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH et al (2019) Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 35(14):i436–i445
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160(1–2):48–61
Turner N, Wong HL, Templeton A, Tripathy S, Whiti Rogers T, Croxford M et al (2016) Analysis of local chronic inflammatory cell infiltrate combined with systemic inflammation improves prognostication in stage II colon cancer independent of standard clinicopathologic criteria. Int J Cancer 138(3):671–678
Kang BW, Seo AN, Yoon S, Bae HI, Jeon SW, Kwon OK et al (2016) Prognostic value of tumor-infiltrating lymphocytes in Epstein-Barr virus-associated gastric cancer. Ann Oncol 27(3):494–501
Bai S, Zhang P, Zhang JC, Shen J, Xiang X, Yan YB et al (2019) A gene signature associated with prognosis and immune processes in head and neck squamous cell carcinoma. Head Neck 41(8):2581–2590
Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R et al (2015) Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol 33(9):983–991
Bradford CR, Kumar B, Bellile E, Lee J, Taylor J, D’Silva N et al (2014) Biomarkers in advanced larynx cancer. Laryngoscope 124(1):179–187
Distel LV, Fickenscher R, Dietel K, Hung A, Iro H, Zenk J et al (2009) Tumour infiltrating lymphocytes in squamous cell carcinoma of the oro- and hypopharynx: prognostic impact may depend on type of treatment and stage of disease. Oral Oncol 45(10):e167–e174
Pretscher D, Distel LV, Grabenbauer GG, Wittlinger M, Buettner M, Niedobitek G (2009) Distribution of immune cells in head and neck cancer: CD8+ T-cells and CD20+ B-cells in metastatic lymph nodes are associated with favourable outcome in patients with oro- and hypopharyngeal carcinoma. BMC Cancer 9:292
Ward MJ, Thirdborough SM, Mellows T, Riley C, Harris S, Suchak K et al (2014) Tumour-infiltrating lymphocytes predict for outcome in HPV-positive oropharyngeal cancer. Br J Cancer 110(2):489–500
Kim HR, Ha SJ, Hong MH, Heo SJ, Koh YW, Choi EC et al (2016) PD-L1 expression on immune cells, but not on tumor cells, is a favorable prognostic factor for head and neck cancer patients. Sci Rep 6:36956
Wang J, Wang S, Song X, Zeng W, Wang S, Chen F et al (2016) The prognostic value of systemic and local inflammation in patients with laryngeal squamous cell carcinoma. Onco Targets Ther 9:7177–7185
Loi S, Drubay D, Adams S, Pruneri G, Francis PA, Lacroix-Triki M et al (2019) Tumor-infiltrating lymphocytes and prognosis: a pooled individual patient analysis of early-stage triple-negative breast cancers. J Clin Oncol 37(7):559–569
Chrisafis G, Wang T, Moissoglu K, Gasparski AN, Ng Y, Weigert R et al (2020) Collective cancer cell invasion requires RNA accumulation at the invasive front. Proc Natl Acad Sci U S A 117(44):27423–27434
Sun Y, He J, Shi DB, Zhang H, Chen X, Xing AY et al (2021) Elevated ZBTB7A expression in the tumor invasive front correlates with more tumor budding formation in gastric adenocarcinoma. J Cancer Res Clin Oncol 147(1):105–115
Zhao C, Wu M, Zeng N, Xiong M, Hu W, Lv W et al (2020) Cancer-associated adipocytes: emerging supporters in breast cancer. J Exp Clin Cancer Res 39(1):156
Trinchieri G (2012) Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol 30:677–706
Kondo M (2016) One niche to rule both maintenance and loss of stemness in HSCs. Immunity 45(6):1177–1179
Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21(3):309–322
Diakos CI, Charles KA, McMillan DC, Clarke SJ (2014) Cancer-related inflammation and treatment effectiveness. Lancet Oncol 15(11):e493-503
Gijsbers K, Gouwy M, Struyf S, Wuyts A, Proost P, Opdenakker G et al (2005) GCP-2/CXCL6 synergizes with other endothelial cell-derived chemokines in neutrophil mobilization and is associated with angiogenesis in gastrointestinal tumors. Exp Cell Res 303(2):331–342
Mantovani A, Cassatella MA, Costantini C, Jaillon S (2011) Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol 11(8):519–531
Mizuno R, Kawada K, Itatani Y, Ogawa R, Kiyasu Y, Sakai Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int J Mol Sci. 2019;20(3)
Augier S, Ciucci T, Luci C, Carle GF, Blin-Wakkach C, Wakkach A (2010) Inflammatory blood monocytes contribute to tumor development and represent a privileged target to improve host immunosurveillance. J Immunol 185(12):7165–7173
