Abstract
Purpose
Current diagnostic and treatment modalities for pancreatic cysts (PCs) are invasive and are associated with patient morbidity. The purpose of this study is to develop and evaluate machine learning algorithms to delineate mucinous from non-mucinous PCs using non-invasive CT-based radiomics.
Methods
A retrospective, single-institution analysis of patients with non-pseudocystic PCs, contrast-enhanced computed tomography scans within 1 year of resection, and available surgical pathology were included. A quantitative imaging software platform was used to extract radiomics. An extreme gradient boosting (XGBoost) machine learning algorithm was used to create mucinous classifiers using texture features only, or radiomic/radiologic and clinical combined models. Classifiers were compared using performance scoring metrics. Shapely additive explanation (SHAP) analyses were conducted to identify variables most important in model construction.
Results
Overall, 99 patients and 103 PCs were included in the analyses. Eighty (78%) patients had mucinous PCs on surgical pathology. Using multiple fivefold cross validations, the texture features only and combined XGBoost mucinous classifiers demonstrated an area under the curve of 0.72 ± 0.14 and 0.73 ± 0.14, respectively. By SHAP analysis, root mean square, mean attenuation, and kurtosis were the most predictive features in the texture features only model. Root mean square, cyst location, and mean attenuation were the most predictive features in the combined model.
Conclusion
Machine learning principles can be applied to PC texture features to create a mucinous phenotype classifier. Model performance did not improve with the combined model. However, specific radiomic, radiologic, and clinical features most predictive in our models can be identified using SHAP analysis.
Graphic abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Pancreatic cysts (PCs) can be described as mucinous (i.e., mucinous cystic neoplasms, intraductal papillary mucinous neoplasm) or non-mucinous (i.e., serous cystic tumors). Mucinous cysts are more likely to develop and harbor high-grade dysplasia (HGD) with an increased risk of malignant transformation [1]. Most guidelines recommend early resection in patients with mucinous PCs although poor diagnostic accuracy on cross-sectional imaging persists [2,3,4,5,6]. Endoscopic ultrasound/fine-needle aspiration (EUS/FNA) and surgical resection can improve diagnostic accuracy at the expense of invasiveness and patient morbidity. A meta-analysis of the diagnostic potential of EUS/FNA in PCs found a pooled sensitivity of 86% and specificity of 95% when determining mucinous character of cyst fluid [7]. Additionally, EUS/FNA can involve morbidity of post-procedural pancreatitis, hemorrhage, infection, and oxygen desaturation [8]. It is crucial to develop tools that enhance detection of mucinous phenotypes and reduces morbidity to appropriately recommend treatment or surveillance strategies.
Radiomics is the high throughput extraction of large sets of quantitative data from imaging studies that can be used to characterize healthy and pathological tissues to inform diagnosis and prognosis [9]. Texture analysis, a subtype of radiomics, quantifies gray-level pixels and voxels in a frequency histogram and their spatial relationships to describe lesion heterogeneity within a 2-dimensional region of interest (ROI) or 3-dimensional volume of interest (VOI) [10, 11]. Computed tomography (CT) texture analysis has demonstrated promise in diagnosing and risk-stratifying patients with PCs [12,13,14,15]. Predictive ability of radiomics models can be enhanced by integrating clinical features in pancreas and non-pancreas tissues [16].
Statistical analysis of radiomics can be further improved with machine learning principles that implement dynamic computer algorithms that automatically improve its performance through experience. Specifically, machine learning builds predictive models around an outcome of interest using provided radiomics training data [17, 18]. Machine learning principles applied to texture features have demonstrated clinical superiority over more traditional statistical methods in bladder and prostate cancer, clear cell renal cell carcinoma, and pancreatic ductal adenocarcinoma [18,19,20].
However, to our knowledge, few studies have investigated the potential of applying machine learning principles to radiomics data to detect mucinous pancreatic cysts from non-mucinous. This study aims to develop and assess a machine learning algorithm that differentiates mucinous cysts using texture features, and texture features combined with other radiomic, radiologic, clinical parameters.
Methods
This retrospective study was HIPAA compliant and was approved by our Institutional Review Board. The requirement for signed informed consent was waived.
Study design and patient selection
A retrospective analysis of patients who underwent resection for a PC at a single institution was conducted. Surgical pathology records were searched for resected PCs between 1995 and 2017. Patients with preoperative contrast-enhanced computed tomography (CECT) scans within one year of resection, and complete surgical pathology commenting on mucinous character and dysplasia were included in this analysis. Pancreatic cysts were defined as mucinous: mucinous cystic neoplasms, intraductal papillary mucinous neoplasms, or indeterminate where mucinous pathology could not be definitively ruled out on histopathological analysis; or non-mucinous: serous cystic tumor, lymphoepithelial cysts, or other (Table 2). Patients with evidence of pancreatic pseudocyst or primarily solid pancreas lesions (e.g., cystic lesions with obvious invasive components, colloid carcinoma, etc.) were excluded. A methodological overview for this study including PC segmentation, data collection, and model building is described in Fig. 1.
