Abstract
In the present study, juvenile striped catfish (Pangasianodon hypophthalmus), a freshwater fish species, have been chronically exposed to a salinity gradient from freshwater to 20 psu (practical salinity unit) and were sampled at the beginning (D20) and the end (D34) of exposure. The results revealed that the intestinal microbial profile of striped catfish reared in freshwater conditions were dominated by the phyla Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia. Alpha diversity measures (observed OTUs (operational taxonomic units), Shannon and Faith’s PD (phylogenetic diversity)) showed a decreasing pattern as the salinities increased, except for the phylogenetic diversity at D34, which was showing an opposite trend. Furthermore, the beta diversity between groups was significantly different. Vibrio and Akkermansia genera were affected differentially with increasing salinity, the former being increased while the latter was decreased. The genus Sulfurospirillium was found predominantly in fish submitted to salinity treatments. Regarding the host response, the fish intestine likely contributed to osmoregulation by modifying the expression of osmoregulatory genes such as nka1a, nka1b, slc12a1, slc12a2, cftr, and aqp1, especially in fish exposed to 15 and 20 psu. The expression of heat shock proteins (hsp) hsp60, hsp70, and hsp90 was significantly increased in fish reared in 15 and 20 psu. On the other hand, the expression of pattern recognition receptors (PRRs) were inhibited in fish exposed to 20 psu at D20. In conclusion, the fish intestinal microbiota was significantly disrupted in salinities higher than 10 psu and these effects were proportional to the exposure time. In addition, the modifications of intestinal gene expression related to ion exchange and stressful responses may help the fish to adapt hyperosmotic environment.
Key points
• It is the first study to provide detailed information on the gut microbiota of fish using the amplicon sequencing method.
• Salinity environment significantly modified the intestinal microbiota of striped catfish.
• Intestinal responses may help the fish adapt to hyperosmotic environment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Striped catfish Pangasianodon hypophthalmus industry has been an important inland aquaculture production in Southeast Asia, particularly in Mekong Delta in Vietnam. In 2019, striped catfish production in Vietnam reached 1.4 million of tons with estimated value of US$2.3 billion (FAO 2020). Nevertheless, the industry is currently facing climatic challenges, especially extensive salinity intrusion caused by the climate change. The saline water was observed up to 40 km far from the river mouth in places and increased salinity level in striped catfish farms up to 9 psu (practical salinity units) (Schmitz et al. 2017a, b, c). Several studies have been conducted to investigate the effects of salinity on physiological striped catfish (Nguyen et al. 2014; Schmitz et al. 2016a,b). However, the intestinal response of fish is largely unknown.
The fish intestine is a complex multifunctional organ that plays crucial roles in nutrient absorption, the immune response, and osmoregulation (Martin et al. 2016; Dehler et al. 2017a). In addition to the gills and kidney, the gastro-intestinal tract contributes to internal osmoregulation through balancing the uptake and maintenance of electrolytes and water when fish migrate from fresh water to seawater (Ringø et al. 2003; Malakpour Kolbadinezhad et al. 2018). When adapting to hyperosmotic environments, fish continuously drink water to compensate water loss that may alter intestinal physiology resulting in changes in physiochemical factors in the intestine, including gastric acidity, digestive enzymes, and bile salts (Usher et al. 1990). These changes are known to affect the intestinal colonization by specific microbes in teleosts (Ringø et al. 2003). Therefore, the changes in internal pH and osmolality could significantly affect the resident and transient intestinal microbes (Ray and Ringø 2014). In fact, previous research on anadromous and euryhaline fish species has shown that salinity could decrease the intestinal microbial richness and evenness and can cause changes in its composition (Zhang et al. 2016; Dehler et al. 2017a). The salinity gradient is an important factor that can shape fish microbial communities, with marked differences in the community composition in freshwater versus seawater fish (Dehler et al. 2017a). Furthermore, the environmental water and diets can also influence the intestinal microbial composition in fish. Hence, it could be predicted that freshwater and seawater fish have different intestinal microbiota due to different compositions in the environment (Herlemann et al. 2011) and, therefore, the fish intestinal microbiota and the respective environmental microbiota should have a similar composition. However, it is known that several bacterial species are specifically abundant in the intestine but not in the water (Schmidt et al. 2015; Dehler et al. 2017b). This suggests that there is a complex mechanism for bacterial colonization of the fish intestine that responds to both temporal environmental changes and host physiology (Schmidt et al. 2015; Dehler et al. 2017a).
Regarding fish responses to environmental salinity changes, fish living in hyperosmotic environments are required to retain water and secrete ions against the concentration gradient (Ronkin et al. 2015). The regulatory mechanisms involve many processes and most of them take place in the gill, kidney, and intestine (Marshall and Grosell 2005). In the euryhaline species Mozambique tilapia (Oreochromis mossambicus), the ability to tolerate hypersaline water may be related to its capacity to upregulate ion exchange in the posterior intestine with significantly increased expressions of Na+-K+-Cl− 2 cotransporter (nkcc2), Na+-K+-Cl− 1 cotransporter (nkcc1), cystic fibrosis transmembrane regulator (cftr) and Na+/K+-ATPase (nka), which are involved in exchanging sodium, potassium, and chloride ions (Li et al. 2014). Another euryhaline fish, silver sea bream (Sparus sarba) acclimated to 33 psu also exhibited an increase of aquaporin 1 (aqp1) transcripts, regulating water and solute transport, when exposed to hyposaline (12 psu) and hypersaline (70 psu) environments (Deane et al. 2011). In striped catfish, osmoregulation is known to increase through upregulation of gill Na+/K+-ATPase activity when the fish are transferred from fresh water (0 psu) to seawater (20 psu) (Schmitz et al. 2016a). In the case of insufficient ionic regulation, salinity fluctuations can lead to osmotic stress in aquatic organisms, eventually causing disruption of protein synthesis and protein damage (Deane et al. 2002). During osmotic stress, the synthesis of heat shock protein (hsp) families increases remarkably; therefore, these proteins are considered to be a biomarker of stress in fish (Hightower 1991; Deane et al. 2011). For instance, hsp70 (heat shock protein 70) expression in gills of tilapia (Sarotherodon melanotheron) significantly increased with hyperosmotic stress. Additionally, hepatic expression of hsp genes, including hsp60 (heat shock protein 60), hsp70, and hsp90 (heat shock protein 90), were lowest in fish reared in isosmotic salinity and highest in those adapted to hypoosmotic and hyperosmotic salinities (Deane et al. 2002). However, not much is known about the physiological changes in the intestine during such osmotic changes.
The interactions between the host and intestinal microbiota play an important role in many aspects of physiology, metabolism, and immune homeostasis in fish (Rawls et al. 2004). Microbial colonization of intestinal mucosal surfaces results in positive effects on gut immune responses, while microbiota imbalance disrupts immunoregulatory functions and may lead to disease development (Pérez et al. 2010). Intestinal epithelial cells are capable of sensing components of the expression of pattern recognition receptors (PRRs), among which toll-like receptors can recognize bacterial lipopolysaccharides and other microbial molecules such as pathogen–associated molecular patterns (Tlaskalova-Hogenova et al. 2005; Hörmannsperger and Haller 2010). The interaction between the receptor and the ligand activates the signalling molecules, resulting in an increase of solute mediators that recruit and regulate the immune and inflammatory responses (Fasano and Shea-Donohue 2005). In sea bass (Dicentrarchus labrax), a downregulation of intestinal inflammatory genes such as interleukin 1 beta (il1β), interleukin 10 (il10), transforming growth factor β (tgfβ) in fish fed with probiotics has been previously shown (Picchietti et al. 2009). In contrast, another probiotic mix upregulated toll-like receptor 1 (tlr1), toll-like receptor 3 (tlr3), il1β, tumor necrosis factor alpha (tnfα), il10, and tgfβ in the gut of zebrafish (Danio rerio) (Gioacchini et al. 2017). Collectively, these studies indicate that there is an alteration of the microbe composition that may lead to locally regulated host inflammatory responses.
In the present study, we investigated alterations in the intestinal microbiota as well as host gene expression in the intestine of striped catfish under different conditions of salinity.
Materials and methods
Experimental conditions
Juvenile striped catfish weighing 24.04 ± 1.02 g/fish were collected from Viet-Uc hatchery, An Giang province, and acclimated in fresh water for 2 weeks before starting the experiment. The juveniles were distributed (40 fish/tank) into 5 experimental recirculating systems with triplicate for each treatment. The final salinities in the different treatments were 0, 5, 10, 15, and 20 psu, and the salinities were gradually increased at a rate of 0, 0.25, 0.5, 0.75, and 1 psu per day, respectively. The increase was done twice daily to reduce the shock to juveniles. Fish were fed ad libitum twice daily with commercial dry pellets (Aquaexcel 7434, Cargill, Vietnam), and the residual feed was removed after 30 m of feeding. All treatments reached the target salinities after 20 experimental days (D20). Then, the fish were continuously reared in the corresponding salinities for 2 weeks (until D34). Water quality parameters, including pH (7.6 ± 0.2), temperature (29.5 ± 0.6 °C), dissolved oxygen (6.7 ± 0.5 mg/l), nitrites (0.2 ± 0.1 mg/l), and total ammonia nitrogen (0.6 ± 0.1 mg/l) were measured and maintained at optimal levels for striped catfish during the experiment.
