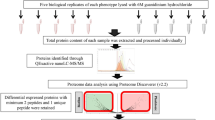
Abstract
Biofilm formation by Mycobacterium fortuitum causes serious threats to human health due to its increased contribution to nosocomial infections. In this study, the first comprehensive global proteome analysis of M. fortuitum was reported under planktonic and biofilm growth states. A label-free Q Exactive Quadrupole-Orbitrap tandem mass spectrometry analysis was performed on the protein lysates. The differentially abundant proteins were functionally characterized and re-annotated using Blast2GO and CELLO2GO. Comparative analysis of the proteins among two growth states provided insights into the phenotypic switch, and fundamental pathways associated with pathobiology of M. fortuitum biofilm, such as lipid biosynthesis and quorum-sensing. Interaction network generated by the STRING database revealed associations between proteins that endure M. fortuitum during biofilm growth state. Hypothetical proteins were also studied to determine their functional alliance with the biofilm phenotype. CARD, VFDB, and PATRIC analysis further showed that the proteins upregulated in M. fortuitum biofilm exhibited antibiotic resistance, pathogenesis, and virulence. Heatmap and correlation analysis provided the biomarkers associated with the planktonic and biofilm growth of M. fortuitum. Proteome data was validated by qPCR analysis. Overall, the study provides insights into previously unexplored biochemical pathways that can be targeted by novel inhibitors, either for shortened treatment duration or for eliminating biofilm of M. fortuitum and related nontuberculous mycobacterial pathogens.
Key points
• Proteomic analyses of M. fortuitum reveals novel biofilm markers.
• Acetyl-CoA acetyltransferase acts as the phenotype transition switch.
• The study offers drug targets to combat M. fortuitum biofilm infections.
Graphical Abstract








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in the manuscript (and its supplemental material). The mass spectrometry (MS) proteomics data are available via the ProteomeXchange Consortium (PRIDE partner repository) with the data set identifier PXD023784. Further information are available from the corresponding author on reasonable request.
References
Aung TT, Beuerman RW (2018) Atypical mycobacterial keratitis: a negligent and emerging thread for blindness. J Rare Dis Res Treat 3:15–20. https://doi.org/10.29245/2572-9411/2018/1.1143
Aung TT, Yam JKH, Lin S, Salleh SM, Givskov M, Liu S, Lwin NC, Yang L, Beuerman RW (2016) Biofilms of pathogenic nontuberculous mycobacteria targeted by new therapeutic approaches. Antimicrob Agents Chemother 60:24–35. https://doi.org/10.1128/AAC.01509-15
Bardouniotis E, Ceri H, Olson ME (2003) Biofilm formation and biocide susceptibility testing of Mycobacterium fortuitum and Mycobacterium marinum. Curr Microbiol 46:28–32. https://doi.org/10.1007/s00284-002-3796-4
Bartos P, Falkinham JO, Pavlik I (2004) Mycobacterial catalases, peroxidases, and superoxide dismutases and their effects on virulence and isoniazid-susceptibility in mycobacteria- a review. Vet Med-Czech 49:161–170. https://doi.org/10.17221/5691-VETMED
Baughn AD, Garforth SJ, Vilcheze C, Jacobs WR Jr (2009) An anaerobic-type α-ketoglutarate ferredoxin oxidoreductase completes the oxidative tricarboxylic acid cycle of Mycobacterium tuberculosis. PLoS Pathog 5:e1000662. https://doi.org/10.1371/journal.ppat.1000662