Methodological overview of applying machine learning algorithms to radiomics. Pancreatic cysts were segmented 3-dimensionally on portal venous contrast-enhanced CT scans. Radiomics data including texture features, and radiologic and clinical features were collected concurrently. Baseline and machine learning algorithms were then applied to our datasets. VOI: volume of interest, XGBoost: extreme gradient boosting, SMOTE: synthetic minority oversampling technique
Image acquisition
All CT scans were acquired with multi-detector CT scanners using multiphasic, intravenous contrast-enhanced protocols. Segmentation of the VOI was conducted using the contrast-enhanced portal venous phase on CT. Imaging parameters consisted of a CECT matrix of 512 × 512 × 16 and most scans using automated or variable tube current. Tube potential ranged 100–140 kV with 85.8% (85/99) of patients receiving scans with 120 kV. Most patients had CECT scans with slice thickness ranging 2.0–5.0 mm (94/99; 94.9% of patients). Four patients (4.0%) had CECT scans with a slice thickness of 1.25 mm and one patient (1.0%) had a CECT slice thickness of 7 mm.
Radiologic data collection and electronic health record review
Radiologic features including but not limited to septations, ductal dilation, and lymphadenopathy from cross-sectional abdominal CECT images were manually collected by a medical student after having received training from a fellowship trained abdominal diagnostic radiologist with 11 years of experience. Additional clinical information was obtained from electronic health records including surgical pathology report details on mucinous phenotype and presence and grade of dysplasia. Categorical radiologic and clinical variables were numerically coded to be incorporated into model building. Databases were created and maintained using REDCap (v10.9.2, Vanderbilt University, TN) and Microsoft Excel (2016, v16.0).
Segmentation and feature extraction
Patients who met the inclusion criteria had their preoperative scans uploaded from the Picture Archiving and Communication Systems program (McKesson, San Francisco, CA) to a commercially available, quantitative imaging software platform, HealthMyne (v5.0, Madison, WI). Pancreatic cyst ROIs were manually segmented by a medical student after having received training from and under the supervision of a fellowship trained abdominal diagnostic radiologist with 11 years of experience. Particularly challenging PC lesions were segmented by both the medical student and radiologist. The medical student and radiologist were blinded to pathologic diagnosis at time of segmentation.
Two-dimensional ROIs were segmented in the axial dimension of the imaging slice with the largest unidimensional diameter. HealthMyne propagated the manually drawn ROI above and below the initial segmentation using validated segmentation algorithms [21]. Specifically, edge boundaries were set using gray-scale pixel intensities of the two-dimensional ROI, and a best-fit and super-sampling algorithm was used to minimize variations imparted by variation in scan slice thickness to generate a three-dimensional VOI encompassing the lesion. Radiomics features were extracted in accordance with the Image Biomarker Standardization Initiative. Radiomic hyperparameters included gray-leveling using a bin width of 20 HU with a bin edge at 0 HU, a Chebychev distance of 1, and considered neighbors in 8 and 26 directions for 2-D and 3-D metrics, respectively. In addition, mesh-based metrics used triangular meshes that align with the user’s manual segmentation. This allows for greater detail to be captured from manually segmenting the PC and aligns with the Image Biomarker Standardization Initiative methodology. HealthMyne requires manual revision and verification prior to extraction of unfiltered radiomics. Segmentation excluded vasculature, free fluid, bowel gas, and intra-abdominal hardware from prior surgeries.
HealthMyne extracted texture feature radiomics data, including first-order (i.e., mean attenuation, kurtosis, entropy, etc.) and gray-level co-occurrence matrix (GLCM) second-order texture features from the three-dimensional VOI. Other non-texture feature radiomics were collected that describe the morphology of the lesion and includes but is not limited to volume, surface area, multi-planar diameters of PCs.
Model building and statistical analyses
Patient demographics and cyst characteristics were tabulated and reported as means and standard deviations, or frequencies and percentages using Microsoft Excel (2016, v16.0). Machine learning models were created using the extreme gradient boosting (XGBoost) library (2020, 1.3.1) for Python (2019, v3.6.9). Scoring metrics were imported from the Scikit-learn (2020, 0.22.1) and Imblearn (2018, 0.4.3) libraries. Receiver operating characteristic (ROC) curves were created using the Matplotlib (2020, 3.2.2) Python library. The code used was entered into a public repository on GitHub, https://github.com/uw-cmg/MeghanPanccystmachine.