Sampling
Three individual juveniles/tank were sampled at D20 and D34 (Supplementary Fig. S1). There were 5 treatments in D20, and 4 treatments in D34 were sampled because all fish exposed 20 psu were dead in the end of the experiment. The fish were first euthanized with 200 mg/L MS-222 (ethyl 3-aminobenzoate methane sulfonate; Sigma, St. Louis, MO, USA). Then, the fish abdominal cavity was opened aseptically. The intestinal samples were separated and transferred to a cryotube containing RNAlater (prepared following the protocol detailed at https://www.protocols.io/view/RNAlater-Recipe-c56y9d). The tubes were stored at − 80 °C until further analysis.
DNA extraction, preparation of 16S rRNA gene amplicon libraries (V3-V4 region), library quantification, and sequencing
DNA was extracted from the intestinal samples using the NucleoMag VET kit (Macherey–Nagel, Düren, Germany) according to the manufacturer’s protocol. The DNA quantities were measured using the Qubit dsDNA BR Assay Kit (Life Technologies, Carlsbad, CA, USA).
Library preparation followed the standard Illumina (San Diego, CA, USA) 16S rRNA metagenomics sequencing protocol (15,044,223 Rev. B). The V3-V4 region of 16S rRNA was amplified using the primers 341F-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and 785R-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC. These primers were shown to have a broad phyletic coverage (Klindworth et al. 2013). The PCR comprised 50 µl reactions, including 10 μl of 5 × KAPA HiFi (Kapa Biosystems, Woburn, USA), 1.5 μl of 10 mM dNTP, 1.5 μl of each forward and reverse primer (1 μM), 1 μl of MgCl2 (2.5 mM), 1 μl Kapa Hifi Hstart (Kapa Biosystems, Woburn, USA), 5 μl of genomic DNA (50 ng), and 28.5 μl of PCR grade water. Thermocycling conditions included an initial denaturation step at 95 °C for 3 min, then 25 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. A final extension step was conducted at 72 °C for 5 min. After the PCR, the products were subsequently purified using AmPure XP beads (Beckman Coulter, High Wycombe, UK) and the concentration was checked using Qubit dsDNA BR Assay Kit. Then, a second PCR amplification was performed to attach dual indices and Illumina sequencing adapters using the Nextera XT Index kit (Illumina, San Diego, CA, USA). The reaction contained 14 µl of DNA elution and PCR grade water, 2.5 μl each of Nextera XT Index 1 Primer (N7XX) and Nextera XT Index 2 Primer (S5XX) (200 nM), 5 μl of 5 × KAPA Hifi, 0.75 μl of 10 mM dNTP, and 0.25 µl of 2 × KAPA HiFi Hstart (0.5 U). The process included an initial denaturation step at 95 °C for 3 min, then 8 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final extension step at 72 °C for 5 min. AMPure XP beads were used to purify PCR products. The 16S rRNA amplicon libraries for each sample were quantified by a fluorometric method using dsDNA binding dyes. Then, 5 μL of the 4 nM DNA library pool was prepared for sequencing on the Illumina MiSeq platform. The library pool was denatured with 0.2 M NaOH and diluted with Pre-chilled HT1 buffer (Illumina, San Diego, CA, USA) to a final concentration of 4 pM. Sequencing was performed with a MiSeq Reagent Kit v3–600 cycle (Illumina, San Diego, CA, USA) following the manufacturer’s protocols. Paired-end sequencing was performed on an Illumina MiSeq platform with 5% PhiX spike-in quality control using PhiX Control Kit v3 (Illumina, San Diego, CA, USA). Raw sequencing data from all samples can be openly accessed on NCBI database with BioProject accession number PRJNA802379.
Data analysis
The data analysis was performed according to the protocol of Lokesh et al. (2019). The FastQ files were used as the input for the UPARSE pipeline (usearch version 11.0.667) (Edgar 2013). First, the forward and reverse reads were merged, with maximum mismatches of 10 and minimum ID percentage of 80%. The primer binding sites were trimmed, and the merged reads were truncated at 400 bp. The resulting sequences were then quality filtered with maximum expected error rate set to 1. Next, the dereplication was performed, and the reads with < 10 sequences were discarded. OTUs (operational taxonomic units) were clustered at a threshold of 97% similarity level and chimeric sequences were discarded. After clustering, the raw reads were mapped to OTUs by searching the reads as queries against the OTU representative sequences. Taxonomic ranks were assigned to OTUs using the SINTAX algorithm using Ribosome Database Project database downloaded at the link of https://drive5.com/sintax/rdp_16s_v18.fa.gz (rdp_16s sequences, version v18). Taxonomic assignments with scores < 0.5 were not considered in the downstream analyses.
Alpha and beta diversities were computed using QIIME2 (Hall and Beiko 2018). Then, one-way ANOVA followed by Tukey’s post hoc tests were used to statistically compare the treatments on the same sampling day and an independent t-test applying a 95% confidence interval was used for the treatments at the same salinity levels from different sampling days in SPSS 20 software (IBM Corporation, Armond, NY, USA). Appropriate transformation was performed when the assumptions of normality and equal variance were not met. Beta diversity was assessed based on the Bray–Curtis distance. Pairwise comparisons between treatments were performed using ANOSIM in QIIME2. The results of Bray–Curtis distance and relative abundance were plotted using R package (version 4.0.5) (Team RC 2013) and phyloseq (version 1.34.0) (McMurdie and Holmes 2013). The significantly abundant OTUs were calculated using Linear Discriminant Analysis Effect Size (LEfSe) (Segata et al. 2011) on Galaxy (version 1.0) with p-value set at 0.05 and LDA log score threshold at 3.5. The LEfSe results were plotted using GraPhlAn (version 1.1.3) (Asnicar et al. 2015). To predict the metagenomes of each sample, the OTU table was used in PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States2) version 2.4.1 with the maximum value of Nearest Sequenced Taxon Index (NSTI) of 0.03 (Douglas et al. 2020). The generated tree was complemented with OTU resulting in hidden-state prediction (hsp) (Louca and Doebeli 2018) using the maximum-parsimony method, generating predicted Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology. The abundance profiles of KEGG orthologys were statistically analysed by LEfSe with p-value set at 0.05 and LDA log score threshold set at 2.5.
RNA extraction and qPCR
Intestinal RNA was extracted using Tri-Reagent solution (Ambion, Thermofisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The RNA concentration and quality were determined by NanoDrop-2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and after migration on agarose gel (1%), respectively. Genomic DNA contamination was removed by digestion using rDNAse I (Thermo Fischer Scientific, Waltham, MA, USA). Then, 1 µg of total RNA was reverse-transcribed using RevertAid RT kit (Thermo Fischer Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The cDNA samples were diluted 1:25 and used to determine gene expressions of nka1a (Na+/K+-ATPase α1a); nka1b (Na+/K+-ATPase α1b), slc12a1 (solute carrier protein family protein 12 group A, member 1); slc12a2 (solute carrier protein family protein 12 group A, member 2); cftr, aqp1, hsp70, hsp90, hsp60, tlr1, and tlr2 (toll-like receptor 2); il1β, il10, and tnfα (tumour necrosis factor alpha); and tgfβ and alpl (alkaline phosphatase) by quantitative PCR (qPCR). The reactions were performed using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA) on QuantStudio 5 (Applied Biosystems, Waltham, MA, USA). The thermal conditions were set at 95 °C for 3 min in the initial denaturation step, followed by 40 cycles at 95 °C for 30 s and 60 °C for 30 s. The primers used in the present study were designed using Primer-BLAST (Ye et al. 2012) from the striped catfish sequence published on GenBank. The efficiency of each gene was tested before analysing. The details concerning the primers are given in Supplementary Table S1. For analysis, sample value and normalization were determined using the relative expression ratio mathematical model (Pfaffl 2001). Briefly, Ct values are transformed according to the equation: Ratio (fold change) = Etarget∆Ct(control−sample)/Ereference∆Ct(control−sample), where Etarget is the qPCR efficiency of the target gene and Ereference is the qPCR efficiency of the reference gene (beta actin actβ) (Pfaffl 2001). To determine the qPCR efficiency, the serial dilution of pooled cDNA from all samples was included in the qPCR reaction. Statistical comparisons were carried out by one-way ANOVA, followed by Tukey’s post hoc test between treatments on the same sampling day and an independent t-test applying a 95% confidence interval was used for the treatments at the same salinity levels in different sampling days in SPSS 20 (IBM Corporation, Armond, NY, USA). Appropriate transformation was performed when the assumptions of normality and equal variance were not met.