Brown-Elliott BA, Wallace RJ Jr (2002) Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev 15:716–746. https://doi.org/10.1128/CMR.15.4.716-746.2002
Capodagli GC, Sedhom WG, Jackson M, Ahrendt KA, Pegan SD (2013) A noncompetitive inhibitor for Mycobacterium tuberculosis’s class IIa fructose 1, 6-bisphosphate aldolase. Biochemistry 53:202–213. https://doi.org/10.1021/bi401022b
Casonato S, Sánchez AC, Haruki H, González MR, Provvedi R, Dainese E, Jaouen T, Gola S, Bini E, Vicente M, Johnsson K, Ghisotti D, Palù G, Hernández-Pando R, Manganelli R (2012) WhiB5, a transcriptional regulator that contributes to Mycobacterium tuberculosis virulence and reactivation. Infect Immun 80:3132–3144. https://doi.org/10.1128/IAI.06328-11
Cole S, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajadream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. https://doi.org/10.1038/31159
Daniel J, Sirakova T, Kolattukudy P (2014) An acyl-CoA synthetase in Mycobacterium tuberculosis involved in triacylglycerol accumulation during dormancy. PLoS One 9:e114877. https://doi.org/10.1371/journal.pone.0114877
de la Paz SM, Gest PM, Guerin ME, Coinçon M, Pham H, Ryan G, Puckett SE, Spencer JS, Gonzalez-Juarrero M, Daher R, Lenaerts AJ, Schnappinger D, Therisod M, Ehrt S, Sygusch J, Jackson M (2011) Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1, 6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli. J Biol Chem 286:40219–40231. https://doi.org/10.1074/jbc.M111.259440
Dokic A, Peterson E, Arrieta-Ortiz ML, Pan M, Di Maio A, Baliga N, Bhatt A (2021) Mycobacterium abscessus biofilms produce an extracellular matrix and have a distinct mycolic acid profile. Cell Surf 7:1–11. https://doi.org/10.1016/j.tcsw.2021.100051
Eoh H, Rhee KY (2013) Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:6554–6559. https://doi.org/10.1073/pnas.1219375110
Falkinham JO 3rd (1996) Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev 9:177–215. https://doi.org/10.1128/CMR.9.2.177
Forrellad MA, Klepp LI, Gioffré A, Garcia JS, Morbidoni HR, Santangelo MDLP, Cataldi AA, Bigi F (2013) Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4:3–66. https://doi.org/10.4161/viru.22329
Franco-Paredes C, Chastain DB, Allen L, Henao-Martínez AF (2018) Overview of cutaneous mycobacterial infections. Curr Trop Med Rep 5:228–232. https://doi.org/10.1007/s40475-018-0161-7
Ganief N, Sjouerman J, Albeldas C, Nakedi KC, Hermann C, Calder B, Blackburn JM, Soares NC (2018) Associating H2O2-and NO-related changes in the proteome of Mycobacterium smegmatis with enhanced survival in macrophage. Emerg Microbes Infect 7:1–17. https://doi.org/10.1038/s41426-018-0210-2
Giuffrè A, Borisov VB, Arese M, Sarti P, Forte E (2014) Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim Biophys Acta - Bioenergetics 1837:1178–1187. https://doi.org/10.1016/j.bbabio.2014.01.016
Gonzalez-Diaz E, Morfin-Otero R, Perez-Gomez HR, Esparza-Ahumada S, Rodriguez-Noriega E (2018) Rapidly growing mycobacterial infections of the skin and soft tissues caused by M. fortuitum and M. chelonae. Curr Trop Med Rep 5:162–169. https://doi.org/10.1007/s40475-018-0150-x
Granato LM, Picchi SC, de Oliveira Andrade M, Takita MA, de Souza AA, Wang N, Machado MA (2016) The ATP-dependent RNA helicase HrpB plays an important role in motility and biofilm formation in Xanthomonas citri subsp. citri. BMC Microbiol 16:55. https://doi.org/10.1186/s12866-016-0655-1
