Abstract
Purpose
To evaluate the efficacy of a vendor-specific deep learning reconstruction algorithm (DLRA) in enhancing image quality and focal lesion detection using three-dimensional T1-weighted gradient-echo images in gadoxetic acid-enhanced liver magnetic resonance imaging (MRI) in patients at a high risk of hepatocellular carcinoma.
Materials and methods
In this retrospective analysis, 83 high-risk patients with hepatocellular carcinoma underwent gadoxetic acid-enhanced liver MRI using a 3-T scanner. Triple arterial phase, high-resolution portal venous phase, and high-resolution hepatobiliary phase images were reconstructed using conventional reconstruction techniques and DLRA (AIRTM Recon DL; GE Healthcare) for subsequent comparison. Image quality and solid focal lesion detection were assessed by three abdominal radiologists and compared between conventional and DL methods. Focal liver lesion detection was evaluated using figures of merit (FOMs) from a jackknife alternative free-response receiver operating characteristic analysis on a per-lesion basis.
Results
DLRA-reconstructed images exhibited significantly improved overall image quality, image contrast, lesion conspicuity, vessel conspicuity, and liver edge sharpness and reduced subjective image noise, ringing artifacts, and motion artifacts compared to conventionally reconstructed images (all P < 0.05). Although there was no significant difference in the FOMs of non-cystic focal liver lesions between the conventional and DL methods, DLRA-reconstructed images showed notably higher pooled sensitivity than conventionally reconstructed images (P < 0.05) in all phases and higher detection rates for viable post-treatment HCCs in the arterial and hepatobiliary phases (all P < 0.05).
Conclusions
Implementing DLRA can enhance the image quality in 3D T1-weighted gradient-echo sequences of gadoxetic acid-enhanced liver MRI examinations, leading to improved detection of viable post-treatment HCCs.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and is associated with high mortality worldwide [1]. Gadoxetic acid-enhanced magnetic resonance imaging (MRI) is widely employed for the noninvasive identification of HCC in high-risk individuals, as advocated by leading guidelines [2,3,4,5]. Gadoxetic acid-enhanced MRI has proven particularly effective in identifying small HCCs (< 2 cm) because of the strong contrast between the lesion and the liver background during the hepatobiliary phase (HBP), resulting in high sensitivity [6]. However, factors such as transient respiratory motion during the arterial phase (AP), low signal-to-noise ratio (SNR), and a limited temporal window of dynamic phases may compromise the image quality and focal lesion detectability of gadoxetic acid-enhanced MRI [7, 8]. Over the years, numerous rapid imaging techniques, such as parallel imaging and compressed sensing, have been developed to enhance the temporal resolution of the dynamic phases [9, 10]. Despite these advancements, the utility of these techniques remains restricted by the intrinsic trade-off between scan duration and image quality parameters, such as SNR and spatial resolution.
Recently, deep learning (DL) algorithms have been applied to MRI, including image acquisition and reconstruction [11,12,13]. Specifically, numerous DL reconstruction algorithms (DLRAs) have been developed to enhance image quality by reducing noise and mitigating artifacts in two-dimensional T2-weighted fast spin-echo, T1-weighted gradient-echo (GRE), and diffusion-weighted images acquired using clinical scanners [14,15,16,17]. DL reconstruction amplifies the image sharpness and clarity by suppressing artifacts such as Gibbs ringing and increasing the SNR [18, 19]. Recently, vendor-specific DL reconstruction was introduced for 3D T1-weighted GRE sequences, which are commonly used in dynamic abdominal MRI scans (AIRTM Recon DL, GE Healthcare, Waukesha, Wisconsin, USA). Based on previous research outcomes that demonstrated the efficacy of DL reconstruction of 2D T1-weighted GRE, 3D T2-weighted sequences in other organs, and 3D T1-weighted MR enterography [20,21,22], we hypothesized that the implementation of DL reconstruction may enhance image quality in 3D T1-weighted GRE sequences of gadoxetic acid-enhanced liver MRI examinations. Consequently, this may improve focal lesion detectability, bearing significant clinical ramifications for early identification and management of HCC.