Results
Differences in the intestinal microbial diversity and composition
Alpha diversity
The intestinal microbiota was clustered in 939 OTUs. Alpha diversity, including observed OTUs, Shannon index, and Faith’s PD (phylogenetic tree) of the intestinal microbiota had a decreasing pattern with the increase in salinity at D20. Notably, the highest values were expressed in the 5 psu (observed OTUs and Shannon index) and 10 psu (Faith’s PD) treatments, which were significantly different compared to the communities of the 15 psu and 20 psu treatments at D20 (p < 0.05) (Fig. 1). The richness at D34 decreased significantly (p < 0.05) in fish exposed to 10 psu compared to the same salinity at D20 (Fig. 1a). Although Shannon index decreased similarly at 5 psu and 10 psu, the changes were not statistically different (Fig. 1b). Interestingly, there was a significant increase in Faith’s PD in saline treatments at D34 compared to the corresponding treatments at D20 (p < 0.05) (Fig. 1c).
Alpha diversity in the intestinal microbiota of striped catfish exposed to different levels of salinity. a Observed operational taxonomic units (OTUs), b Shannon index, and c Faith’s PD (phylogenetic diversity). Results are given as mean ± SD. Different lower-case and upper-case letters denote statistically significant differences between treatments at D20 and D34, respectively (p < 0.05). Asterisks denote significantly higher values in the treatment compared to the corresponding values on the other sampling days (p < 0.05)
Beta diversity
To visualize the intestinal microbiota impaired under different salinity conditions, a Bray–Curtis similarity-based ordination analysis (PCoA) was performed. The ordination at D20 showed that the samples in 20 psu were completely separated from those in the other treatments (Fig. 2a). This was confirmed with the p-value < 0.01 and R-value > 0.7 (ANOSIM) (Table 1). Furthermore, the pairwise comparison between 15 psu and the other treatments showed similar patterns, with the most significant difference found between 5 and 15 psu with a p-value of 0.001 and an R-value of 0.655. The remaining pairwise comparisons varied between 0.001 and 0.009 for the p-value, and 0.323 and 0.462 for the R-value. At D34, there were significant differences in all pairwise comparisons (p < 0.01 and R > 0.6), except between the 5 and 10 psu treatments and 10 and 15 psu (Fig. 2b). The differences in beta diversities under each salinity treatment between two different sampling points (D20 and D34) were significant in all groups, except for the groups with 0 psu (Supplementary Fig. S2 and Supplementary Table S2).
Relative abundance of the prominent taxa in the intestinal microbiota of striped catfish
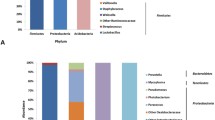
It is clear that Bacteroidetes was the dominant phylum in all treatment groups at D20 (Fig. 3) with the highest abundance (73.7%) in the 20 psu treatment. This was followed by Firmicutes (23.3%) and then Verrucomicrobia (14.7%) in the freshwater treatment. Firmicutes was also the predominant phylum in fish exposed to 5, 10, and 15 psu with 20.0, 20.4 and 25.4%, respectively. Campilobacterota increased significantly up to 27.5% in the 10 psu group. In addition, a gradual decrease in Verrucomicrobia was observed with increasing salinity, from 14.7% in fresh water (0 psu) to 1% at 20 psu. In the 20 psu treatment group, Proteobacteria (20%) were the most abundant after Bacteroidetes (73.7%) (Fig. 3a). At D34, there was a significant increase in the Campilobacterota phylum in the 5 and 10 psu groups (46.0% and 37.4%, respectively), which became one of the most abundant phyla in the intestinal microbiota. In addition, the proportion of Proteobacteria as the predominant bacteria in the gut microbiota changed in fish treated with 10 and 15 psu, at 38.8% and 30%, respectively (Fig. 3b).
At D20, Akkermansia was the most dominant genus in the gut microbiota of fish kept in freshwater conditions (13.4%) (Fig. 4a), followed by several different Bacteroides species, Helicobacter (4.2%), and Cetobacterium (1.5%). In fish exposed to 5 psu, Helicobacter was the most abundant genus (9.5%), followed by several Bacteroides species and Cetobacterium (4.9%). Under 10 psu, Sulfurospirillum accounted for 20.4% and was the predominant genus in the intestinal microbiota. When treated with 15 psu, the Bacteroides species were still predominant in the microbiota, but a significant increase in Vibrio and Parabacteroides was observed up to 6.7 and 5.4%, respectively. The most notable change in the 20 psu treatment was that the Bacteroides_OTU1 accounted for the greatest proportion of the microbiota at 70%, followed by the Vibrio genus (8%). The fish at D34 had a similar diversity of gut microbiota as those at D20, except for the increase in the genus Treponema (5.3%). Sulfurospirillum was the prominent genus in fish in 5 psu, accounting for over 36% of the community, followed by Helicobacter at 9.1%. In the 10 and 15 psu treatments, the genera Sulfurospirillum and Vibrio dominated (27.1 and 14.0%, respectively). In the 15 psu group, Bacteroides_OTU1 was dominant (12.7%). In addition, the proportion of the genera Cetobacterium (3.5 and 5.7% at 10 and 15 psu, respectively) and Romboutsia (2.1 and 2.5% at 10 and 15 psu, respectively) increased with salinity (Fig. 4b).
Differential abundance of the OTUs in intestinal microbiota of striped catfish
LEfSe was used to determine the intestinal biomarkers (OTUs) in striped catfish exposed to different levels of salinity (Fig. 5a). At the phylum level, Verrucomicrobia, mainly reflecting the abundance of OTUs of the genera Akkermansia and Lentisphaerae, were biomarkers in the 0 psu treatment. In addition, Fusobacter, mainly represented by the genus Cetobacterium, were differentially abundant in the 5 psu treatment, while Tenericutes and Campilobacterota were biomarkers in 10 psu. The latter was mainly comprised of the genera Sulfurospirilum and Helicobacter. The abundance of Treponema genus, belonging to the Spirochaetes phylum, was significantly higher in the 0 and 5 psu treatments than for the other treatments. The 15 psu treatment was significantly enriched in the Firmicutes phylum, reflecting the abundance of the Clostridia class, although the lower taxonomic levels shared significant abundance in all treatments except 20 psu. The Bacteroidia clade, which belongs to the Bacteriodetes phylum, was significantly abundant in the 20 psu treatment, but the Bacteroides genus shared high abundances in all the different salinities. Moreover, the most significant abundance of Vibrio genus belonging to the class Gammaproteobacteria were found in the 20 psu treatment, indicating that the genus is a biomarker in fish subject to a saline environment.
Cladogram showing differentially abundant taxa in the intestinal microbiota of striped catfish at D20 (a) and D34 (b) for the various salinity conditions. Linear discriminant analysis effect size (LEfSe) was used to validate the statistical significance and the effect size of the different abundances of taxa in the treatments compared to the control
At D34, the phyla Bacteroidetes, Verrucomicrobia, and Spirochaetes, reflecting the abundance of the genera Bacteroides, Akkermansia, and Treponema, respectively, were significantly more abundant in the freshwater group (Fig. 5b). In the 5 psu treatment, the phyla Letisphaerae and Campilobacterota had the highest abundances, the latter being reflected in the highly abundant genera Sulfurospirillum and Helicobacter. Although the phylum Proteobacteria was a biomarker in the 10 psu treatment, it reflected only the Gammaproteobacteria class with the representation of the Vibrio genus. The point of interest in the 15 psu treatment was the high abundance of Bacteroides_OTU 1.
Presumptive metabolic potential of the intestinal microbiota of striped catfish
The OTU table was used as the input file for PICRUSt2, and accuracy was calculated using the NSTI value. The results according to LEfSe showed that there were 20 significantly different abundances of KEGG pathways at level 2 in which the treatment of 20 psu had the highest number (9) of significantly enriched pathways, such as carbohydrate metabolism, lipid metabolism, cell motility, and membrane transport (Table 2). This was followed by 6, 4, and 1 enriched pathways at 0, 5 and 15 psu, respectively, at D20. At D34, the 0 psu treatments had the highest number of enriched pathways with 9 of 18 total pathways, while the 5, 10, and 15 psu treatments were significantly abundant with 5, 3, and 1 pathways, respectively.