Gupta RS, Lo B, Son J (2018) Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol 9:67. https://doi.org/10.3389/fmicb.2018.00067
Hall-Stoodley L, Keevil CW, Lappin-Scott HM (1998) Mycobacterium fortuitum and Mycobacterium chelonae biofilm formation under high and low nutrient conditions. J Appl Microbiol 85:60S-69S. https://doi.org/10.1111/j.1365-2672.1998.tb05284.x
He W, Wang Y, Liu W, Zhou CZ (2007) Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1. BMC Struct Biol 7:38. https://doi.org/10.1186/1472-6807-7-38
Honda JR, Virdi R, Chan ED (2018) Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front Microbiol 9:1–11. https://doi.org/10.3389/fmicb.2018.02029
Jaiswal AK, Husaini SHA, Kumar A, Subbarao N (2018) Designing novel inhibitors against Mycobacterium tuberculosis FadA5 (acetyl-CoA acetyltransferase) by virtual screening of known anti-tuberculosis (bioactive) compounds. Bioinformation 14:327. https://doi.org/10.6026/97320630014327
Jatana N, Jangid S, Khare G, Tyagi AK, Latha N (2011) Molecular modeling studies of fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis-a potential target for the development of antitubercular drugs. J Mol Model 17:301–313. https://doi.org/10.1007/s00894-010-0727-3
Jeong W, Cha MK, Kim IH (2000) Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem 275:2924–2930. https://doi.org/10.1074/jbc.275.4.2924
Johansen MD, Herrmann JL, Kremer L (2020) Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol 18:392–407. https://doi.org/10.1038/s41579-020-0331-1
Karuppiah V, Thistlethwaite A, Dajani R, Warwicker J, Derrick JP (2014) Structure and mechanism of the bifunctional CinA enzyme from Thermus thermophilus. J Biol Chem 289:33187–33197. https://doi.org/10.1074/jbc.M114.608448
Kentache T, Abdelkrim AB, Jouenne T, Dé E, Hardouin J (2017) Global dynamic proteome study of a pellicle-forming Acinetobacter baumannii strain. Mol Cell Proteomics 16:100–112. https://doi.org/10.1074/mcp.M116.061044
Khoshkholgh-Sima B, Sardari S, Mobarakeh JI, Khavari-Nejad RA (2015) In-silico metabolome target analysis towards PanC-based antimycobacterial agent discovery. Iran J Pharm Sci 14:203
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Lee YV, Wahab HA, Choong YS (2015) Potential inhibitors for isocitrate lyase of Mycobacterium tuberculosis and non-M. tuberculosis: a summary. BioMed Res Int 2015. https://doi.org/10.1155/2015/895453
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lixa C, Mujo A, Anobom CD, Pinheiro AS (2015) A structural perspective on the mechanisms of quorum sensing activation in bacteria. An Acad Bras Cienc 87:2189–2203. https://doi.org/10.1590/0001-3765201520140482
Meehan CJ, Barco RA, Loh YH, Cogneau S, Rigouts L (2021) Reconstituting the genus Mycobacterium. Int J Syst Evol Microbiol 71:1–10. https://doi.org/10.1099/ijsem.0.004922
Mukhopadhyay S, Nair S, Ghosh S (2012) Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol Rev 36:463–485. https://doi.org/10.1111/j.1574-6976.2011.00302.x
Muñoz-Egea MC, Carrasco-Antón N, Esteban J (2020) State-of-the-art treatment strategies for nontuberculous mycobacteria infections. Expert Opin Pharmacother 21:969–981. https://doi.org/10.1080/14656566.2020.1740205
Nagmoti MB, Kulgod SY, Narang R, Mulla RG (2019) Diagnosis and management of postlaparotomy wound infection caused by Mycobacterium fortuitum. Int J Mycobacteriol 8:400–402. https://doi.org/10.4103/ijmy.ijmy_93_19