Therefore, this retrospective study aimed to investigate the effectiveness of a vendor-specific DLRA for 3D T1-weighted GRE sequences in improving image quality and focal lesion detectability of gadoxetic acid-enhanced liver MRI of patients at a high risk of HCC.
Materials and methods
This retrospective study was approved by the Institutional Review Board of Seoul National University Hospital (IRB No. H-2206-176-1336), which waived the requirement for informed consent.
Patients
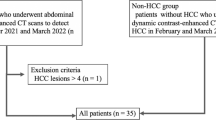
Between September 2021 and May 2022, 106 consecutive patients underwent gadoxetic acid-enhanced dynamic liver MRI using a 3-T scanner, and DLRA was applied to the acquired images. Among these patients, we enrolled those who fulfilled the following inclusion criteria: (a) patients at risk of developing HCC and (b) age ≥ 18 years. Patients were excluded if (a) more than five focal liver lesions (FLLs) were found and (b) liver MRI was not consistent with the routine protocol for any reason. Thus, 23 patients were excluded because of no risk factor for HCC (n = 3) or equal or more than five FLLs (n = 20). Finally, 83 patients (male, n = 49; median age 65.8 [interquartile range 60.0–72.0] years) were included in this study (Fig. 1; Table 1).
MRI acquisition
All examinations were performed using a 3-T scanner (SIGNA Premier; GE Healthcare). Liver MRI examination consisted of the following sequences: heavily T2-weighted imaging (T2WI), fat-suppressed T2WI, precontrast T1-weighted imaging (T1WI), postcontrast T1WI (arterial, portal venous, transitional, and hepatobiliary phases), dual-echo imaging, and diffusion-weighted imaging with three b-values (50, 400, and 800 s/mm2; Table 2). A standard dose of gadoxetic acid (0.025 mmol/kg; Primovist/Eovist; Bayer Healthcare, Berlin, Germany) was administered with a 25 mL saline flush at a rate of 1 mL/s. Dynamic phases, including triple AP, portal venous phase (PVP, 60 s after contrast injection), transitional phase (3 min), and HBP (20 min), were obtained using a fat-suppressed 3D T1-weighted GRE sequence (LAVA, GE Healthcare) with standard resolution after the injection. Furthermore, during PVP and HBP, high-resolution axial images with a 2-mm slice thickness and 1-mm reconstruction interval were routinely obtained using a 3D T1-weighted GRE sequence immediately after standard-resolution imaging.
Deep learning reconstruction algorithm
The DLRA used in this study was a vendor-provided prototype version of the AIRTM Recon DL 3D (GE Healthcare) [14], which is now commercially available. This image reconstruction technique includes a deep convolution neural network to reduce noise, reduce Gibbs ringing, and improve image sharpness in all three directions. The MR images were reconstructed offline from the raw k-space data at a user-specified denoising level. In addition, among the liver MR images, triple AP, high-resolution PVP, and high-resolution HBP images were reconstruct ed using DL reconstruction at a 75% denoising level [22] and used for comparison between conventional and DL reconstruction.
Image analysis
All 83 pairs of MRI datasets (conventional and DL reconstructions) were evaluated by three abdominal radiologists with 5–7 years of experience in interpreting abdominal MR images. Triple AP, high-resolution PVP, and high-resolution HBP images were reconstructed using the conventional reconstruction technique and DLRA. Precontrast T1W and heavily T2W images reconstructed with the conventional method were added to both sets to determine whether enhancement existed and to exclude cysts. Therefore, conventional and DL image sets included axial precontrast T1W, heavily T2W, and postcontrast triple AP, high-resolution PVP, and high-resolution HBP images reconstructed with either conventional or DL reconstruction, respectively.
Paired sets of MR images generated with conventional and DL reconstruction methods were provided in random order. The image sets were anonymous and randomly assigned to folders A or B to avoid any bias. The reviewers were blinded to both the reconstruction method and clinical information except that the patients were at risk of developing HCC. A minimum 4-week washout period was used between the evaluations of folders A and B.
Image quality
For each image set, the degree of contrast enhancement of the hepatic vessels (right hepatic artery on AP, right portal vein on PVP, and right hepatic vein on HBP images), conspicuity of the hepatic vessels, liver edge sharpness, ringing artifacts, susceptibility artifacts, motion artifacts, subjective image noise levels, and overall image quality were evaluated for each phase. The detailed scale information used for image evaluation is provided in Table E1.