Gene expression
Expression of genes related to osmoregulation
The expression of nka1a and nka1b showed a significant increase with salinity at D20 and D34. In addition, a remarkable increase in mRNA levels of nka1b was observed after fish were maintained at the targeted salinities of 5, 10, and 15 psu for two weeks (Fig. 6a, b). A similar pattern was observed for the solute carrier family genes slc12a1 and slc12a2, which encode nkcc2 and nkcc1, respectively (Fig. 6c, d). Notably, the mRNA level of slc12a2 was the lowest at 5 and 10 psu, whereas it was the highest at 20 psu at D20 (Fig. 6d). Moreover, slc12a1 showed a gradual increase with salinity at D20 (Fig. 6c). At D34, the expression of both genes was significantly higher in the 15 psu group than in the other groups. Fish exposed to salinity of 5, 10, and 15 psu at D34 had significantly higher mRNA levels for these genes than the corresponding fish at D20. The transcripts of nka1a, nka1b, slc12a1, and slc12a2 were lowest on D20 at 5 psu compared to the other conditions. With respect to cystic fibrosis transmembrane conductance regulator (cftr) and aquaporin 1 (aqp1), the highest levels of the genes were observed in the 0 psu treatment and the lowest in the 20 psu treatment at D20. Similarly, the mRNA levels of cftr and aqp1 were significantly higher in fish exposed to fresh water than in fish exposed to 10 and 15 psu (Fig. 6e, f). Notably, both gene expressions significantly decreased on D34 compared to D20 when treated with 10 psu.
Fold changes in the expression of genes related to osmoregulation in the intestine of striped catfish exposed to different salinity levels. a nka1a, b nka1b, c slc12a1, d slc12a2, e cftr, and f aqp1. Results are expressed as mean ± SEM. Different lower-case and upper-case letters denote statistically significant differences between treatments at D20 and D34, respectively (p < 0.05). Asterisks denote significantly higher values in the treatment compared to the corresponding values on the other sampling day (p < 0.05)
Heat shock protein family
The highest expression of heat shock protein 70 (hsp70) was found in the 15 and 20 psu groups, and was significantly different from that of the freshwater group at D20. However, the changes recovered after 2 weeks of exposure to salinity (Fig. 7a). A similar pattern was observed in the mRNA levels of heat shock protein 90 and 60 at D20, where there was a significant increase with salinity. Moreover, the increased responses were prolonged until D34 when treated with 15 psu (Fig. 7b, c). In general, salinity greater than 10 psu had a significant effect on the expression of hsp genes in the initial period of exposure and these responses decreased with exposure time.
Fold change expression of genes in the heat shock protein family in the intestine of striped catfish exposed to different salinities. a hsp70, b hsp90, and c hsp60. Results are given as mean ± SEM. Different lower-case and upper-case letters denote statistically significant differences between treatments on D20 and D34, respectively (p < 0.05). Asterisks denote significantly higher values in the treatment compared to the corresponding values on the other sampling days (p < 0.05)
Intestinal pattern recognition receptor and its associated pathways (TLRs)
In general, the expression of genes involved in PRRs, such as tlr1, tlr2, il1β, tnfα, il10, and tgfβ, showed the lowest expression in fish exposed to 20 psu at D20 (Fig. 8). Notably, the expression of tlr1 was significantly lower in fish exposed to 20 psu than that in the lower salinity levels, while the statistical significance of tlr2 was found only between the treatments of 5 psu and 20 psu (p < 0.05) (Fig. 8a, b). Pro-inflammatory genes such as il1β and tnfα were remarkably downregulated in the 20 psu treatment compared to the others (p < 0.05) (Fig. 8c, d). Moreover, il1β expression was significantly lower in fish exposed to 15 psu compared to the others at D34. For anti-inflammatory responses, tgfβ transcripts were the lowest at 20 psu and significantly different from 0, 5, and 10 psu treatments. For il10 expression, there was no statistical difference between treatments at D20, although it was the lowest in the 20 psu treatment, whereas at D34, it was highest in the freshwater treatment and significantly different compared to that of 10 and 15 psu (Fig. 8e, f). Moreover, il10 gene expression was significantly decreased in the 10 psu treatment after 2 weeks of exposure to salinity. The expression of a gene related to lipopolysaccharide detoxification, alkaline phosphastase, was examined (Fig. 8g). It showed a gradual increase with the salinity gradient, and was statistically higher in the 20 psu treatment than in the 0, 5, and 10 psu treatments. There were no remarkable differences in the expression of this gene after two weeks of exposure to salinity.
Fold change expression of intestinal pattern recognition receptors (PRRs) in the intestine of striped catfish exposed to different salinity levels. a tlr1, b tlr2, c il1β, d tnfα, e il10, f tgf, and g alpl. Results are given as mean ± SEM. Different lower-case and upper-case letters indicate statistically significant differences between treatments on D20 and D34, respectively (p < 0.05). Asterisks indicate that the values for each treatment are significantly higher compared to the corresponding values on the other sampling days (p < 0.05)
Discussion
Fish intestine responds to changes in the osmotic conditions
In teleost fishes, the gut plays an important role in water balance in marine environments but its role in osmoregulation is less important in freshwater fish, which are hyperosmotic to the environment (Takei and Hwang 2016). Entry into a saline environment results in a 10- to 50-fold increase in drinking volume to compensate for water losses in fish (Grosell 2006). The oesophageal-gastrointestinal tract absorbs more than 95% of the NaCl in seawater during ingestion and this absorption facilitates water absorption (Grosell, 2010). The major components of the absorption are basolateral Na+/K+-ATPase and apical Na+-K+-2Cl− (Grosell 2006). In the present study, the genes involved in ion transporters, especially nka1a and slc12a1 were generally upregulated at salt levels of 15 and 20 psu for short-term exposure (D20) and even at 10 psu for long-term exposure (D34). The genes nka1a and nka1b are two similar paralogues of nka1 that encode the α-subunit of Na+/K+-ATPase (Lind et al. 2013). Na+/K+-ATPase, known to be an active pump, represents the primary driving force for activation of many ion transport systems in osmoregulatory organs, including the intestinal tract in teleost fish (Yang et al. 2018), and its upregulated expression indicates a higher need for ion uptake or excretion (Lin et al. 2003; Yang et al. 2009; Whittamore 2012). Also, slc12a1 coding nkcc2 plays a dominant role in intestinal NaCl absorption in euryhaline fish (Takei 2021). The upregulation of nka1 and slc12a1 was also found in a study of the euryhaline fish Mozambique tilapia and sea bream when transferred from hypo- to hyperosmotic environments (Gregório et al. 2013; Li et al. 2014). Salinities between freshwater and 10 psu generally did not upregulate the expressions of these osmoregulatory genes, even fish subjected to 5 and 10 psu showed a significant decrease in slc12a2 expressions compared to that in freshwater in the early exposure. The regulations indicate that fish may have a tendency to reduce ion exchange salinities close to their isosmotic point which is estimated to be 9 psu in striped catfish (Nguyen et al. 2014). In mammals, Cl− secretion is regulated by cftr on the apical membrane and by nkcc1 on the basolateral membrane (Takei 2021). In the present study, the upregulated expression of slc12a2 in the highest salinity treatment, i.e., 20 psu at D20 and 15 psu at D34, may demonstrate their effective activities in Cl− regulation; however, cftr expression showed downregulation. A decline in cftr transcript expression during seawater adaption was also found in the intestine of killifish (Fundulus heteroclitus) and European eel (Anguilla anguilla) (Marshall and Singer 2002; Takei 2021), while the opposite pattern was found in different euryhaline fish species (Li et al. 2014). The downregulated expression is possibly explained by the fact that the CFTR of striped catfish intestine is localized in the basolateral membrane for Cl− absorption, as suggested in previous research (Marshall and Singer 2002; Takei 2021). The transport of NaCl parallels that of water, moving from the intestinal lumen into the body fluid through the epithelium when luminal fluid is almost isotonic to the body fluid (Li et al. 2005). Water absorption by transcellular passage is greatly facilitated by aquaporins (AQPs) which showed an upregulated expression in European and Japanese eels (Anguilla japonica) acclimated to seawater (Aoki et al. 2003; Martinez et al. 2005). However, the expression of aqp1 decreased when striped catfish were exposed to salinity in the current study. A similar result was found in medaka (Oryzias latipes), which may be explained by the movement of AQP1a from the apical membrane to the subapical area and the possible transport of water via paracellular routes after fish enter seawater (Madsen et al. 2014). Additionally, the duration of exposure to salinity also affected the osmoregulation process in the gut. In general, these genes were upregulated in fish exposed to increased salinity in a manner proportional to the exposure time. Similar findings were reported in Sacramento splittail (Pogonichthys macrolepid) and killifish, showing an increase of Na+/K+-ATPase activity during seawater acclimation periods (Scott et al. 2008; Mundy et al. 2020). These results confirm the osmoregulatory ability of the intestinal tract in striped catfish during saline exposure through differential regulation of Na+/K+-ATPase activity. However, the exact mechanism of changes in Cl− secretion and water absorption in striped catfish under the influence of salinity requires further research.