Niesteruk A, Jonker HR, Richter C, Linhard V, Sreeramulu S, Schwalbe H (2018) The domain architecture of PtkA, the first tyrosine kinase from Mycobacterium tuberculosis, differs from the conventional kinase architecture. J Biol Chem 293:11823–11836. https://doi.org/10.1074/jbc.RA117.000120
Ortíz-Pérez A, Martín-de-Hijas N, Alonso-Rodríguez N, Molina-Manso D, Fernández-Roblas R, Esteban J (2011) Importance of antibiotic penetration in the antimicrobial resistance of biofilm formed by non-pigmented rapidly growing mycobacteria against amikacin, ciprofloxacin and clarithromycin. Enferm Infecc Microbiol Clin 29:79–84. https://doi.org/10.1016/j.eimc.2010.08.016
Pecsi I, Hards K, Ekanayaka N, Berney M, Hartman T, Jacobs WR, Cook GM (2014) Essentiality of succinate dehydrogenase in Mycobacterium smegmatis and its role in the generation of the membrane potential under hypoxia. Mbio 5. https://doi.org/10.1128/mBio.01093-14
Peterson EJ, Bailo R, Rothchild AC, Arrieta-Ortiz M, Kaur A, Pan M, Mai D, Cooper C, Aderem A, Bhatt A, Baliga NS (2018) An essential mycolate remodeling program for mycobacterial adaptation in host cells. BioRxiv 354431. https://doi.org/10.1101/354431
Poonam, Yennamalli RM, Bisht GS, Shrivastava R (2019) Ribosomal maturation factor (RimP) is essential for survival of nontuberculous mycobacteria Mycobacterium fortuitum under in vitro acidic stress conditions. 3 Biotech 9:1–10. https://doi.org/10.1007/s13205-019-1659-y
Rasmussen JJ, Vegge CS, Frøkiær H, Howlett RM, Krogfelt KA, Kelly DJ, Ingmer H (2013) Campylobacter jejuni carbon starvation protein A (CstA) is involved in peptide utilization, motility and agglutination, and has a role in stimulation of dendritic cells. J Med Microbiol 62:1135–1143. https://doi.org/10.1099/jmm.0.059345-0
Raynaud C, Etienne G, Peyron P, Laneelle MA, Daffe M (1998) Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 144:577–587. https://doi.org/10.1099/00221287-144-2-577
Rosen BC, Dillon NA, Peterson ND, Minato Y, Baughn AD (2017) Long-chain fatty acyl coenzyme A ligase FadD2 mediates intrinsic pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61. https://doi.org/10.1128/AAC.02130-16
Sankhe GD, Dixit NM, Saini DK (2018) Activation of bacterial histidine kinases: insights into the kinetics of the cis autophosphorylation mechanism. mSphere 3. https://doi.org/10.1128/mSphere.00111-18
Sarathy JP, Dartois V, Lee EJD (2012) The role of transport mechanisms in Mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals 5:1210–1235. https://doi.org/10.3390/ph5111210
Saunders AH, Griffiths AE, Lee KH, Cicchillo RM, Tu L, Stromberg JA, Krebs C, Booker SJ (2008) Characterization of quinolinate synthases from Escherichia coli, Mycobacterium tuberculosis, and Pyrococcus horikoshii indicates that [4Fe-4S] clusters are common cofactors throughout this class of enzymes. Biochemistry 47:10999–11012. https://doi.org/10.1021/bi801268f
Sharbati S, Schramm K, Rempel S, Wang H, Andrich R, Tykiel V, Kunisch R, Lewin A (2009) Characterisation of porin genes from Mycobacterium fortuitum and their impact on growth. BMC Microbiol 9:31. https://doi.org/10.1186/1471-2180-9-31
Sharma A, Gupta P, Kumar R, Bhardwaj A (2016a) dPABBs: a novel in silico approach for predicting and designing anti-biofilm peptides. Sci Rep 6:1–13. https://doi.org/10.1038/srep21839