The overall goal of our models was to develop and assess machine learning classifiers in delineating PCs with a mucinous from non-mucinous phenotype. Baseline classifiers were built and included minority, majority, random guesser, and stratified guesser models. The minority and majority classifiers always predict the least and most frequent predetermined outcome in the training set, which are non-mucinous and mucinous PCs, respectively, in our dataset (Table 2). The random guesser model makes predictions uniformly at random. The stratified guesser model generates predictions randomly based on the training set’s class distribution. The random and stratified guesser models calculated scoring metrics that were obtained from averaging results over 10,000 runs. XGBoost is an open-source software package that implements a gradient boosted machine learning algorithm of decision trees to arrive at a specified outcome, mucinous from non-mucinous PCs in our study [22]. XGBoost uses gradient boosting, a machine learning method that combines an ensemble of weak "learners" into a single strong learner in a highly efficient and flexible manner and is generally considered to yield accurate and well-performing models with characteristics that help avoid overfitting and allow for low-bias predictions [22]. The XGBoost machine learning algorithm was applied to data as received, and to training data augmented with synthetic minority oversampling technique (SMOTE). The synthetic minority oversampling technique was applied to our training data set to oversample the minority class to generate artificial data points and account for imbalance between the majority and minority classes. Additional machine learning models (e.g., linear, support-vector machine, random forest, multi-lay perception, and k-means clustering, etc.) were explored, and their performances compared to XGBoost (Supplemental Table 1).
One model consisted of 35 first- and second-order texture feature radiomics: texture features only model. A second combined model consisting of 95 total features included texture feature radiomics, non-texture feature radiomic (i.e., volume, surface area, etc.), radiologic (i.e., septations, lymphadenopathy, etc.), and clinical variables (i.e., age, history of pancreatitis, etc.) (Online Appendix A). Pre-processing of our data sets included normalizing variables using a minimum–maximum scaler to represent data on a scale from 0 to 1 using the Scikit-learn package (2020, 0.22.1). Weak learners were present in the texture features only and combined models. In developing the mucinous classifier models, 8 and 11 weak learners were used in the decision trees for the texture features only and combined models, respectively. Machine learning models calculated scoring metrics that were obtained from averaging results over 500 runs. Each run provided the results of the left-out validation data from fivefold cross validation applied to training and test set splits. Thus, assessment is based on every data point being predicted exactly 500 times while being excluded from the training set. Metrics are determined for each fivefold cross validation and then statistics (e.g., mean and standard deviations) are calculated for the distributions from the 500 distinct fivefold cross validations. All metric values given are means and all errors are standard deviations over the 500 distinct fivefold cross validations. This cross validation scheme was used for the XGBoost and all baseline classifiers when assessing metrics. When SMOTE is used it is applied only to included training data during cross validation and not the excluded validation data. The XGBoost algorithms developed for the mucinous classifier used a positive class weight scaling of 0.25, maximum depth of 3, and did not include oversampling parameters. The remainder of the decision tree parameters were left at their defaults and included booster set to Gbtree; verbosity set to 1; validate parameters set to “true”; n-thread set to maximum number of thread available; disable default evaluation metrics set to “false”; number pbuffer set to number of training instances; and number feature set to maximum dimension of the feature. All features were evaluated without prior assessment of their individual performances with predictive modeling to prevent data leakage.
Scoring metrics included sensitivity, specificity, precision, accuracy, and area under the curve (AUC) and have been previously described in Gurbani, et al. 2021 [23]. Additional composite metrics that combine precision and recall (F1-score), and sensitivity and specificity (Geometric mean; G-mean) were determined by these models.
G-mean: A metric that measures the classification accuracy in an imbalanced dataset
F1-score: A metric that measures a test’s accuracy by determining the harmonic mean of precision and recall.
Statistical significance of a model was assessed using a permutation test. In a permutation test, a statistic (e.g., G-mean from fivefold cross validation) is determined for a model fit to the original unpermuted data and 1000 random permutations of the target data (1001 values), where the permutation assures that any natural correlation between the target values and their features are removed. The p-value is the fraction of cases in the distribution of the statistic over all 1001 cases where the statistic value from the model fit to the permuted data is greater than or equal to that obtained from the model fit to the unpermuted data. P < 0.05 was considered to be statistically significant. We assessed two statistics, specifically, G-mean and AUC from fivefold cross validation. Overall model performance of the mucinous classifiers was described and compared using scoring metrics and AUC of ROC curves. Again, all metrics were calculated on predictions for validation data excluded from the training data using the approach described above.