Heat shock proteins are known as stress proteins and extrinsic chaperones and are produced in all cellular organisms exposed to stressors (Roberts et al. 2010). HSPs are a useful biomarker indicating the stress status of teleost fish (Roberts et al. 2010). The increase in HSPs has been found in freshwater and euryhaline fishes such as common carp (Cyprinus carpio) and salmon (Salmo salar) under salinity stress, although it is not systematic in all organs (De Wachter et al. 1998; Pan et al. 2000). In the current study, the increase in all tested hsp transcripts were exhibited in the highly osmotic environments, such as 15 and 20 psu, although the levels decreased slightly with the duration of exposure to salinity. The upregulation of HSPs plays an important role in preventing protein misfolding under stress conditions (Winklhofer et al. 2008). Interestingly, there was a highly similar pattern between HSP gene expression and the major reactive genes of the osmoregulation process, i.e., nka1a and slc12a1, especially on D20. This observation is supported by results in other species showing a positive correlation between the expression of hsp70 and nka under hyperosmotic stress (Deane and Woo 2004; Larsen et al. 2008; Tine et al. 2010). However, Schmitz et al. (2016a) found that HSP70 expression in the kidney of juvenile striped catfish did not vary significantly with the salinity gradient from fresh water to 20 psu. Since a significantly lower survival is observed at 20 psu, it may be hypothesized that juvenile striped catfish have mechanisms to respond to hyperosmotic conditions, through hsp, nka, and slc12a expression changes, but these are not sufficient for adaptation to 20 psu.
Osmotic stress alters the intestinal microbial diversity
The effect of salinity on striped catfish has been investigated in previous studies; however, most of them were focused on physiological and immunological responses (Nguyen et al., 2014; Schmitz et al. 2016a,b). To date, no attention has been given to how the intestinal microbiota changes under high salinity conditions in this species. Additionally, the intestinal microbiota in striped catfish has not been investigated by amplicon sequencing-based methods, except a study with general phylum information by Sutriana et al (2018).
We obtained 4.5 million raw sequence reads from the Miseq runs, distributed across 74 samples and 4,066,509 quality filtered sequences were clustered into 939 OTUs (63,486 ± 23,401, mean ± SD with the min and max values of 2,849 and 90,739 sequences, respectively). This allowed for rarefaction and even sampling of 6000 sequences per sample. After rarefaction, 73 of 81 samples were retained for further steps. The present study revealed that the alpha diversity indicators of the intestinal microbiota were significantly decreased in salinities higher than 10 psu at D20, which is in accordance with the previous studies on euryhaline species (Schmidt et al. 2015; Dehler et al. 2017a). The salinity gradient is an important barrier for bacterial colonization, and the significant decrease in microbial species richness and diversity in saline treatments might reflect this evolutionary barrier (Dehler et al. 2017a). Furthermore, the increase of water uptake and salt excretion in order to regulate ions in the intestinal tract may lead to changes in the niche and contribute to alterations in the microbiota structure and composition (Whittamore 2012; Dehler et al. 2017a). In fact, changes in ionic regulation with increasing salinity were confirmed through the modified expression of genes related to osmoregulatory processes, such as nka1a, slc12a1, aqp1, and cftr, which could reflect the physiological changes observed in the current study. These genes generally showed significant differences in 15 and 20 psu treatment groups, in which higher nka1a and slc12a1 and lower aqp1 and cftr compared to the lower salinity treatments were observed. The physiological changes (possibly including pH, osmolality, passage rate and residence time, which are different in fresh water and seawater) significantly affect microbiota establishment (Ray and Ringø 2014). Additionally, phylogenetic diversity estimated by Faith’s PD significantly increased with salinity after two weeks of exposure to highly saline conditions. Faith’s PD is calculated based on the sum of branch lengths of a phylogenetic tree connecting all species in the community (Faith 1992); therefore, there is a strong positive correlation between species richness and phylogenetic diversity. However, our results showed an opposite trend between phylogenetic diversity and richness at D34. This finding might be explained by the fact that although the number of OTUs was reduced, the phylogenetic differences of remaining OTUs in salinity treatments were extremely significant and this reflected the destabilized microbiota under saline conditions. Furthermore, the results of the Bray–Curtis index, followed by the ANOSIM test, showed significant differences in the intestinal microbiota structure between treatments. Notably, differences in diversity increased with duration of saline exposure, with medium salinity treatments (i.e., 5 psu and 10 psu) showing the greatest changes. Lokesh et al. (2019) reported that the diversity of intestinal microbiota in Atlantic salmon decreased significantly after 3 weeks of exposure to saline conditions. Striped catfish exposed to 10 psu showed the greatest difference in diversity according to exposure duration in the current study. Taken together, these results suggest that the salinity gradient and exposure time can disturb the intestinal community in striped catfish.
Osmotic stress alters the intestinal microbiota taxonomic composition
In the present study, we found that Bacteroidetes, Firmicutes, Verrucomicrobia, and Proterobacteria phyla were predominant in the intestinal microbiota of striped catfish in freshwater. This is consistent with previous studies on striped catfish (Sutriana et al. 2018) and channel catfish Ictalurus punctatus (Bledsoe et al. 2016). Fish exposed to 20 psu had a significantly lower diversity, in which Bacteroides_OTU1 and Vibrio accounted for around 80%. The low diversity combined with the loss of some Bacteroides species and the increase in the Proteobacteria phylum may reflect a dysbiosis status of the intestinal tract (Humphreys 2020). The Bacteroides genus was dominant in the intestinal microbiota in all treatment groups at the beginning of exposure to salinity, however its abundance decreased in all groups by the end of the experiment, except for Bacteroides_OTU1. Although its significantly higher relative abundance at 20 psu possibly reflected low diversity in the treatment, this OTU was also abundant at 15 psu, suggesting a high salinity tolerance. Although Bacteriodes is considered the dominant genus in the endogenous microbiota of freshwater fish (Gómez and Balcázar 2008), some strains prefer estuarine and riverine environments (Langenheder et al. 2003). Bacteroides species are anaerobic, bile-resistant, non-spore-forming, Gram-negative rods, and the genus accounts for approximately 25% of anaerobes in the healthy human colon (Salyers 1984; Wexler 2007). They perform a variety of functions in the human gut that may be related to carbohydrate metabolism, secretion of toxins, and interaction with the host immune system by controlling pathogens (Wexler 2007). The common presence of Bacteroides species under different osmotic conditions indicates their functional importance in the striped catfish gut. However, further studies are needed to determine their functional role, especially Bacteroides_OTU1, which has a high tolerance to the high salinity environment. Similarly, Verrucomicrobia, mostly represented by the Akkermansia genus, were significantly affected by the salinity gradient and exposure time. Previous studies have shown its abundance in the intestine of freshwater common carp, cichlid fish, and marine rabbit fish (Siganus guttatus) (Baldo et al. 2019; Chang et al. 2020; Le et al. 2020). In humans, salinity in faeces was associated with reduced Akkermansia abundance, independent of geography, age, and obesity status (Seck et al. 2019). Although the study of this bacterial genus in aquatic animals is limited, it is known as a mucin degrader in human and mice intestine and plays a key role in reducing systemic inflammation, improving glucose metabolism and the barrier function of the gut (Everard et al. 2013; Xu et al. 2020). Therefore, Akkermansia has been considered as a promising probiotic due to its ability to ameliorate host immune responses and metabolic functions (Gómez-Gallego et al. 2016; Zhang et al. 2019). In the present study, the relative abundance of Akkermansia decreased in fish intestines exposed to salinity, suggesting that the functional barrier of the intestine might be disrupted by osmotic conditions. It is clear that the increase of Proteobacteria with increasing osmotic conditions was mainly represented in the abundance of Vibrio spp. The observation is not surprising because Vibrio spp. abundance in the gut microbiota has been shown to increase in Nile tilapia (Oreochromis niloticus), Chinook salmon (Oncorhynchus tshawytscha), and Atlantic salmon after transfer to saline environments (Zhang et al. 2016; Lokesh et al. 2019; Zhao et al. 2020). Although Vibrio could be found in the intestinal tract of freshwater fish (Mishra et al. 2010; Givens et al. 2014), it was almost absent in freshwater conditions in our study, representing less than 0.002% of abundance. It may be assumed that Vibrio is not an inherent bacterium in striped catfish gut and that they initially entered the intestinal tract with the increased drinking of water in the saline environment. In one of the Vibrio species, Vibrio vulnificus, the optimal salinity for survival was shown to vary between 5 and 10 psu and this strain develops well up to 25 psu (Randa et al. 2004), which is consistent with the abundance of the bacteria in high salinity environments. The abundance of Campilobacterota, mainly represented by the genera Helicobacter and Sulfurospirillum, was significantly affected by the salinity gradient, the former being inhibited in saline conditions higher than 10 psu and the latter being stimulated in all salinities after two weeks of exposure. Helicobacter pylori was observed in faecal samples of farmed and wild tilapia from different water types and locations, while this bacterium was absent in all other tested fish species, even when collected from the same sources (Abdel-Moein et al. 2015). It is likely that the presence of this bacterium is more related to fish species-specific genetic traits than to environmental factors. Our results are in accordance with the findings of Gancz et al. (2008) who showed that a strain of H. pylori, known as a human pathogen, could not grow at a concentration of 15 g/L of sodium chloride. Sulfurospirillum, known as a free-living organism, produces hydrogen, which is an important electron supplier in an anoxic environment (Kruse et al. 2018). This genus also plays a key role in the metabolic cycle of sulphur and nitrogen (Ross et al. 2016). It is positively correlated with dissolved oxygen and negatively correlated with biological and chemical oxygen demands (Tyagi et al. 2021). Fish gut microbiota contain relatively few free-living environmental bacteria (Wong and Rawls 2012). Sulfurospirillum spp. was present in the freshwater treatment during our experiment with a relative abundance around 0.5%, indicating a relationship between the surrounding environment and the intestinal microbiota. Its increase in the intestine may be explained by the increased drinking of water by fish when transferred to a highly osmotic environment. However, the functions of Sulfurospirillum in the intestine of aquatic animals are still largely unknown.