Sharma R, Keshari D, Singh KS, Yadav S, Singh SK (2016b) MRA_1571 is required for isoleucine biosynthesis and improves Mycobacterium tuberculosis H37Ra survival under stress. Sci Rep 6:1–12. https://doi.org/10.1038/srep27997
Sharma A, Vashistt J, Shrivastava R (2021) Response surface modeling integrated microtiter plate assay for Mycobacterium fortuitum biofilm quantification. Biofouling 37:830–843. https://doi.org/10.1080/08927014.2021.1974846
Sharma A, Vashistt J, Shrivastava R (2022a) Knockdown of the type-II fatty acid synthase gene hadC in Mycobacterium fortuitum does not affect its growth, biofilm formation, and survival under stress. Int J Mycobacteriology 11:159–166. https://doi.org/10.4103/ijmy.ijmy_46_22
Sharma A, Vashistt J, Shrivastava R (2022b) Mycobacterium fortuitum fabG4 knockdown studies: implication as pellicle and biofilm specific drug target. J Basic Microbiol 62:1504–1513. https://doi.org/10.1002/jobm.202200230
Sharma A, Nadda AK, Shrivastava R (2023) Mycobacterium tuberculosis DapA as a target for antitubercular drug design. In: Brahmachari G (ed) Biotechnology of microbial enzymes, 2nd edn. Elsevier, pp 279–296
Shrivastava P, Navratna V, Silla Y, Dewangan RP, Pramanik A, Chaudhary S, Rayasam GV, Kumar A, Gopal B, Ramachandran S (2016) Inhibition of Mycobacterium tuberculosis dihydrodipicolinate synthase by alpha-ketopimelic acid and its other structural analogues. Sci Rep 6:1–7. https://doi.org/10.1038/srep30827
Singh SP, Bora TC, Bezbaruah RL (2012) Molecular interaction of novel compound 2-methylheptyl isonicotinate produced by Streptomyces sp. 201 with dihydrodipicolinate synthase (DHDPS) enzyme of Mycobacterium tuberculosis for its antibacterial activity. Indian J Microbiol 52:427–432. https://doi.org/10.1007/s12088-012-0252-4
Singh A, Varela C, Bhatt K, Veerapen N, Lee OY, Wu HHT, Besra GS, Minnikin DE, Fujiwara N, Teramoto K, Bhatt A (2016a) Identification of a desaturase involved in mycolic acid biosynthesis in Mycobacterium smegmatis. PLoS One 11:e0164253. https://doi.org/10.1371/journal.pone.0164253
Singh P, Rao RN, Reddy JRC, Prasad RBN, Kotturu SK, Ghosh S, Mukhopadhyay S (2016b) PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci Rep 6:1–16. https://doi.org/10.1038/srep21624
Sood S (2016) Establishment and validation of Mycobacterium fortuitum model for Mycobacterium tuberculosis persistent infection. http://hdl.handle.net/10603/149190. Accessed 10 June 2023
Sousa S, Bandeira M, Carvalho PA, Duarte A, Jordao L (2015) Nontuberculous mycobacteria pathogenesis and biofilm assembly. Int J Mycobacteriol 4:36–43. https://doi.org/10.1016/j.ijmyco.2014.11.065
Standish AJ, Morona R (2014) The role of bacterial protein tyrosine phosphatases in the regulation of the biosynthesis of secreted polysaccharides. Antioxid Redox Sign 20:2274–2289. https://doi.org/10.1089/ars.2013.5726
Taylor RC, Brown AK, Singh A, Bhatt A, Besra GS (2010) Characterization of a β-hydroxybutyryl-CoA dehydrogenase from Mycobacterium tuberculosis. Microbiology 156:1975–1982. https://doi.org/10.1099/mic.0.038802-0
Varughese P, Helbecque DM, McRae KB, Eidus L (1974) Comparison of strip and Ziehl-Neelsen methods for staining acid-fast bacteria. Bull World Health Org 51:83–91
Wang F, Langley R, Gulten G, Wang L, Sacchettini JC (2007) Identification of a type III thioesterase reveals the function of an operon crucial for Mtb virulence. Chem Biol 14:543–551. https://doi.org/10.1016/j.chembiol.2007.04.005