Detectability of focal liver lesions
Reviewers were asked to detect up to four FLLs per patient, excluding arterioportal shunts, post-treatment changes, and cysts, and to record the conspicuity score of each suspected lesion in each of the three phases (AP, PVP, and HBP) on a 4-point scale (Table E1). They also were requested to indicate the location and image slices of suspected lesions for localization.
Reference standards
Two experienced abdominal radiologists (J.H.Y. and J.H.K., with 15 and eight years of experience in interpreting abdominal MRI, respectively), who did not participate in the review session, evaluated the entire MRI sequence and follow-up images to identify and characterize FLLs in consensus. A total of 89 FLLs were identified and categorized according to the Liver Imaging Reporting and Data System (LI-RADS) version 2018 [23] and LI-RADS 2017 Treatment Response algorithm [24].
Statistical analysis
The Mann–Whitney U test was used to compare the image quality scores, which were averaged across the three readers, between the conventional and DL methods. The performance of each reader in detecting FLLs in the AP, PVP, and HBP was evaluated using figures of merit (FOMs) from a jackknife alternative free-response receiver operating characteristic (JAFROC) analysis (version 4.2.1) on a per-lesion basis. Comparisons of the FOMs were performed using the Hillis improvement [25] of the method described by Dorfman et al. [26] with the modeling assumption of random reader-random cases. Per-lesion sensitivity was calculated as the number of correctly localized lesions divided by the total number of lesions. The false-positive interpretation rate was defined as the number of false-positive interpretations divided by the total number of MRI scans. McNemar’s test was used to compare the sensitivities of the conventional and DL methods for individual readers. The generalized estimating equation was used to compare the pooled sensitivities, lesion conspicuity scores, and rates of false-positive interpretations between the conventional and DL methods.
Interreader agreement levels were evaluated using Gwet’s AC1 [27], as the prevalence of a trait and the bias of raters affect the kappa statistic. For example, if there is an imbalance in the number of positive or negative ratings, the kappa coefficient decreases. Similarly, if there is a consistent variation in the ratings made by raters, the kappa coefficient may be artificially high. In contrast, Gwet's AC1 is not affected by trait prevalence or rater bias, thus providing more reliable results [27]. For the interpretation of the chance-corrected agreement, we used the criteria suggested by Landis and Koch [28]: 0.00–0.20, slight agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, substantial agreement; 0.81–1.00, almost perfect agreement.
Results
Patient demographics and FLL characteristics
The clinical characteristics of the patients are summarized in Table 1. In total, 89 FLLs (median diameter, 1.0 cm; interquartile range, 0.8–1.2 cm) were included in this study. Detailed information on the FLLs is presented in Table 1.
Image quality parameter comparisons
DL-reconstructed AP, PVP, and HBP images showed compared to conventionally reconstructed images significantly better overall image quality, image contrast, vessel conspicuity, liver edge sharpness, significantly less subjective image noise, ringing artifacts, and motion artifacts (P < 0.05 for all; Table 3; Fig. 2). In terms of susceptibility artifacts, no difference in AP and PVP images was found between conventional and DL methods (3.02 ± 0.46 vs. 3.01 ± 0.40 and 3.02 ± 0.47 vs. 3.02 ± 0.48; P = 0.505 and 0.886, respectively). However, for HBP, the DL method resulted in significantly fewer artifacts than the conventional method (3.21 ± 0.53 vs. 3.11 ± 0.46; P = 0.037; Table 3). In addition, the DL-reconstructed AP, PVP, and HBP images showed a better conspicuity score for FLLs than conventionally reconstructed images (P < 0.05; Table 3; Fig. 3).