Presumptive metabolic pathways changed under an osmotic environment
Global changes in putative functions in intestinal microbiota were examined using PICRUSt2. In the first phase of salinity exposure, beside carbohydrate and lipid metabolism, the 20 psu treatment was enriched in pathways related to the metabolism of a number of amino acids, such as arginine, proline, trytophane, tyrosine, and glutathione (Supplementary Table S3). Furthermore, the energy production from nitrogen, sulphur, and oxidative phosphorylation process were enhanced in fish subject to 20 psu (Supplementary Table S3). A study in intestinal Atlantic salmon supports these findings that the metabolism of carbohydrates, lipids, and amino acids is enriched during seawater adaptation (Dehler et al. 2017a). Bacteria can use amino acids and fatty acids as an energy source for their growth (Kazakov et al. 2009). They also restore glutathione, which bacteria use to protect their cells from external stressors such as low pH, cold, oxidation, and osmoregulation (Masip et al. 2006). These changes in metabolic pathways may be evidence that bacteria in the striped catfish gut tend to obtain their energy from different sources, such as carbohydrates, lipids, amino acids, and inorganic compounds, to maintain their metabolism during salinity adaptation. Interestingly, a proteomic study of striped catfish kidney also showed that amino acid metabolism pathways, including amino acid synthesis, branched amino acid degradation, and metabolism of 13 amino acids, had higher abundance in fish exposed to salt stress (Schmitz et al. 2017a, b, c). This could explain the interaction between host and gut microbiota in response to salt stress through changes in energy metabolism. Interestingly, the signaling pathways mainly related to stress responses, such as membrane transport, cell motility, and signal transduction (two component system) were significantly higher in abundance in fish exposed to 20 psu (D20) and 10 psu (D34) compared to the others (Table 2 and Supplementary Table S3). The two-component system is the main way microorganisms perceive and regulate their expression profile under environmental changes (Teschler et al. 2017; Xu et al. 2017). External signals are detected and transmitted by the sensor histidine kinase component of the system. This protein senses the signals and then autophosphorylates a conserved histidine residue of the phosphotransferase domain. Subsequently, the phosphoryl group is transferred to the response regulator, which may activate gene transcription in response to the specific signals from environmental stimuli (Fabret et al. 1999; Liu et al. 2019). The process may also upregulate gene expression encoding a bacterial secretion pathway (Gallique et al. 2017). Bacterial motility is an important ability to explore suitable conditions via regulation of energy demand and plays a key role in halotolerance (Mitchell and Kogure 2006; Chen et al. 2017). Bacterial motility and chemotaxis are influenced by changes in the osmotic environment in species such as Vibrio anguillarum, Aeromonas hydrophila, and Vibrio salmonicida, which showed differential motility with salinity gradient (Larsen et al. 2004; Karlsen et al. 2008; Jahid et al. 2015). Based on these results, we hypothesize that intestinal bacteria might respond to the saline environment through a two-component pathway and upregulated expression of genes involved in their motility to adapt to osmotic stress during long-term exposure. The most intensive responses were seen in the 20 psu condition in the first phase of exposure to salinity, but 10 psu in the longer period.
Host-microbiota interactions under osmotic stress
The interaction between host and microbiota plays an important role in many aspects of fish health, such as metabolic activity and immune status (Pérez et al. 2010). In the present study, the interaction was investigated via PRR changes and the presence of dominant bacteria in the intestine. The mucosal immune system discriminates between pathogenic and commensal bacteria via the PRR pathway, which recognizes bacterial antigens and activates signalling cascades that regulate the immune response (Fasano and Shea-Donohue 2005). In the present study, a highly saline environment of 20 psu resulted in a significant decrease in the expression of tlr1 and tlr2, which followed the downregulation of inflammatory genes such as il1β, tnfα, il10, and tgfβ at D20. Schmitz et al. (2017a) found similar results in striped catfish, in which the expression of toll-like receptors was decreased in the kidneys under conditions of 20 psu. Interestingly, a previous study found that Vibrio infection increased the expression of inflammatory genes (Wu et al. 2015). In addition, the presence of Akkermansia in freshwater has been shown to suppress inflammatory responses through increasing the anti-inflammatory cytokine il10 to prevent the activation of pro-inflammatory cytokines like il12 (Demirci et al. 2019). However, the saline treatments in the current study showed a high abundance of the genus Vibrio and low abundance of the genus Akkermansia, but their inflammatory responses were unchanged and even decreased at 20 psu. A plausible explanation could be the increase in the expression of alkaline phosphatase in the intestine (Bates et al. 2007). This plays a crucial role in the detoxification of lipopolysaccharide and prevention of intestinal inflammation, and enhances mucosal tolerance to the resident microbiota (Bates et al. 2007). However, the effect of hyperosmotic stress diminished over time, and responses tended to recover at D34, except for il1β and il10. These results suggest that striped catfish may regulate excessive inflammation due to invasive bacteria by responding to high lipopolysaccharide stress. However, due to the complex intestinal microbiota, the exact mechanism remains unknown.
In conclusion, this study revealed microbiota and gene expression alterations in striped catfish intestine under different saline conditions. A detailed profile of the intestinal microbiota in striped catfish was elucidated using the 16S rRNA sequencing method. The results were strongly concordant with those of other freshwater fish, with a predominance of the phyla Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia. Regarding host responses to salinity, the intestine contributed to the regulation of the osmoregulatory and adaptive stress processes; however, this may be insufficient at 20 psu resulting in intestinal dysbiosis as well as high mortality. The osmoregulation was more intensive while the stress indicators returned to normal after a 2-week exposure to salinity. These osmotic changes in the intestine significantly reduced the diversity of the microbiota. However, extended salt exposure destabilized the intestinal microbiota, leading to an increase in phylogenetic diversity with the salt gradient. Despite the predominance of Bacteroides OTUs in all treatments, their abundances were mostly significantly decreased after extended exposure to salinity, except Bacteroides_OTU1. The roles of this OTU need to be clarified due to its high tolerance to salinity. Akkermansia, known to be a probiotic, had a lower abundance at the higher salinities and longer exposures. Sulfurospirillium and Vibrio were the most abundant in high salinity treatments at the end of the experiment, indicating a strong correlation between evironmental conditions and fish intestinal bacterial communities. Regarding predicted metabolic pathways, when treated with 20 psu the fish obtained higher energy from various sources that might be needed to cope with osmotic stress. Additionally, the intestinal microbiota at 20 psu was most sensitive to environmental changes in the first phase of exposure to salinity, while this was 10 psu for the extended salt exposure. The salinity gradient of up to 10 psu did not negatively affect local immune responses. However, the exact mechanism needs further research, especially the interaction with the microbiota.
Data availability
The datasets supporting the conclusions of the article are included within the article and its supplementary materials. The sequencing raw data was deposited in the NCBI Sequence Read Archive database (accession: PRJNA759359).