Wang S, Yang Y, Zhao Y, Zhao H, Bai J, Chen J, Zhou Y, Wang C, Li Y (2016) Sub-MIC tylosin inhibits Streptococcus suis biofilm formation and results in differential protein expression. Front Microbiol 7:384. https://doi.org/10.3389/fmicb.2016.00384
Wang J, Guo H, Cao C, Zhao W, Kwok L, Zhang H, Zhang W (2018) Characterization of the adaptive amoxicillin resistance of Lactobacillus casei Zhang by proteomic analysis. Front Microbiol 9:1–9. https://doi.org/10.3389/fmicb.2018.00292
Weimar JD, DiRusso CC, Delio R, Black PN (2002) Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. J Biol Chem 277:29369–29376. https://doi.org/10.1074/jbc.M107022200
Williams MM, Yakrus MA, Arduino MJ, Cooksey RC, Crane CB, Banerjee SN, Hilborn ED, Donlan RM (2009) Structural analysis of biofilm formation by rapidly and slowly growing nontuberculous mycobacteria. Appl Environ Microbiol 75:2091–2098. https://doi.org/10.1128/AEM.00166-09
Wright DG, Castore R, Shi R, Mallick A, Ennis DG, Harrison L (2017) Mycobacterium tuberculosis and Mycobacterium marinum non-homologous end-joining proteins can function together to join DNA ends in Escherichia coli. Mutagenesis 32:245–256. https://doi.org/10.1093/mutage/gew042
Yahya MFZR, Hamid UMA, Norfatimah MY, Kambol R (2014) In silico analysis of essential tricarboxylic acid cycle enzymes from biofilm-forming bacteria. Trends Bioinform 7:19–26. https://doi.org/10.3923/tb.2014.19.26
Yang Y, Luo M, Zhou H, Li C, Luk AWS, Zhao GP, Fung K, Ip M (2019) Role of two-component system response regulator BceR in the antimicrobial resistance, virulence, biofilm formation, and stress response of group B Streptococcus. Front Microbiol 10:10. https://doi.org/10.1101/340679
Yokoyama T, Choi KJ, Bosch AM, Yeo HJ (2009) Structure and function of a Campylobacter jejuni thioesterase Cj0915, a hexameric hot dog fold enzyme. Biochim Biophys Acta - Proteins Proteom 1794:1073–1081. https://doi.org/10.1016/j.bbapap.2009.03.002
Zhang L, Kent JE, Whitaker M, Young DC, Herrmann D, Aleshin AE, Ko YH, Cingolani G, Saad JS, Moody DB, Marassi FM (2022) A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis. Nat Commun 13:2255. https://doi.org/10.1038/s41467-022-29873-6
Zubieta C, Arkus KA, Cahoon RE, Jez JM (2008) A single amino acid change is responsible for evolution of acyltransferase specificity in bacterial methionine biosynthesis. J Biol Chem 283:7561–7567. https://doi.org/10.1074/jbc.M709283200
Acknowledgements
The authors would like to acknowledge the administration of Jaypee University of Information Technology, Waknaghat, Solan (Himachal Pradesh), India for providing the research facility, and a fellowship to Dr. Ayushi Sharma. The authors are also grateful to Central Drug Research Institute (CDRI), Lucknow, India, for providing the mycobacterial strain.
Author information
Authors and Affiliations
Contributions
AS and RS conceived and designed research. AS conducted experiments. AS, SB, and NK analyzed data. JV and RS supervised the study. AS wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sharma, A., Bansal, S., Kumari, N. et al. Comparative proteomic investigation unravels the pathobiology of Mycobacterium fortuitum biofilm. Appl Microbiol Biotechnol 107, 6029–6046 (2023). https://doi.org/10.1007/s00253-023-12705-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12705-y