Gadoxetic acid-enhanced MRI of a 66-year-old male patient with chronic hepatitis B. The conventional arterial (A), portal venous (B), and hepatobiliary (C) phases in the top row show worse vessel conspicuity (arrows), image contrast, liver edge sharpness, and increased ringing artifact (arrows) compared with the DL-reconstructed arterial (D), portal venous (E), and hepatobiliary (F) phases in the bottom row
Comparison of FLL detectability between conventional and DL reconstruction
In all three AP, PVP, and HBP images, the conventional and DL methods did not show significant differences in JAFROC FOMs (0.603 vs. 0.611, 0.603 vs. 0.635, and 0.648 vs. 0.682; P = 0.847, 0.467, and 0.617, respectively; Table 4). However, the DL reconstruction of AP, PVP, and HBP images demonstrated significantly higher pooled sensitivity than the conventional method (24.3% [65/267] vs. 21.7% [58/267], 30.7% [82/267] vs. 24.7% [66/267], and 41.9% [112/267] vs. 36.3% [97/267], respectively; P < 0.05). Table 4 summarizes the FLL detection performance of each reviewer.
Comparison of HCC diagnosis between conventional and DL reconstruction
The detection rates of LR-TR viable lesions in DL-reconstructed AP and HBP images were significantly higher than those in the corresponding conventionally reconstructed images (37.5% [18/48] vs. 22.9% [11/48] and 52.1% [25/48] vs. 39.6% [19/48], respectively; P < 0.05; Table 5; Fig. 3). In addition, DL-reconstructed PVP images showed a higher lesion detection rate for LR-3 than conventionally reconstructed images (24.0% [18/75] vs. 16.0% [12/75]; P = 0.031). In all other LI-RADS categories, no significant difference in lesion detection rates was observed between the conventional and DL methods (Table 5).
Gadoxetic acid-enhanced MRI of a 64-year-old female patient with chronic hepatitis B. The conventional arterial (A) and hepatobiliary (B) phases in the top row show worse LR-TR viable lesion (arrows) conspicuity compared with the DL-reconstructed arterial (C) and hepatobiliary (D) phases in the bottom row
Interreader agreement for image quality evaluation
Interreader agreements for image contrast, vessel conspicuity, liver edge sharpness, subjective image noise, ringing artifact, susceptibility artifact, motion artifact, and overall image quality in AP, PVP, and HBP ranged from moderate to almost perfect (Gwet’s AC1 range, 0.516–0.969). In terms of lesion conspicuity, a moderate-to-substantial agreement was found in the AP and PVP (Gwet’s AC1 range, 0.626–0.861), whereas agreement was poor to moderate in the HBP (Gwet’s AC1 range, 0.150–0.593; Table E2).
Discussion
In this study, we demonstrated that by implementing vendor-specific DLRA in 3D T1-weighted GRE sequences for gadoxetic acid-enhanced liver MR examinations of high-risk HCC patients, image quality and sensitivity in detecting non-cystic FLLs and LR-TR viable lesions were notably improved. DLRA substantially improved the overall image quality, image contrast, vessel conspicuity, and liver edge sharpness, while reducing subjective image noise, ringing artifacts, and motion artifacts. The enhancement in image quality with the DLRA can be attributed to the inherent ability of DL algorithms to address the noise and artifacts present in the imaging data. By operating on raw and complex-valued k-space data, these algorithms can suppress ringing artifacts, reduce noise, and enhance image sharpness and clarity [18]. The observed enhancements in image quality parameters are consistent with prior research that has shown the efficacy of DLRA in improving the image quality for a range of MRI sequences. These include 2D T1-weighted GRE sequences, 3D T1-weighted MR enterography, and T2-weighted sequences of various organs [15,16,17, 20,21,22].
Notably, in our study, the detection of LR-TR viable lesions in the AP and HBP was significantly improved with DLRA application. This finding is clinically relevant, as locoregional treatments are frequently used for early- and intermediate-stage HCCs. Furthermore, DL-reconstructed 3D T1-weighted GRE images displayed higher conspicuity scores for FLLs, ascribed to increased contrast and liver edge sharpness, as well as reduced noise and artifacts. The enhanced image sharpness and clarity and reduced noise and artifacts contributed to better identification of LR-TR viable lesions and higher conspicuity scores for FLLs. Given that the early detection of LR-TR viable lesions may enable additional treatment to achieve complete necrosis of HCC lesions and improve survival, this finding has substantial clinical value. Our results suggest that DLRA can enhance the diagnostic accuracy of gadoxetic acid-enhanced liver MRI for early-stage HCC detection in high-risk individuals, potentially improving patient management and outcomes. Future multicenter prospective studies with larger cohorts are required to validate and generalize these findings.
