Abstract
Purpose
To assess the performance of a machine learning model trained with contrast-enhanced CT-based radiomics features in distinguishing benign from malignant solid renal masses and to compare model performance with three abdominal radiologists.
Methods
Patients who underwent intra-operative ultrasound during a partial nephrectomy were identified within our institutional database, and those who had pre-operative contrast-enhanced CT examinations were selected. The renal masses were segmented from the CT images and radiomics features were derived from the segmentations. The pathology of each mass was identified; masses were labeled as either benign [oncocytoma or angiomyolipoma (AML)] or malignant [clear cell, papillary, or chromophobe renal cell carcinoma (RCC)] depending on the pathology. The data were parsed into a 70/30 train/test split and a random forest machine learning model was developed to distinguish benign from malignant lesions. Three radiologists assessed the cohort of masses and labeled cases as benign or malignant.
Results
148 masses were identified from the cohort, including 50 benign lesions (23 AMLs, 27 oncocytomas) and 98 malignant lesions (23 clear cell RCC, 44 papillary RCC, and 31 chromophobe RCCs). The machine learning algorithm yielded an overall accuracy of 0.82 for distinguishing benign from malignant lesions, with an area under the receiver operating curve of 0.80. In comparison, the three radiologists had significantly lower accuracies (p = 0.02) ranging from 0.67 to 0.75.
Conclusion
A machine learning model trained with CT-based radiomics features can provide superior accuracy for distinguishing benign from malignant solid renal masses compared to abdominal radiologists.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Solid renal masses are commonly detected on medical imaging studies either in the workup of hematuria or as an incidental finding [1]. Such solid renal masses include malignant entities, such as renal cell carcinoma (RCC), but also benign lesions, such as oncocytomas and angiomyolipomas (AMLs) [2, 3]. Unfortunately, radiologists have limited sensitivity and specificity for distinguishing benign from malignant entities via cross-sectional imaging (Fig. 1) [4].
The clinical management of solid renal masses is variable across institutions. Some institutions employ ultrasound-guided percutaneous biopsy for the diagnosis of renal masses. Biopsy is an invasive procedure and can fail to provide a diagnosis in up to 20% of cases [5]. Complications from biopsy, such as bleeding and tumor seeding, are rare but do occur [6]. Nevertheless, many institutions forego biopsy and proceed directly to surgical resection, which is within the standard of care [7]. A study by Frank et al. demonstrated in a cohort of 2770 patients that 12.8% of resected solid renal masses were benign entities [8]; the proportion of benign entities was greater for small lesions, approximately 19.9% for tumors < 4 cm, 22.0% for < 3 cm, 22.4% for < 2 cm, and 46.3% for < 1 cm. A reliable method for distinguishing benign from malignant solid renal masses is desired to reduce the need for biopsy and mitigate the unnecessary resection of benign entities.
Radiomics is a field of image analysis and interpretation that extracts quantitative imaging features, and which has the potential to identify complex patterns beyond that which is detectable by the human eye [9, 10]. Radiomics features can be correlated with clinical information in an attempt to predict patient prognosis, response to therapy, or tumor biology from the images alone. The application of radiomics to solid renal masses may provide a means to distinguish benign from malignant lesions.
Machine learning algorithms can be used to process radiomics data [11] and construct a model to predict the likelihood of benignity versus malignancy. Multiple studies have been conducted using radiomics and machine learning to compare RCCs versus oncocytomas or RCCs versus fat-poor angiomyolipomas [12]; however, these comparisons are of limited utility in clinical workflow, as only a single benign entity instead of the corpus (fat-poor angiomyolipomas + oncocytomas) of benign entities is evaluated, and such a focused conundrum is not encountered clinically. Furthermore, several studies have used radiomics and machine learning to distinguish the histologic subtypes of RCC, i.e., clear cell, chromophobe, and papillary subtypes [13, 14]. Yet, such a study is also incongruent with clinical workflow, as it assumes that benignity is already excluded. Some studies with radiomics and machine learning have focused on the more clinically applicable problem of comparing the aggregate of benign (fat-poor angiomyolipomas and oncocytomas) lesions to the aggregate of malignant (clear cell, chromophobe, and papillary RCCs) lesions [12]. However, many of these studies have either a small sample size and/or the inclusion of masses that are obviously malignant and therefore not a diagnostic dilemma.