Shapely additive explanation (SHAP) analysis was conducted to demonstrate how features within the XGBoost mucinous classifier impact the model output. SHAP analysis has the capacity to quantify continuous and categorical variables present in the texture features only and combined models. The SHAP values were calculated for a full fit of all features to complete data sets without any cross validation. SHAP analysis was not pursued in the training cohorts for each cross validation as SHAP analysis targets obtaining the best understanding of the feature impacts for the whole data set [24]. Features listed higher on the left vertical axis indicated stronger importance on the overall model outcome. Feature values are color-coded with red data points indicating higher values and blue data points indicating lower values. The SHAP values on the horizontal axis represent relation to mucinous prediction of each PC. More positive SHAP values indicate directionality toward mucinous and negative SHAP values suggest directionality toward non-mucinous prediction of PCs. Given the limited data and the fact that XGBoost provides a robust feature-selection process that well-informs SHAP analysis, additional feature selection approaches (e.g., filter-based, wrapping-based, embedding-based, etc.) were not pursued.
Additional machine learning algorithms were applied to mucinous PCs to create a classifier that distinguishes cysts with HGD from cysts without HGD. Baseline models used for comparisons included minority, majority, random guesser, and stratified guesser models. XGBoost was also explored to evaluate their performance compared to baseline models. In developing the HGD classifier models, 5 and 9 weak learners were used in the decision trees for the texture features only and combined models, respectively. The XGBoost algorithms developed for the HGD classifier used a positive class weight scaling of 1.45 and 2.47, and maximum depth of 4 and 5 for the texture features only and combined models, respectively. The remainder of the decision tree parameters were left at their defaults. Accuracy, F1-score, and G-mean values were determined to compare performance of models.
Results
Patient cohorts
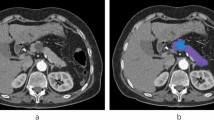
Overall, 99 patients and 103 cysts were included for analysis (Fig. 2). The majority of patients were female (70.7%) and white (96.0%). The mean age of patients was 62 ± 14 years (Table 1). More patients had mucinous PCs (78%) compared to non-mucinous PCs (22%). The most common mucinous lesions were intraductal papillary mucinous neoplasms (IPMNs) (69%) and the most common non-mucinous lesions were serous cystic tumors (74%) (Table 2). Representative CECT cross-sectional imaging of a patient with mucinous and non-mucinous lesions are reported in Fig. 3. Twenty-six (32%) of the 80 mucinous cysts had HGD reported on surgical pathology.
Representative images of pancreatic cysts on the portal venous phase used for segmentation and radiomics extraction in two different patients. A branch-duct intraductal papillary mucinous neoplasm of the pancreatic head denoted in axial and coronal sections by white arrows (a, b), and a serous cystadenoma in the pancreatic head exhibited in axial and coronal sections by white arrow heads (c, d)
In the overall patient cohort, the average maximum axial diameter of PCs was 4.2 ± 3.1 cm, the average surface area of PCs was 71.3 ± 157.5 cm2, and the average volume of PCs was 88.1 ± 386.8 cm3. Mucinous PCs with HGD had an average maximum axial diameter of 5.6 ± 4.7 cm, average surface area of 140.8 ± 285.7 cm2, and average volume of 246.0 ± 735.6 cm3.
Performance of mucinous classifiers
The performances of the baseline and machine learning mucinous classifier models were evaluated using independent and composite scoring metrics. The minority baseline classifier produced a sensitivity and specificity of 0.0 and 1.0, respectively, and the majority baseline classifier produced a sensitivity and specificity of 1.0 and 0.0, respectively. Furthermore, both baseline classifiers generated an AUC of 0.50. The random and stratified mucinous classifiers yielded sensitivities of 0.50 ± 0.06 and 0.78 ± 0.05, specifities of 0.50 ± 0.10 and 0.22 ± 0.09, and AUCs of 0.50 ± 0.06 and 0.50 ± 0.05, respectively. Applying XGBoost to the texture features only data produced a sensitivity of 0.75 ± 0.12, specificity of 0.63 ± 0.22, F1-score of 0.80 ± 0.08, G-mean of 0.66 ± 0.15, and AUC of 0.72 ± 0.14. The XGBoost combined model produced a sensitivity of 0.77 ± 0.11, specificity of 0.61 ± 0.22, F1-score of 0.82 ± 0.07, G-mean of 0.67 ± 0.15, and AUC of 0.73 ± 0.14. We observed that XGBoost alone outperformed XGBoost with SMOTE applied to the training data. When SMOTE was applied to our training data, the XGBoost texture features only model generated a sensitivity of 0.55 ± 0.15, specificity of 0.67 ± 0.21, F1-score of 0.66 ± 0.12, G-mean of 0.59 ± 0.12, and AUC of 0.61 ± 0.11. When SMOTE was applied to our training data, the XGBoost combined model generated a sensitivity of 0.63 ± 0.14, specificity of 0.70 ± 0.23, F1-score of 0.72 ± 0.11, G-mean of 0.64 ± 0.14, and AUC of 0.66 ± 0.12. Furthermore, the XGBoost performances of the texture features only and combined mucinous classifier models were compared using ROC curve analysis (Fig. 4). A complete report of scoring metrics for each mucinous classifier model is given in Table 3.