References
Abdel-Moein KA, Saeed H, Samir A (2015) Novel detection of Helicobacter pylori in fish: a possible public health concern. Acta Trop 152:141–144. https://doi.org/10.1016/j.actatropica.2015.09.005
Aoki M, Kaneko T, Katoh F, Hasegawa S, Tsutsui N, Aida K (2003) Intestinal water absorption through aquaporin 1 expressed in the apical membrane of mucosal epithelial cells in seawater-adapted Japanese eel. J Exp Biol 206:3495–3505. https://doi.org/10.1242/jeb.00579
Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N (2015) Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029
Baldo L, Riera JL, Salzburger W, Barluenga M (2019) Phylogeography and ecological niche shape the cichlid fish gut microbiota in Central American and African Lakes. Front Microbiol 10:2372
Bates JM, Akerlund J, Mittge E, Guillemin K (2007) Intestinal Alkaline Phosphatase Detoxifies Lipopolysaccharide and Prevents Inflammation in Zebrafish in Response to the Gut Microbiota. Cell Host Microbe 2:371–382. https://doi.org/10.1016/j.chom.2007.10.010
Bledsoe JW, Peterson BC, Swanson KS, Small BC (2016) Ontogenetic characterization of the intestinal microbiota of channel catfish through 16S rRNA gene sequencing reveals insights on temporal shifts and the influence of environmental microbes. PLoS ONE 11:e0166379–e0166379. https://doi.org/10.1371/journal.pone.0166379
Chang X, Wang X, Feng J, Su X, Liang J, Li H, Zhang J (2020) Impact of chronic exposure to trichlorfon on intestinal barrier, oxidative stress, inflammatory response and intestinal microbiome in common carp (Cyprinus carpio L.). Environ Pollut 259:113846. https://doi.org/10.1016/j.envpol.2019.113846
Chen Y-H, Lu C-W, Shyu Y-T, Lin S-S (2017) Revealing the saline adaptation strategies of the halophilic bacterium Halomonas beimenensis through high-throughput omics and transposon mutagenesis approaches. Sci Rep 7:13037. https://doi.org/10.1038/s41598-017-13450-9
De Wachter B, Scholliers A, Blust R (1998) Semiquantitative immunoblot detection of 70 kDa stress proteins in the carp Cyprinus carpio. Bull Environ Contam Toxicol 60:37–44. https://doi.org/10.1007/s001289900588
Deane EE, Woo NYS (2004) Differential gene expression associated with euryhalinity in sea bream (Sparus sarba). Am J Physiol Integr Comp Physiol 287:R1054–R1063
Deane EE, Kelly SP, Luk JCY, Woo NYS (2002) Chronic salinity adaptation modulates hepatic heat shock protein and insulin-like growth factor I expression in black sea bream. Mar Biotechnol 4:193–205
Deane E, Luk J, Woo NYS (2011) Aquaporin 1a expression in gill, intestine, and kidney of the euryhaline silver sea bream. Front Physiol 2:39
Dehler CE, Secombes CJ, Martin SAM (2017) Seawater transfer alters the intestinal microbiota profiles of Atlantic salmon (Salmo salar L.). Sci Rep 7:13877. https://doi.org/10.1038/s41598-017-13249-8
Dehler CE, Secombes CJ, Martin SAM (2017) Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture 467:149–157. https://doi.org/10.1016/j.aquaculture.2016.07.017
Demirci M, Tokman HB, Uysal HK, Demiryas S, Karakullukcu A, Saribas S, Cokugras H, Kocazeybek BS (2019) Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol Immunopathol (madr) 47:365–371. https://doi.org/10.1016/j.aller.2018.12.009
Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. https://doi.org/10.1038/s41587-020-0548-6
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD (2013) Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci 110:9066 LP – 9071. https://doi.org/10.1073/pnas.1219451110
Fabret C, Feher VA, Hoch JA (1999) Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol 181:1975–1983. https://doi.org/10.1128/JB.181.7.1975-1983.1999
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. https://doi.org/10.1016/0006-3207(92)91201-3
FAO (2020) GLOBEFISH Highlights January 2020 issue, with Jan. – Sep. 2019 Statistics – A quarterly update on world seafood markets. Globefish Highlights no. 1–2020. Rome. https://doi.org/10.4060/ca7968en
Fasano A, Shea-Donohue T (2005) Mechanisms of disease: the role of intestinal barrier function in the pathogenesis of gastrointestinal autoimmune diseases. Nat Clin Pract Gastroenterol Hepatol 2:416–422. https://doi.org/10.1038/ncpgasthep0259
Gallique M, Bouteiller M, Merieau A (2017) The type VI secretion system: a dynamic system for bacterial communication? Front Microbiol 8:1454
Gancz H, Jones KR, Merrell DS (2008) Sodium chloride affects Helicobacter pylori growth and gene expression. J Bacteriol 190:4100–4105. https://doi.org/10.1128/JB.01728-07
Gioacchini G, Rossi G, Carnevali O (2017) Host-probiotic interaction: new insight into the role of the endocannabinoid system by in vivo and ex vivo approaches. Sci Rep 7:1261. https://doi.org/10.1038/s41598-017-01322-1
Givens CE, Bowers JC, DePaola A, Hollibaugh JT, Jones JL (2014) Occurrence and distribution of Vibrio vulnificus and Vibrio parahaemolyticus – potential roles for fish, oyster, sediment and water. Lett Appl Microbiol 58:503–510. https://doi.org/10.1111/lam.12226
Gómez GD, Balcázar JL (2008) A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol Med Microbiol 52:145–154. https://doi.org/10.1111/j.1574-695X.2007.00343.x
Gómez-Gallego C, Pohl S, Salminen S, De Vos WM, Kneifel W (2016) Akkermansia muciniphila: a novel functional microbe with probiotic properties. Benef Microbes 7:571–584
Gregório SF, Carvalho ESM, Encarnação S, Wilson JM, Power DM, Canário AVM, Fuentes J (2013) Adaptation to different salinities exposes functional specialization in the intestine of the sea bream (Sparus aurata L.). J Exp Biol 216:470–479. https://doi.org/10.1242/jeb.073742
Grosell M (2006) Intestinal anion exchange in marine fish osmoregulation. J Exp Biol 209:2813–2827. https://doi.org/10.1242/jeb.02345
Grosell M (2010) The role of the gastrointestinal tract in salt and water balance. In: Grosell M (ed) Fish physiology. Elsevier, pp 135–164
Hall M, Beiko RG (2018) 16S rRNA gene analysis with QIIME2. In: Beiko RG, Hsiao W, Parkinson J (eds) Microbiome analysis. Springer New York, pp 113–129. https://doi.org/10.1007/978-1-4939-8728-3_8
Herlemann DPR, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. https://doi.org/10.1038/ismej.2011.41
Hightower LE (1991) Heat shock, stress proteins, chaperones, and proteotoxicity. Cell 66:191–197
Hörmannsperger G, Haller D (2010) Molecular crosstalk of probiotic bacteria with the intestinal immune system: clinical relevance in the context of inflammatory bowel disease. Int J Med Microbiol 300:63–73. https://doi.org/10.1016/j.ijmm.2009.08.006
Humphreys C (2020) Intestinal Permeability. In: Pizzorno JE, Murray MT (eds) Textbook of Natural Medicine, 5th edn. Churchill Livingstone, St. Louis (MO), pp 166–177
Jahid IK, Mizan MFR, Ha AJ, Ha S-D (2015) Effect of salinity and incubation time of planktonic cells on biofilm formation, motility, exoprotease production, and quorum sensing of Aeromonas hydrophila. Food Microbiol 49:142–151. https://doi.org/10.1016/j.fm.2015.01.016
Karlsen C, Paulsen SM, Tunsjø HS, Krinner S, Sørum H, Haugen P, Willassen N-P (2008) Motility and flagellin gene expression in the fish pathogen Vibrio salmonicida: Effects of salinity and temperature. Microb Pathog 45:258–264. https://doi.org/10.1016/j.micpath.2008.06.002
Kazakov AE, Rodionov DA, Alm E, Arkin AP, Dubchak I, Gelfand MS (2009) Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J Bacteriol 191:52–64. https://doi.org/10.1128/JB.01175-08
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1–e1. https://doi.org/10.1093/nar/gks808
Kruse S, Goris T, Westermann M, Adrian L, Diekert G (2018) Hydrogen production by Sulfurospirillum species enables syntrophic interactions of Epsilonproteobacteria. Nat Commun 9:4872. https://doi.org/10.1038/s41467-018-07342-3
Langenheder S, Kisand V, Wikner J, Tranvik LJ (2003) Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol Ecol 45:189–202. https://doi.org/10.1016/S0168-6496(03)00149-1
Larsen MH, Blackburn N, Larsen JL, Olsen JE (2004) Influences of temperature, salinity and starvation on the motility and chemotactic response of Vibrio anguillarum. Microbiology 150:1283–1290. https://doi.org/10.1099/mic.0.26379-0
Larsen PF, Nielsen EE, Koed A, Thomsen DS, Olsvik PA, Loeschcke V (2008) Interpopulation differences in expression of candidate genes for salinity tolerance in winter migrating anadromous brown trout (Salmo trutta L.). BMC Genet 9:1–9