The purpose of this study was to investigate the use of radiomics and a machine learning model to distinguish benign from malignant solid renal masses on contrast-enhanced pre-operative CT data. The cohort used in this study is sourced from our database of patients who underwent partial nephrectomy and required intra-operative ultrasound. As a result, most of the lesions tended to be small (< 4 cm) and ambiguous on pre-operative imaging, and therefore a formidable challenge for a radiomics and machine learning approach. Furthermore, for comparison three radiologists assessed the cohort of renal masses and attempted to label the masses as benign or malignant. The performances of the machine learning model and the radiologists were compared.
Materials and methods
This Health Insurance Portability and Accountability Act—compliant study was approved by our institutional review board. The need for written informed consent was waived given the retrospective nature of the study.
Study population
The patient population included in this study was comprised of patients who required intra-operative ultrasound guidance during a partial nephrectomy between January 2002 and March 2020. Intra-operative ultrasound is routinely requested for localization of renal masses and to delineate lesion margins for surgical planning of all partial nephrectomies performed at our institution. Inclusion criteria were as follows: adults ≥ 18 years of age; patients who underwent partial nephrectomy for a renal mass; diagnostic pathology reported from the resected mass, with a final diagnosis of angiomyolipoma, oncocytoma, clear cell RCC, papillary RCC, or chromophobe RCC; and those patients who had a pre-operative contrast-enhanced CT irrespective of scanner vendor or imaging parameters, such as peak voltage or available slice thickness. Our institution does not routinely perform ultrasound-guided biopsy of renal masses and therefore the initial tissue diagnosis is via the resected samples. None of the angiomyolipomas had gross fat on cross-sectional imaging and therefore could not be identified via this criterion.
Image acquisition
Patients underwent contrast-enhanced CT examinations via a variety of scanner manufacturers and imaging protocols, depending on whether the patients were scanned within our own institution or referred from an outside facility. Individual were scanned with a multidetector CT from either GE Healthcare (n = 42), Philips (n = 10), Siemens (n = 85), or Toshiba (n = 11) at 100–140 kVp with a variable tube current (mA). The thinnest available slices were used for analysis, and ranged from 0.625 to 5 mm. All subjects received iohexol 300 mgI/mL for contrast.
Analysis
CT datasets were imported into a radiomics research prototype (syngoVia Frontier, Siemens Healthineers, Forchheim, Germany) [15]. Volumetric segmentation of each lesion was performed semi-automatically by the software, with small manual adjustments performed as needed by a radiologist (AW) with 4 years of experience. The segmentations encompassed the entire mass to the margins and included any calcifications, cystic components, or areas of central hypoattenuation if present. The entire segmented volumes were used for analysis. Segmentations were confirmed by an abdominal fellowship-trained radiologist with 15 years of experience (AK). Radiomics features were computed from the built-in PyRadiomics framework and were subsequently exported from the software. Radiomics features included 28 gray-level co-occurrence matrix features, 16 gray-level size zone matrix features, 16 gray-level run length matrix features, and 19 first order features.
The maximum cross-sectional diameter of each lesion was computed from the segmentations. These maximum cross-sections were averaged for the entire cohort and also among the groups of benign and malignant lesions. Cross-sectional size was compared between the benign and malignant lesions using an unpaired equal variance Student’s t-test (p < 0.05).
Machine learning predictive modeling
The 148 renal lesions were grouped into benign (angiomyolipoma and oncocytoma) and malignant (clear cell, papillary, and chromophobe RCC) categories. Data were divided into a 70/30 train/test split [16] in a random fashion with stratification based on class labels.
Model building was performed on the segmented lesions. A random forest machine learning classifier [17, 18] was implemented in Python via the sklearn toolkit and was validated with fivefold cross-validation on the training set. Receiver operating characteristic (ROC) and precision-recall curves were created, and summary statistics were computed for the model performance, including sensitivity (recall), specificity, positive predictive value (precision), negative predictive value, and accuracy, along with area under the curve (AUC), Matthews correlation coefficient, Youden’s J statistic, and a weighted F1 score.
Radiomic feature ranking for the random forest model was performed to determine which features were the greatest contributors to model performance. Feature importances were computed.
Reader evaluation
The cohort of renal masses was randomized such that the list of subjects was not grouped by pathology. This randomized list of 148 subjects was provided to three abdominal radiologists for independent review. The readers were aware that each subject had a renal mass but were blinded to the existence of follow-up imaging, subsequent surgery, and pathologic diagnosis. The three radiologists (Readers 1, 2, 3) had 40 years (RBJ), 15 years (AK), and 2 years (LS) of experience, respectively. Each radiologist labeled the cases as either benign or malignant renal masses. The number of true negative, true positive, false negative, and false positive cases was determined for each reader. The sensitivity (recall), specificity, positive predictive value (precision), negative predictive value, accuracy, and F1 score were computed for each reader.