In our study, we employed triple AP imaging, acquiring three independent 3D datasets during a single breath-hold, and applied high acceleration factors to mitigate transient respiratory motion in gadoxetic acid-enhanced liver MRI. This is crucial because the occurrence of such motion in gadoxetic acid-enhanced liver MRI is not negligible, with reported incidence rates ranging from 3.2 to 26.7% [10, 29]. Because the depiction of AP hyperenhancement is crucial for HCC diagnosis [2], the acquisition of high-quality 3D GRE images during AP with high temporal and spatial resolution may benefit high-risk HCC patients. Various techniques, such as parallel imaging, view sharing, and compressed sensing, have been employed to achieve rapid AP image acquisition while preserving spatial resolution [8,9,10]. Nonetheless, these techniques may encounter challenges related to low SNR, artifacts, and motion-related problems. We discovered that DLRA significantly reduced image noise and artifacts while improving image resolution using a super-resolution algorithm [18, 19, 22]. By enabling a higher acceleration factor in the 3D T1-weighted GRE sequences, the breath-hold time might be reduced without sacrificing image quality. Recent research has also demonstrated that DLRA methods decrease 2D T1-weighted GRE sequence acquisition time while improving image quality. Our study findings indicate that triple AP imaging using 3D T1-weighted GRE sequences with DLRA and high acceleration factors can assist in the diagnosis of HCC, especially with gadoxetic acid.
Regarding interreader agreement, all items, except for lesion conspicuity score in the HBP (which ranged from poor to moderate), demonstrated moderate to almost perfect agreement. The HBP is known to be the most sensitive phase for lesion detection among the dynamic phases of liver MRI. Therefore, in terms of lesion conspicuity scores, it is believed that reader preferences (sensitivity vs. specificity) may have a relatively more pronounced impact on HBP compared to other phases.
Our study has some limitations. First, the retrospective nature of the study and the inclusion of a single-center cohort may have limited the generalizability of our findings. Second, we utilized a vendor-specific DLRA, which may restrict the applicability of our results to other MRI systems and DL algorithms. Third, the relatively small sample size and number of lesions may constrain the statistical power of our analyses. Forth, subtraction images were not included in our study data sets. Given that subtraction images could be beneficial for the evaluation of non-cystic focal liver lesions, it would be valuable to conduct future studies that incorporate subtraction images.
In conclusion, utilizing vendor-specific DLRA for 3D T1-weighted GRE sequences in gadoxetic acid-enhanced liver MRI can greatly enhance the image quality and improve the detection of FLLs, especially LR-TR viable lesions. The clinical adoption of DLRA for 3D T1-weighted GRE in liver MRI could potentially aid in the diagnosis and treatment of HCC.
Data availability
The datasets generated or analyzed during the study are not publicly available due to the patients’ personal medical information but are available from the corresponding author on reasonable request.
References
El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–27. https://doi.org/10.1056/NEJMra1001683.
Chernyak V, Fowler KJ, Kamaya A, Kielar AZ, Elsayes KM, Bashir MR, et al. Liver Imaging Reporting and Data System (LI-RADS) Version 2018: Imaging of Hepatocellular Carcinoma in At-Risk Patients. Radiology. 2018;289(3):816–30. https://doi.org/10.1148/radiol.2018181494.
European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. https://doi.org/10.1016/j.jhep.2018.03.019.
Omata M, Cheng AL, Kokudo N, Kudo M, Lee JM, Jia J, et al. Asia–Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatology International. 2017;11(4):317–70. https://doi.org/10.1007/s12072-017-9799-9.
Korean Liver Cancer A, National Cancer Center K. 2022 KLCA-NCC Korea Practice Guidelines for the Management of Hepatocellular Carcinoma. Korean Journal of Radiology. 2022;23(12):1126–240. https://doi.org/10.3348/kjr.2022.0822.
Roberts LR, Sirlin CB, Zaiem F, Almasri J, Prokop LJ, Heimbach JK, et al. Imaging for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. Hepatology. 2018;67(1):401–21. https://doi.org/10.1002/hep.29487.