Performance of high-grade dysplasia classifiers
The performances of baseline and machine learning mucinous classifier models for the detection of HGD in mucinous PCs were assessed using independent and composite scoring metrics. The G-mean is of particular importance given the imbalance of a small patient sample. The G-mean of the minority, majority, random, and stratified HGD classifiers yielded values of 0.0, 0.0, 0.50 ± 0.06, and 0.46 ± 0.07, respectively. The accuracy of the minority, majority, random, and stratified HGD classifiers yielded values of 0.33, 0.67, 0.50 ± 0.06, and 0.56 ± 0.05, respectively. The sensitivity of the minority, majority, random, and stratified HGD classifiers yielded values of 1.0, 0.0, 0.50 ± 0.10, and 0.32 ± 0.09, respectively. The specificity of the minority, majority, random, and stratified HGD classifiers yielded values of 0.0, 1.0, 0.50 ± 0.07, and 0.67 ± 0.06, respectively. An XGBoost of the texture features only model produced a G-mean, accuracy, sensitivity, and specificity of 0.50 ± 0.18, 0.62 ± 0.10, 0.41 ± 0.21, and 0.72 ± 0.14, respectively. An XGBoost of the combined model produced a G-mean, accuracy, sensitivity, and specificity of 0.56 ± 0.16 and 0.62 ± 0.12, 0.51 ± 0.22, 0.68 ± 0.15, respectively. A complete report of scoring metrics for each model is described in Table 4. Performance of the XGBoost combined model was evaluated using ROC curve analysis demonstrating an AUC of 0.59 ± 0.13 (Fig. 5).
Performance comparison of machine learning models
Additional machine learning models were constructed for mucinous and HGD classifiers and included linear, support-vector machine, random forest, multi-layer perceptrons, and k-nearest neighbor (Supplemental Table 1). None of these models outperformed the XGBoost models for the mucinous and HGD classifiers. A complete report of scoring metrics for machine learning models is reported in Supplemental Table 1.
Variable identification
Shapely additive explanation analyses were conducted to delineate which features were the most important in constructing the XGBoost mucinous classifiers. Root mean square was the most important feature followed by mean attenuation and kurtosis that were identified by SHAP analysis in constructing the texture features only XGBoost mucinous classifier. Again, root mean square was the most important feature followed by cyst location and mean attenuation that were identified by SHAP analysis in constructing the combined XGBoost mucinous classifier. Using PC mean attenuation as an example, SHAP analysis identified mean attenuation as an important feature in each XGBoost model. Pancreatic cysts in our data set had mean attenuations ranging from 14.6 to 171.4 HU (median 41.4 HU). Cysts with lower mean attenuation values correlated or anti-correlated to SHAP values (Fig. 6a). The top 10 features for both the texture features and combined models are outlined in descending order in Fig. 6a, b.
Shapely additive explanations (SHAP) analyses of XGBoost mucinous classifier texture features only model (a) and combined model (b). Features most important in building XGBoost models on the y-axis in descending order of importance. SHAP values on the x-axis describes directionality of each feature toward mucinous (+ SHAP value) or non-mucinous lesions (− SHAP value). High feature values are indicated in red and low feature values are indicated in blue and are unique to the range of values for each feature noted on the y-axis. GLCM: gray-level co-occurrence matrix; HU: Hounsfield units
Discussion
This retrospective analysis demonstrated that machine learning principles applied to radiomics, clinical parameters, and surgical pathology can be used to create a mucinous classifier of PCs. The machine learning mucinous classifiers outperformed the baseline mucinous classifiers on G-mean and AUC scoring metrics, which we believe are the metrics best suited to assess the model quality and potential for useful predictions. Performance was comparable between XGBoost texture feature only and combined models. Shapely additive explanation analysis demonstrated that trends in important model-building variables can be identified. However, overall this remains a challenging task with only moderate performance of the best model.
Our patient cohort reflected that of the general population of patients who undergo surgical resection for PCs and the analyses conducted used a robust approach. The composition of cyst subtypes in our study reflects that of expected composition and does not bias toward lesion type [25]. All the cysts in our cohort underwent surgical resection with corresponding pathologic analysis. Another benefit of our approach is that we applied state of the art machine learning approaches with careful cross validation to minimize data leakage. Specifically, XGBoost was used as it allows non-linear fitting and avoids both over-fitting and bias. The accuracy of these models was further supported by averaging model output over 500 runs with fivefold cross validation. Our approach provided robust data and applied careful machine learning to build a classifier that can help identify PCs with a mucinous phenotype.