Results
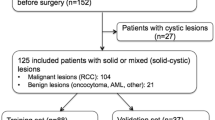
From the database, our search criteria yielded 236 patients. Of these, 88 patients did not have pre-operative contrast-enhanced CT; these patients had an MRI, ultrasound, or non-contrast CT instead of a contrast-enhanced CT, or the imaging was unavailable due to referral from an outside institution (Fig. 2). As a result, 148 patients were included in the study (87 male, 61 female; mean age ± standard deviation = 57.5 ± 12.1 years; age range = 25–87 years). Of these 148 patients, 23 had AMLs (15.5%), 27 had oncocytomas (18.2%), 23 had clear cell RCC (15.5%), 44 had papillary RCC (29.7%), and 31 had chromophobe RCC (20.9%). Each of the 148 patients had a single renal mass. None of the masses demonstrated tumor in vein or other local invasion.
In maximum dimension, the overall cohort of lesions was on average 3.1 ± 1.5 cm (range 1.2–11.6 cm). Benign lesions were on average significantly smaller than malignant lesions [p = 0.02; 2.7 ± 1.1 cm (range 1.2–5.6 cm) vs. 3.3 ± 1.6 cm (range 1.2–11.6 cm)].
The random forest machine learning classifier for distinguishing benign from malignant solid renal masses yielded an overall accuracy of 0.82 (Fig. 3), with an AUC of 0.80 (Fig. 4). The model had a sensitivity of 0.87, a specificity of 0.71, a positive predictive value of 0.87, and a negative predictive value of 0.29 (Table 1). The Matthews correlation coefficient was 0.59, the Youden’s J statistic was 0.58, and the weighted F1 score was 0.82. On average, cross-validation analysis of the model demonstrated a test accuracy of 0.72.
An analysis of radiomic feature ranking for the random forest model demonstrated that wavelet transforms were overwhelmingly the greatest contributing features for the model. Nine of the top ten features were wavelet transform variations with importances ranging from 0.0064 to 0.0089 (Table 2).
The three abdominal radiologists analyzed the cohort of renal masses for benignity versus malignancy and yielded overall accuracies ranging from 0.67 to 0.75 (Table 1) compared to 0.82 for the machine learning model (p = 0.02). The sensitivities of the radiologists ranged from 0.85 to 0.98 and were therefore similar to or greater than the sensitivity of the machine learning model (0.87). The specificity tended to be low among the radiologists, ranging from 0.27 to 0.33 compared to 0.71 for the machine learning model. The overall F1 score was similar among the radiologists (0.78–0.84) compared to the machine learning model (0.82).
Discussion
Solid renal masses are commonly encountered by radiologists in clinical practice. For fat-poor solid renal masses, cross-sectional imaging provides limited accuracy and reliability for distinguishing benign from malignant lesions [4], and as a result most solid renal masses are further evaluated via biopsy or surgical resection [7]. Reliable imaging-based diagnosis of solid renal masses is sorely needed in clinical practice. Our study demonstrated that CT-based radiomics fed into a machine learning model can differentiate benign from malignant solid renal masses with an overall accuracy of 0.82 and an AUC of 0.80. The performance of the model exceeded the performances of three abdominal radiologists who span from early to mid to late career (overall accuracies ranging from 0.67 to 0.75).
Radiomics-based machine learning models may be used as a non-invasive tool for characterizing renal masses and therefore may be beneficial to clinical workflow [19]. A newly identified renal mass can be evaluated with a trained machine learning model, and the model can provide a probability of benignity versus malignancy. The provided probability can be weighed against the patient’s comorbidities in deciding whether active surveillance, biopsy, or resection is the optimal course of action.