Condeelis J, Pollard JW (2006) Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 124(2):263–266
Gaertner F, Massberg S (2019) Patrolling the vascular borders: platelets in immunity to infection and cancer. Nat Rev Immunol 19(12):747–760
Michael JV, Wurtzel JGT, Mao GF, Rao AK, Kolpakov MA, Sabri A et al (2017) Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth. Blood 130(5):567–580
Wuerdemann N, Putz K, Eckel H, Jain R, Wittekindt C, Huebbers CU, et al. LAG-3, TIM-3 and VISTA Expression on Tumor-Infiltrating Lymphocytes in Oropharyngeal Squamous Cell Carcinoma-Potential Biomarkers for Targeted Therapy Concepts. Int J Mol Sci. 2020;22(1)
Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC et al (2013) Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39(4):782–795
Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G (2017) The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 14(12):717–734
Deng L, Huang S, Chen B, Tang Y, Huang F, Li D et al (2020) Tumor-linked macrophages promote HCC development by mediating the CCAT1/Let-7b/HMGA2 signaling pathway. Onco Targets Ther 13:12829–12843
Wang D, Jiao Z, Ji Y, Zhang S (2020) Elevated TUBA1A might indicate the clinical outcomes of patients with gastric cancer, being associated with the infiltration of macrophages in the tumor immune microenvironment. J Gastrointestin Liver Dis 29(4):509–522
Liu R, Liao YZ, Zhang W, Zhou HH (2020) Relevance of immune infiltration and clinical outcomes in pancreatic ductal adenocarcinoma subtypes. Front Oncol. 10:575264
Jairath NK, Farha MW, Jairath R, Harms PW, Tsoi LC, Tejasvi T. Prognostic value of intratumoral lymphocyte-to-monocyte ratio and M0 macrophage enrichment in tumor immune microenvironment of melanoma. Melanoma Manag. 2020;7(4):MMT51
Peres Lde P, da Luz FA, Pultz Bdos A, Brigido PC, de Araujo RA, Goulart LR et al (2015) Peptide vaccines in breast cancer: the immunological basis for clinical response. Biotechnol Adv 33(8):1868–1877
Wang W, Xiong Y, Ding X, Wang L, Zhao Y, Fei Y et al (2019) Cathepsin L activated by mutant p53 and Egr-1 promotes ionizing radiation-induced EMT in human NSCLC. J Exp Clin Cancer Res 38(1):61
Suzuki H, Sinclair L, Soderstrom PA, Lorusso G, Davies P, Ferreira LS et al (2017) A nuclear sandbank beyond the proton drip line. Phys Rev Lett. 119(19):192503
Han ML, Zhao YF, Tan CH, Xiong YJ, Wang WJ, Wu F et al (2016) Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin 37(12):1606–1622
Cai LS, Chen QX, Fang SY, Lian MQ, Lian MJ, Cai MZ (2020) ETV4 promotes the progression of gastric cancer through regulating KDM5D. Eur Rev Med Pharmacol Sci 24(5):2442–2451
Shen X, Hu K, Cheng G, Xu L, Chen Z, Du P et al (2019) KDM5D inhibit epithelial-mesenchymal transition of gastric cancer through demethylation in the promoter of Cul4A in male. J Cell Biochem 120(8):12247–12258
Komura K, Yoshikawa Y, Shimamura T, Chakraborty G, Gerke TA, Hinohara K et al (2018) ATR inhibition controls aggressive prostate tumors deficient in Y-linked histone demethylase KDM5D. J Clin Invest 128(7):2979–2995
Acknowledgement
We would like to show sincere appreciation to the editors and reviewers for critical comments on this article.
Funding
This work was supported by Postdoctoral Scientific Research Developmental Fund of Heilongjiang Province (LBH-Q18088).
Author information
Authors and Affiliations
Contributions
XW, KC, and EG conceptualized the project and all data analysis and wrote the first draft of the manuscript. XM, CA, LG, CZ, JG, XY, JS, and WY contributed to the collection of specimens, the processing, analysis, and interpretation of data. XL and SM contributed to guide the data analysis, and manuscript writing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
We had obtained ethical approval from the ethics committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, X., Cao, K., Guo, E. et al. Assessment of immune status of laryngeal squamous cell carcinoma can predict prognosis and guide treatment. Cancer Immunol Immunother 71, 1199–1220 (2022). https://doi.org/10.1007/s00262-021-03071-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-021-03071-7