Similar studies have also successfully used machine learning principles applied to radiomics to help predict PC subtypes. A recent study investigated the utility of multivariate analysis to differentiate between mucinous cystic neoplasms from macrocystic serous cystadenomas [26]. An imbalance-adjusted logistic regression was used to build radiologic, radiomics, and combined models using data obtained from 3-D image analysis. The radiologic model consisted of cyst features easily identified by the manual interpretation of the CECT scan like location, shape, and wall enhancement, and generated an AUC of 0.775. The radiomics model, that includes mathematical descriptors of lesion heterogeneity and pixel/voxel organization, included an 18-feature full fit optimization that consisted of the highest AUC values and generated an AUC of 0.989. A combined model further improved performance with an AUC of 0.994. Model performance in the Xie et al. 2020 study is likely higher compared to our results because an optimal feature set was used without test/validation data. Our methodological approach evaluated features without prior assessment of performance and may contribute to differences in machine learning model performance. Additionally, there is value in creating models that delineate mucinous cystic neoplasms from macrocystic serous adenomas. However, in our approach, machine learning models were built to specifically predict mucinous phenotype regardless of PC subtype making our results more generalizable to what is encountered in every day clinical practice.
Models that independently use radiomics, radiologic, or clinical data may not acknowledge important cyst and patient features that predispose patients to mucinous or malignant lesions. Using combined models that are multimodal may improve model performance in predicting mucinous PCs. In Xie 2020, the model that combined radiologic (AUC of 0.775) and radiomics features (AUC of 0.989) had the strongest performance with an AUC of 0.994. Related studies have demonstrated that the addition of molecular and/or genetic markers may enhance performance of predictive models [13, 27]. Another study used texture analysis and miRNA genomic classifier data to delineate between low- and high-grade dysplasia of IPMNs [13]. The performance of a radiomics model (AUC of 0.77) improved with the addition of miRNA genomic data to yield an AUC of 0.92. In our study, our combined model that included radiomic, radiologic, and clinical variables did not significantly outperform our texture features only model (Fig. 4). Our models included data that could easily be obtained for retrospective analysis. More advanced data from cyst fluid analysis like mRNA and other cyst biomarkers could not be obtained for study inclusion, retrospectively. Cyst location, the measured cyst short axis in the coronal plane, and patient age were the most important radiologic and clinical features used to build the combined mucinous classifier model. These features are not referenced in guidelines for the management of patients with PCs to aid in surgical decision-making [2, 3, 28, 29]. While radiologic and clinical worrisome features and high-risk stigmata of PCs can help identify patients with mucinous PCs, the addition of these features to our radiomics data likely did not improve performance given low incidence in our patient cohort, and their effect was likely diluted by an abundance of features with unknown clinical significance.
The utility of a machine learning algorithm to help detect presence of HGD in the subset of patients with mucinous cysts was conducted. Based on the AUC p-values, the XGBoost model performed similarly to chance (p-values > 0.2), suggesting poor clinical utility of this model (Fig. 5). This poor result is not necessarily an indication that the present modeling approach is inadequate and may simply be due to small sample size. Further evaluation in a larger cohort is warranted.
Non-XGBoost machine learning models were investigated to compare performance. When identifying strong-performing models two standards are considered: (1) the model needs to perform better than baseline models (e.g., random guesser) and (2) the model needs to show good performance across scoring metrics and their statistical significance. For instance, the mucinous classifier for the XGBoost combined model yields a G-mean of 0.67 ± 0.15 and p-value of < 0.001, which demonstrates its strong performance. For the HGD classifier, we did not identify any models that perform better than XGBoost. The best performance was the 3-Nearest Neighbor model with a G-mean of 0.57 ± 0.20 and a p-value of 0.001, which is significantly better than the baseline models but worse than XGBoost on both metrics. Overall, we find that XGBoost outperforms other machine learning models in creating a classifier that helps predict mucinous PCs.
This study has several limitations to address. First, our retrospective study was conducted at a single institution with a small patient sample limiting the generalizability of our XGBoost models. A selection bias was introduced with the inclusion criteria selecting for resected cysts, which implies they are more likely than a sample of all PCs to have suspicious features warranting resection. This was done so that we would have a confirmed pathologic diagnosis, which is only available in surgically resected cysts. While commonplace, the heterogeneity in CT image acquisition in our patient cohort has the potential to impact texture measures. Lastly, our XGBoost models were built to classify PCs with or without a mucinous phenotype and did not factor in further delineating PC subtypes from one another (i.e., MCN from IPMN). Further prospective, multi-institutional studies to assess external validity of these results are warranted. Additional investigations comparing machine learning models across different image analysis/radiomics platforms are required to standardize the clinical application of machine learning models.