A number of studies have been performed to differentiate renal masses using radiomics derived from CT and MR images [12, 20]. In a recent study by Nassiri et al. [21], in a large cohort of 684 subjects, their overall CT-based radiomics machine learning model yielded an AUC of 0.84, and when evaluating the sub-selection of small renal masses, the AUC was 0.77. The results of the Nassiri study are overall similar to the performance of our machine learning model. A study by Deng et al. [22] performed an analysis of CT-based radiomics features (i.e., texture analysis) to differentiate benign from malignant renal masses in a cohort of 501 subjects; no machine learning algorithm was employed. Their radiomics features yielded AUCs ranging from 0.58 to 0.62. A similar analysis using MRI-based radiomics in 125 subjects [14] achieved an AUC of 0.73. Uhlig et al. [23] in a relatively small cohort of 94 patients achieved similar performance compared to our study for distinguishing benign from malignant masses via a CT-based radiomics random forest machine learning model—achieving an AUC of 0.83 and a radiologist AUC of 0.68. A study by Sun et al. [24] developed a CT-based radiomics machine learning model and also compared the model performance to radiologists; however the evaluation did not compare the conglomerate of benign versus malignant masses, but rather compared the ability to differentiate specific pathologic entities (such as clear cell RCC versus AMLs and oncocytomas). Such comparisons are not applicable in clinical practice, as they assume that the other pathologic entities are already excluded.
Our study has several strengths and unique aspects compared to prior published works. Given that our cohort was sourced from cases requiring intraoperative ultrasound, the tumors tended to be small (average 3.1 ± 1.5 cm) and diagnostically indeterminate on cross-sectional imaging. Since most of the tumors were relatively small, all patients specifically underwent partial laparoscopic nephrectomy; larger tumors or tumors that are frankly malignant would have proceeded for radical nephrectomy instead. None of the cases included in our cohort were obviously malignant, such as demonstrating frank invasion or necrosis. All included AMLs were without gross fat and therefore could not be definitely diagnosed by imaging alone. Furthermore, comparison of machine learning model performance to radiologist performance in distinguishing benign from malignant solid renal masses has been limited in the literature.
This study had several limitations. The data included for training the machine learning model was acquired with a variety of scanners, slice thicknesses, and peak voltages. Such variability has been shown to affect model performance [25]. Additionally, CT technology evolved over the 18-year time period from which our data were acquired, such as the implementation of iterative reconstruction methods with resultant reduced image noise. However, this variability in imaging parameters and scanner technology does allow for a more generalizable model. Further attempts to account for this variability and assess the generalizability would require additional training and/or assessment of the machine learning model from an outside institution. Although the segmentations were confirmed by a senior radiologist, the lesion segmentation was not repeated due to time constraints, which may affect the results. The machine learning model in this study was trained on contrast-enhanced CT images; not all patients can receive contrast and therefore non-contrast examinations cannot be evaluated with our model. The cohort in this study only included AMLs, oncocytomas, and the three most common RCC subtypes. As a result, the performance of the model is unknown if it were to encounter other entities, such as metastasis, lymphoma, abscess, or rare RCC subtypes. Furthermore, the machine learning algorithm used in this study was solely defined with a random forest classifier. A support vector machine (SVM) approach was considered, given that SVM is intrinsically two-class, whereas random forest is intrinsically suited for multiclass problems. However, five pathologies are included in the study cohort, and while they are grouped into a binary problem of benignity versus malignancy, inherently each pathologic entity has a potentially unique radiomic signature that is more appropriately classified with a random forest approach. A five-class machine learning model was considered instead of the binary benign versus malignant classifier presented; however, the number of cases for each of the five pathologic entities was considered too small for a five-class model, particularly given the need to split the cases into training and testing sets. The machine learning algorithm trained in this study specifically targeted lesions that proceeded to surgical resection and required intraoperative ultrasound. As a result, there is inherent and intentional selection bias, as lesions that were grossly aggressive, AMLs with macroscopic fat, or lesions not taken for surgical resection were excluded in the training of the model. A larger and multicenter cohort would likely improve model performance and generalizability. On a similar note, the radiologists’ performance is likely biased, as the lesions included in this study were relatively small; there is likely a higher pretest probability for benignity than would be expected if all renal masses were evaluated without exclusion criteria. The radiologists’ accuracies presented here are likely artifactually low compared to what would be expected for all renal masses in general because the cohort did not include fat-containing solid masses or cystic renal masses, which are more easily recognizable as benign.
In conclusion, our study demonstrated that a machine learning model trained from CT-based radiomics features can differentiate benign from malignant fat-poor solid renal masses with a high degree of accuracy, and which exceeds the performance of abdominal radiologists.
References
O’Connor, S.D., et al., Incidental finding of renal masses at unenhanced CT: prevalence and analysis of features for guiding management. AJR Am J Roentgenol, 2011. 197(1): p. 139-45.
Collins, S., J. McKiernan, and J. Landman, Update on the epidemiology and biology of renal cortical neoplasms. J Endourol, 2006. 20(12): p. 975-85.