Jang EB, Kim DW, Choi SH, Hong SB, Park T, Ko Y, et al. Transient severe motion artifacts on gadoxetic acid-enhanced MRI: risk factor analysis in 2230 patients. Eur Radiol. 2022;32(12):8629–38. https://doi.org/10.1007/s00330-022-08885-2.
Yoon JH, Lee JM, Yu MH, Kim EJ, Han JK. Triple Arterial Phase MR Imaging with Gadoxetic Acid Using a Combination of Contrast Enhanced Time Robust Angiography, Keyhole, and Viewsharing Techniques and Two-Dimensional Parallel Imaging in Comparison with Conventional Single Arterial Phase. Korean Journal of Radiology. 2016;17(4):522–32.https://doi.org/10.3348/kjr.2016.17.4.522.
Nam JG, Lee JM, Lee SM, Kang HJ, Lee ES, Hur BY, et al. High Acceleration Three-Dimensional T1-Weighted Dual Echo Dixon Hepatobiliary Phase Imaging Using Compressed Sensing-Sensitivity Encoding: Comparison of Image Quality and Solid Lesion Detectability with the Standard T1-Weighted Sequence. Korean Journal of Radiology. 2019;20(3):438–48. https://doi.org/10.3348/kjr.2018.0310.
Kim JH, Yoon JH, Bae JS, Park S, Han S, Lee JM. Multiarterial Phase Acquisition in Gadoxetic Acid-Enhanced Liver MRI for the Detection of Hypervascular Hepatocellular Carcinoma in High-Risk Patients: Comparison of Compressed Sensing Versus View Sharing Techniques. Invest Radiol. 2023;58(2):139–47. https://doi.org/10.1097/RLI.0000000000000910.
Montalt-Tordera J, Muthurangu V, Hauptmann A, Steeden JA. Machine learning in Magnetic Resonance Imaging: Image reconstruction. Phys Med. 2021;83:79–87. https://doi.org/10.1016/j.ejmp.2021.02.020.
Gore JC. Artificial intelligence in medical imaging. Magn Reson Imaging. 2020;68:A1–A4. https://doi.org/10.1016/j.mri.2019.12.006.
Mazurowski MA, Buda M, Saha A, Bashir MR. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J Magn Reson Imaging. 2019;49(4):939–54. https://doi.org/10.1002/jmri.26534.
Lebel RM. Performance characterization of a novel deep learning-based MR image reconstruction pipeline. ArXiv. 2020; http://arxiv.org/abs/2008.06559.
Wessling D, Herrmann J, Afat S, Nickel D, Almansour H, Keller G, et al. Application of a Deep Learning Algorithm for Combined Super-Resolution and Partial Fourier Reconstruction Including Time Reduction in T1-Weighted Precontrast and Postcontrast Gradient Echo Imaging of Abdominopelvic MR Imaging. Diagnostics (Basel). 2022;12(10). https://doi.org/10.3390/diagnostics12102370.
Almansour H, Herrmann J, Gassenmaier S, Lingg A, Nickel MD, Kannengiesser S, et al. Combined Deep Learning-based Super-Resolution and Partial Fourier Reconstruction for Gradient Echo Sequences in Abdominal MRI at 3 Tesla: Shortening Breath-Hold Time and Improving Image Sharpness and Lesion Conspicuity. Acad Radiol. 2022. https://doi.org/10.1016/j.acra.2022.06.003.
Hahn S, Yi J, Lee HJ, Lee Y, Lim YJ, Bang JY, et al. Image Quality and Diagnostic Performance of Accelerated Shoulder MRI With Deep Learning-Based Reconstruction. AJR Am J Roentgenol. 2022;218(3):506–16. https://doi.org/10.2214/AJR.21.26577.
Lee DH, Park JE, Nam YK, Lee J, Kim S, Kim YH, et al. Deep learning-based thin-section MRI reconstruction improves tumour detection and delineation in pre- and post-treatment pituitary adenoma. Sci Rep. 2021;11(1):21302. https://doi.org/10.1038/s41598-021-00558-2.
Wang X, Ma J, Bhosale P, Ibarra Rovira JJ, Qayyum A, Sun J, et al. Novel deep learning-based noise reduction technique for prostate magnetic resonance imaging. Abdom Radiol (NY). 2021;46(7):3378–86. https://doi.org/10.1007/s00261-021-02964-6.