In conclusion, our study demonstrates that machine learning principles can be applied to radiomics data of PCs to help detect mucinous phenotypes. While this information does not obviate the need for other diagnostic testing, it may help risk stratify patients with PCs. We also demonstrate that integration of radiologic and clinical features with texture feature radiomics data does not improve performance of our mucinous classifier. However, unique radiomic, radiologic, and clinical features were important in building our machine learning mucinous classifiers. These results highlight the potential of machine learning algorithms applied to high-throughput PC radiomics features in helping to detect mucinous cyst phenotype in patients and deserves further study to improve and validate such models.
Data availability
Data was collected with HealthMyne (v5.0, Madison, WI).
Code availability
Git Hub Repository, https://github.com/uw-cmg/MeghanPanccystmachine.
References
Stark A, Donahue TR, Reber HA, Hines OJ (2016) Pancreatic Cyst Disease: A Review. JAMA 315(17):1882–1893. https://doi.org/10.1001/jama.2016.4690
Tanaka M, Fernández-Del Castillo C, Kamisawa T, Jang JY, Levy P, Ohtsuka T, Salvia R, Shimizu Y, Tada M, Wolfgang CL (2017) Revisions of international consensus Fukuoka guidelines for the management of IPMN of the pancreas. Pancreatology 17(5):738–753. https://doi.org/10.1016/j.pan.2017.07.007
Vege SS, Ziring B, Jain R, Moayyedi P (2015) Clinical Guidelines Committee; American Gastroenterology Association. American gastroenterological association institute guideline on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 148(4):819–822; quize12–3. https://doi.org/10.1053/j.gastro.2015.01.015
Wu J, Wang Y, Li Z, Miao H (2019) Accuracy of Fukuoka and American Gastroenterological Association Guidelines for predicting advanced neoplasia in pancreatic cyst neoplasm: A meta-analysis. Ann Surg Oncol 26(13):4522-4536. https://doi.org/10.1245/s10434-019-07921-8
Cho CS, Russ AJ, Loeffler AG, Rettammel RJ, Oudheusden G, Winslow ER, Weber SM (2013) Preoperative classification of pancreatic cystic neoplasms: the clinical significance of diagnostic inaccuracy. Ann Surg Oncol 20(9):3112-3119. https://doi.org/10.1245/s10434-013-2986-6
Jones MJ, Buchanan AS, Neal CP, Dennison AR, Metcalfe MS, Garcea G (2013) Imaging of indeterminate pancreatic cystic lesions: a systematic review. Pancreatology 13(4):436-442. https://doi.org/10.1016/j.pan.2013.05.007
Rift CV, Scheie D, Toxvaerd A, Kovacevic B, Klausen P, Vilmann P, Hansen CP, Lund EL, Hasselby JP (2021) Diagnostic accuracy of EUS-guided through-the-needle-biopsies and simultaneously obtained fine needle aspiration for cytology from pancreatic cysts: A systematic review and meta-analysis. Pathology-Research and Practice 220(1):153368-153381. https://doi.org/10.1016/j.prp.2021.153368
Zhu H, Jiang F, Zhu J, Du Y, Jin Z, Li Z (2017). Assessment of morbidity and mortality associated with endoscopic ultrasound-guided fine-needle aspiration for pancreatic cystic lesions: A systematic review and meta-analysis. Dig Endosc 29(1):667-675. https://doi.org/10.1111/den.12851
Gillies RJ, Kinahan PE, Hricak H (2016). Radiomics: Images are more than pictures, they are data. Radiology 278(2):563-577. https://doi.org/10.1148/radiol.2015151169
Haralick RM, Shanmugam K, Dinstein I (1973). Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics. SMC-3:610–621.