Reuter, V.E. and J.C. Presti, Jr., Contemporary approach to the classification of renal epithelial tumors. Semin Oncol, 2000. 27(2): p. 124-37.
Kang, S.K., et al., Solid renal masses: what the numbers tell us. AJR Am J Roentgenol, 2014. 202(6): p. 1196-206.
Lebret, T., et al., Percutaneous core biopsy for renal masses: indications, accuracy and results. J Urol, 2007. 178(4 Pt 1): p. 1184-8; discussion 1188.
Alle, N., et al., Percutaneous image-guided core biopsy of solid renal masses: analysis of safety, efficacy, pathologic interpretation, and clinical significance. Abdom Radiol (NY), 2018. 43(7): p. 1813-1819.
Campbell, S.C., et al., Guideline for management of the clinical T1 renal mass. J Urol, 2009. 182(4): p. 1271-9.
Frank, I., et al., Solid renal tumors: an analysis of pathological features related to tumor size. J Urol, 2003. 170(6 Pt 1): p. 2217-20.
de Leon, A.D., P. Kapur, and I. Pedrosa, Radiomics in Kidney Cancer: MR Imaging. Magn Reson Imaging Clin N Am, 2019. 27(1): p. 1-13.
Gillies, R.J., P.E. Kinahan, and H. Hricak, Radiomics: Images Are More than Pictures, They Are Data. Radiology, 2016. 278(2): p. 563-77.
Jordan, M.I. and T.M. Mitchell, Machine learning: Trends, perspectives, and prospects. Science, 2015. 349(6245): p. 255-60.
Muhlbauer, J., et al., Radiomics in renal cell carcinoma-a systematic review and meta-analysis. Cancers (Basel), 2021. 13(6): p.1-15.
Uhlig, J., et al., Radiomic features and machine learning for the discrimination of renal tumor histological subtypes: a pragmatic study using clinical-routine computed tomography. Cancers (Basel), 2020. 12(10): p.1-13.
Said, D., et al., Characterization of solid renal neoplasms using MRI-based quantitative radiomics features. Abdom Radiol (NY), 2020. 45(9): p. 2840-2850.
Wels, M., et al. General purpose radiomics for multi-modal clinical research. SPIE Medical Imaging. Proceedings Volume 10950, 2019
Mehta, P., et al., A high-bias, low-variance introduction to Machine Learning for physicists. Phys Rep, 2019. 810: p. 1-124.
Raman, S.P., et al., CT texture analysis of renal masses: pilot study using random forest classification for prediction of pathology. Acad Radiol, 2014. 21(12): p. 1587-96.
Kruppa, J., et al., Probability estimation with machine learning methods for dichotomous and multicategory outcome: applications. Biom J, 2014. 56(4): p. 564-83.
Yu, H., et al., Texture analysis as a radiomic marker for differentiating renal tumors. Abdom Radiol (NY), 2017. 42(10): p. 2470-2478.
Lubner, M.G., Radiomics and Artificial Intelligence for Renal Mass Characterization. Radiol Clin North Am, 2020. 58(5): p. 995-1008.
Nassiri, N., et al., A Radiomic-based Machine Learning Algorithm to Reliably Differentiate Benign Renal Masses from Renal Cell Carcinoma. Eur Urol Focus, 2022. 8(4): p. 988-994.
Deng, Y., et al., Usefulness of CT texture analysis in differentiating benign and malignant renal tumours. Clin Radiol, 2020. 75(2): p. 108-115.
Uhlig, J., et al., Discriminating malignant and benign clinical T1 renal masses on computed tomography: A pragmatic radiomics and machine learning approach. Medicine (Baltimore), 2020. 99(16): p. e19725.
Sun, X.Y., et al., Radiologic-Radiomic Machine Learning Models for Differentiation of Benign and Malignant Solid Renal Masses: Comparison With Expert-Level Radiologists. AJR Am J Roentgenol, 2020. 214(1): p. W44-W54.
van Timmeren, J.E., et al., Radiomics in medical imaging-“how-to” guide and critical reflection. Insights Imaging, 2020. 11(1): p. 91.
Acknowledgements
Support via a Radiological Society of North America Resident Research Grant as well as the Etta Kalin Moskowitz Fund Research Award.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wentland, A.L., Yamashita, R., Kino, A. et al. Differentiation of benign from malignant solid renal lesions using CT-based radiomics and machine learning: comparison with radiologist interpretation. Abdom Radiol 48, 642–648 (2023). https://doi.org/10.1007/s00261-022-03735-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-022-03735-7