Sun S, Tan ET, Mintz DN, Sahr M, Endo Y, Nguyen J, et al. Evaluation of deep learning reconstructed high-resolution 3D lumbar spine MRI. Eur Radiol. 2022;32(9):6167–77. https://doi.org/10.1007/s00330-022-08708-4.
Chazen JL, Tan ET, Fiore J, Nguyen JT, Sun S, Sneag DB. Rapid lumbar MRI protocol using 3D imaging and deep learning reconstruction. Skeletal Radiol. 2023. https://doi.org/10.1007/s00256-022-04268-2.
Son JH, Lee Y, Lee HJ, Lee J, Kim H, Lebel MR. LAVA HyperSense and deep-learning reconstruction for near-isotropic (3D) enhanced magnetic resonance enterography in patients with Crohn's disease: utility in noise reduction and image quality improvement. Diagn Interv Radiol. 2023. https://doi.org/10.4274/dir.2023.232113.
Liver Imaging Reporting and Data System [Internet]. [cited February 7, 2023]. Available from: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/LI-RADS.
Kielar A, Fowler KJ, Lewis S, Yaghmai V, Miller FH, Yarmohammadi H, et al. Locoregional therapies for hepatocellular carcinoma and the new LI-RADS treatment response algorithm. Abdom Radiol (NY). 2018;43(1):218–30. https://doi.org/10.1007/s00261-017-1281-6.
Hillis SL. A comparison of denominator degrees of freedom methods for multiple observer ROC analysis. Statistics in Medicine. 2007;26(3):596–619. https://doi.org/10.1002/sim.2532.
Dorfman DD, Berbaum KS, Metz CE. Receiver operating characteristic rating analysis. Generalization to the population of readers and patients with the jackknife method. Invest Radiol. 1992;27(9):723–31.
Wongpakaran N, Wongpakaran T, Wedding D, Gwet KL. A comparison of Cohen's Kappa and Gwet's AC1 when calculating inter-rater reliability coefficients: a study conducted with personality disorder samples. BMC Medical Research Methodology. 2013;13:61. https://doi.org/10.1186/1471-2288-13-61.
Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–74.
Kim DW, Choi SH, Park T, Kim SY, Lee SS, Byun JH. Transient Severe Motion Artifact on Arterial Phase in Gadoxetic Acid-Enhanced Liver Magnetic Resonance Imaging: A Systematic Review and Meta-analysis. Invest Radiol. 2022;57(1):62–70. https://doi.org/10.1097/RLI.0000000000000806.
Acknowledgments
Statistical analyses were supported by Medical Research Collaborating Center (MRCC) of Seoul National University Hospital.
Funding
This work was supported by GE Healthcare (Project Number: 0620216290).
Author information
Authors and Affiliations
Contributions
Conceptualization: J.M.L. Data curation: J.H.K. Formal analysis: J.H.K. Funding acquisition: N/A. Investigation: S.W.K, J.P, S.H.B. Methodology: J.M.L., J.H.Y., J.H.K. Project administration: J.M.L. Resources: J.M.L. Software: N/A. Supervision: J.M.L., J.H.Y. Validation: N/A. Visualization: J.H.K. Writing-original draft: J.M.L., J.H.K. Writing-review and editing: J.M.L, J.H.Y, J.H.K.
Corresponding author
Ethics declarations
Conflict of interest
We received technical support from GE Healthcare for the submitted work. J.M.L. has received grants from Bayer Healthcare, Canon Healthcare, Philips Heathcare, GE Healthcare, CMS, Guerbet, Samsung Medison, and Bracco. J.M.L. has received personal fees from Bayer Healthcare, Siemens Healthineer, Samsung Medison, Guerbet, and Philips Healthcare. J.H.Y. has received honorarium from Bayer Healthcare and personal fee from Philips Healthcare. For the remaining authors none were declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, J.H., Yoon, J.H., Kim, S.W. et al. Application of a deep learning algorithm for three-dimensional T1-weighted gradient-echo imaging of gadoxetic acid-enhanced MRI in patients at a high risk of hepatocellular carcinoma. Abdom Radiol 49, 738–747 (2024). https://doi.org/10.1007/s00261-023-04124-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-023-04124-4