Espinasse M, Pitre-Champagnat S, Charmettant B, Bidault F, Volk A, Balleyguier C, Lassau N, Caramella C (2020). CT texture analysis challenges: influence of acquisition and reconstruction parameters: A comprehensive review. Diagnostics (Basel, Switzerland) 10(5):258-266. https://doi.org/10.3390/diagnostics10050258
Hanania AN, Bantis LE, Feng Z, Wang H, Tamm EP, Katz MH, Maitra A, Koay EJ (2016). Quantitative imaging to evaluate malignant potential of IPMNs. Oncotarget 7(52):85766–85784. https://doi.org/10.18632/oncotarget.11769
Permuth JB, Choi J, Balarunathan Y, et al. (2016). Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget 7(52):85785–85797. https://doi.org/10.18632/oncotarget.11768
Attiyeh MA, Chakraborty J, Gazit L, et al. (2019). Preoperative risk prediction for intraductal papillary mucinous neoplasms by quantitative CT image analysis. HPB 21(2):212-218. https://doi.org/10.1016/j.hpb.2018.07.016
Chakraborty J, Midya A, Gazit L, Attiyeh M, Langdon-Embry L, Allen PJ, Do RKG, Simpson AL (2018). CT radiomics to predict high-risk intraductal papillary mucinous neoplasms of the pancreas. Med Phys 45(11):5019-5029. https://doi.org/10.1002/mp.13159
Wang XX, Ding Y, Wang SW, Dong D, Li HL, Chen J, Hu H, Lu C, Tian J, Shan XH (2020). Intratumoral and peritumoral radiomics analysis for preoperative Lauren classification in gastric cancer. Cancer Imaging 20(1):83-92. https://doi.org/10.1186/s40644-020-00358-3
Jordan MI, Mitchell TM (2015). Machine learning: Trends, perspectives, and prospects. Science 349(6245):255-260. https://doi.org/10.1126/science.aaa8415
Suarez-Ibarrola R, Hein S, Reis G, Gratzke C, Miernik A (2020). Current and future applications of machine and deep learning in urology: a review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World J Urol 38(10):2329-2347. https://doi.org/10.1007/s00345-019-03000-5
Bektas CT, Kocak B, Yardimci AH, Turkcanoglu MH, Yucetas U, Koca SB, Erdim C, Kilickesmez O (2019). Clear cell renal cell carcinoma: Machine learning-based quantitative computed tomography texture analysis for prediction of Fuhrman nuclear grade. Eur Radiol 29(3):1153-1163. https://doi.org/10.1007/s00330-018-5698-2
Qiu W, Duan N, Chen X, Ren S, Zhang Y, Wang Z, Chen R (2019) Pancreatic ductal adenocarcinoma: Machine learning-based quantitative computed tomography texture analysis for prediction of histopathological grade. Cancer Manag Res 11(1):9253-9264. https://doi.org/10.2147/CMAR.S218414
Zhang C, Chen T (2001). Efficient feature extraction for 2D/3D objects in mesh representation. Proceedings 2001 International Conference on Image Processing (Cat. No.01CH37205) 1(3):935–938.
Chen T, Guestrin C (2016). XGBoost: A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 1(1):785–794.
Gurbani S, Morgan D, Jog V, Dreyfuss L, Shen M, Das A, Abel EJ, Lubner MG (2021). Evaluation of radiomics and machine learning in identification of aggressive tumor features in renal cell carcinoma (RCC) [published online ahead of print, 2021 Apr 15]. Abdom Radiol (NY) 2021; 1–11. https://doi.org/10.1007/s00261-021-03083-y
Lundberg SM, Lee S (2017). A unified approach to interpreting model predictions. 31st conference on Neural Information Processing Systems 1(1):1–10.
Pezzilli R, Buscarini E, Pollini T, et al. (2020) Epidemiology, clinical features and diagnostic work-up of cystic neoplasms of the pancreas: Interim analysis of the prospective PANCY survey. Dig Liver Dis 52(5):547-554. https://doi.org/10.1016/j.dld.2020.02.003
Xie H, Ma S, Guo X, Zhang X, Wang X (2020). Preoperative differentiation of pancreatic mucinous cystic neoplasm from macrocystic serous cystic adenoma using radiomics: Preliminary findings and comparison with radiological model. Eur J Radiol 122(1):108747-108753. https://doi.org/10.1016/j.ejrad.2019.108747
Springer S, Masica DL, Dal Molin M, et al. (2019). A multimodality test to guide the management of patients with a pancreatic cyst. Sci Transl Med 11(501):1-29. https://doi.org/10.1126/scitranslmed.aav4772
Masica DL, Dal Molin M, Wolfgang CL, et al. (2017). A novel approach for selecting combination clinical markers of pathology applied to a large retrospective cohort of surgically resected pancreatic cysts. J Am Med Inform Assoc 24(1):145-152. https://doi.org/10.1093/jamia/ocw069
Elta GH, Enestvedt BK, Sauer BG, Lennon AM (2018). ACG clinical guideline: Diagnosis and management of pancreatic cysts. Am J Gastroenterol 113(4):464-479. https://doi.org/10.1038/ajg.2018.14
Funding
ML4MI internal funding.
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding author
Ethics declarations
Conflict of interest
M Lubner: Prior Grant funding from Ethicon, Philips. The remaining authors do not have any disclosures. This data was previously presented as a scientific abstract at the virtual American Roentgen Ray Society meeting 2021; and as a scientific poster at the virtual Society of Abdominal Radiology meeting 2021.
Ethics approval
This study was approved by our Institutional Review board and was found to be HIPAA complaint. Thus, requirement for informed consent was waived.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Awe, A.M., Vanden Heuvel, M.M., Yuan, T. et al. Machine learning principles applied to CT radiomics to predict mucinous pancreatic cysts. Abdom Radiol 47, 221–231 (2022). https://doi.org/10.1007/s00261-021-03289-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-021-03289-0