Laverty G, Skadhauge E (2012) Adaptation of teleosts to very high salinity. Comp Biochem Physiol Part A Mol Integr Physiol 163:1–6. https://doi.org/10.1016/j.cbpa.2012.05.203
Le D, Nguyen P, Nguyen D, Dierckens K, Boon N, Lacoere T, Kerckhof F-M, De Vrieze J, Vadstein O, Bossier P (2020) Gut microbiota of migrating wild rabbit fish (Siganus guttatus) larvae have low spatial and temporal variability. Microb Ecol 79:539–551. https://doi.org/10.1007/s00248-019-01436-1
Li J, Dai Z, Jana D, Callaway DJE, Bu Z (2005) Ezrin controls the macromolecular complexes formed between an adapter protein Na+/H+ exchanger regulatory factor and the cystic fibrosis transmembrane conductance regulator*. J Biol Chem 280:37634–37643. https://doi.org/10.1074/jbc.M502305200
Li Z, Lui EY, Wilson JM, Ip YK, Lin Q, Lam TJ, Lam SH (2014) Expression of key ion transporters in the gill and esophageal-gastrointestinal tract of euryhaline Mozambique tilapia Oreochromis mossambicus acclimated to fresh water, seawater and hypersaline water. PLoS One 9:e87591
Lin YM, Chen CN, Lee TH (2003) The expression of gill Na, K-ATPase in milkfish, Chanos chanos, acclimated to seawater, brackish water and fresh water. Comp Biochem Physiol Part A Mol Integr Physiol 135:489–497
Lind U, Alm Rosenblad M, Wrange A-L, Sundell KS, Jonsson PR, André C, Havenhand J, Blomberg A (2013) Molecular characterization of the α-subunit of Na+/K+ ATPase from the euryhaline barnacle Balanus improvisus reveals multiple genes and differential expression of alternative splice variants. PLoS ONE 8:e77069–e77069. https://doi.org/10.1371/journal.pone.0077069
Liu C, Sun D, Zhu J, Liu W (2019) Two-Component Signal Transduction Systems: A Major Strategy for Connecting Input Stimuli to Biofilm Formation. Front Microbiol 9:3279
Lokesh J, Kiron V, Sipkema D, Fernandes JMO, Moum T (2019) Succession of embryonic and the intestinal bacterial communities of Atlantic salmon (Salmo salar) reveals stage-specific microbial signatures. Microbiologyopen 8:e00672. https://doi.org/10.1002/mbo3.672
Louca S, Doebeli M (2018) Efficient comparative phylogenetics on large trees. Bioinformatics 34:1053–1055
Madsen SS, Bujak J, Tipsmark CK (2014) Aquaporin expression in the Japanese medaka (Oryzias latipes) in freshwater and seawater: challenging the paradigm of intestinal water transport? J Exp Biol 217:3108–3121. https://doi.org/10.1242/jeb.105098
Malakpour Kolbadinezhad S, Coimbra J, Wilson JM (2018) Osmoregulation in the Plotosidae catfish: Role of the salt secreting dendritic organ. Front Physiol 9:761
Marshall WS, Grosell M (2005) Ion transport, osmoregulation, and acid-base balance. In: Evans DH, Clairborne JB (eds) The physiology of fishes. CRC Press, Boca Raton, pp 177–230
Marshall WS, Singer TD (2002) Cystic fibrosis transmembrane conductance regulator in teleost fish. Biochim Biophys Acta - Biomembr 1566:16–27. https://doi.org/10.1016/S0005-2736(02)00584-9
Martin SAM, Dehler CE, Król E (2016) Transcriptomic responses in the fish intestine. Dev Comp Immunol 64:103–117. https://doi.org/10.1016/j.dci.2016.03.014
Martinez A-S, Cutler CP, Wilson GD, Phillips C, Hazon N, Cramb G (2005) Regulation of expression of two aquaporin homologs in the intestine of the European eel: effects of seawater acclimation and cortisol treatment. Am J Physiol Integr Comp Physiol 288:R1733–R1743. https://doi.org/10.1152/ajpregu.00747.2004
Masip L, Veeravalli K, Georgiou G (2006) The many faces of glutathione in bacteria. Antioxid Redox Signal 8:753–762. https://doi.org/10.1089/ars.2006.8.753
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217
Mishra P, Samanta M, Mohanty S, Maiti NK (2010) Characterization of Vibrio species isolated from freshwater fishes by ribotyping. Indian J Microbiol 50:101–103. https://doi.org/10.1007/s12088-010-0010-4
Mitchell JG, Kogure K (2006) Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol Ecol 55:3–16
Mundy PC, Jeffries KM, Fangue NA, Connon RE (2020) Differential regulation of select osmoregulatory genes and Na+/K+-ATPase paralogs may contribute to population differences in salinity tolerance in a semi-anadromous fish. Comp Biochem Physiol Part A Mol Integr Physiol 240:110584. https://doi.org/10.1016/j.cbpa.2019.110584
Nguyen PTH, Do HTT, Mather PB, Hurwood DA (2014) Experimental assessment of the effects of sublethal salinities on growth performance and stress in cultured tra catfish (Pangasianodon hypophthalmus). Fish Physiol Biochem 40:1839–1848. https://doi.org/10.1007/s10695-014-9972-1
Pan F, Zarate JM, Tremblay GC, Bradley TM (2000) Cloning and characterization of salmon hsp90 cDNA: Upregulation by thermal and hyperosmotic stress. J Exp Zool 287:199–212. https://doi.org/10.1002/1097-010X(20000801)287:3%3c199::AID-JEZ2%3e3.0.CO;2-3
Pérez T, Balcázar JL, Ruiz-Zarzuela I, Halaihel N, Vendrell D, de Blas I, Múzquiz JL (2010) Host–microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol 3:355
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. https://doi.org/10.1093/nar/29.9.e45
Picchietti S, Fausto AM, Randelli E, Carnevali O, Taddei AR, Buonocore F, Scapigliati G, Abelli L (2009) Early treatment with Lactobacillus delbrueckii strain induces an increase in intestinal T-cells and granulocytes and modulates immune-related genes of larval Dicentrarchus labrax (L.). Fish Shellfish Immunol 26:368–376. https://doi.org/10.1016/j.fsi.2008.10.008
Randa MA, Polz MF, Lim E (2004) Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl Environ Microbiol 70:5469–5476. https://doi.org/10.1128/AEM.70.9.5469-5476.2004
Rawls JF, Samuel BS, Gordon JI (2004) Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A 101:4596–4601. https://doi.org/10.1073/pnas.0400706101
Ray AK, Ringø E (2014) The gastrointestinal tract of fish. In: Merrifield D, Ringø E (eds) Aquaculture nutrition: gut health, probiotics and prebiotics. Wiley-Blackwell Publishing, Oxford, UK, pp 1–13
Ringø E, Olsen RE, Mayhew TM, Myklebust R (2003) Electron microscopy of the intestinal microflora of fish. Aquaculture 227:395–415. https://doi.org/10.1016/j.aquaculture.2003.05.001
Roberts RJ, Agius C, Saliba C, Bossier P, Sung YY (2010) Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis 33:789–801. https://doi.org/10.1111/j.1365-2761.2010.01183.x
Ronkin D, Seroussi E, Nitzan T, Doron-Faigenboim A, Cnaani A (2015) Intestinal transcriptome analysis revealed differential salinity adaptation between two tilapiine species. Comp Biochem Physiol Part D Genomics Proteomics 13:35–43. https://doi.org/10.1016/j.cbd.2015.01.003
Ross DE, Marshall CW, May HD, Norman RS (2016) Comparative genomic analysis of Sulfurospirillum cavolei MES reconstructed from the metagenome of an electrosynthetic microbiome. PLoS ONE 11:e0151214–e0151214. https://doi.org/10.1371/journal.pone.0151214
Salyers AA (1984) Bacteroides of the human lower intestinal tract. Annu Rev Microbiol 38:293–313. https://doi.org/10.1146/annurev.mi.38.100184.001453
Schmidt VT, Smith KF, Melvin DW, Amaral-Zettler LA (2015) Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol Ecol 24:2537–2550. https://doi.org/10.1111/mec.13177
Schmitz M, Douxfils J, Mandiki SNM, Morana C, Baekelandt S, Kestemont P (2016) Chronic hyperosmotic stress interferes with immune homeostasis in striped catfish (Pangasianodon hypophthalmus, S.) and leads to excessive inflammatory response during bacterial infection. Fish Shellfish Immunol 55:550–558. https://doi.org/10.1016/j.fsi.2016.06.031
Schmitz M, Mandiki SNM, Douxfils J, Ziv T, Admon A, Kestemont P (2016) Synergic stress in striped catfish (Pangasianodon hypophthalmus, S.) exposed to chronic salinity and bacterial infection: Effects on kidney protein expression profile. J Proteomics 142:91–101. https://doi.org/10.1016/j.jprot.2016.04.046
Schmitz M, Baekelandt S, Bequet S, Kestemont P (2017) Chronic hyperosmotic stress inhibits renal Toll-Like Receptors expression in striped catfish (Pangasianodon hypophthalmus, Sauvage) exposed or not to bacterial infection. Dev Comp Immunol 73:139–143. https://doi.org/10.1016/j.dci.2017.03.020
Schmitz M, Baekelandt S, Tran Thi LK, Mandiki SNM, Douxfils J, Nguyen TQ, Do Thi Thanh H, Kestemont P (2017) Osmoregulatory and immunological status of the pond-raised striped catfish (Pangasianodon hypophthalmus S.) as affected by seasonal runoff and salinity changes in the Mekong Delta. Vietnam Fish Physiol Biochem 43:39–49. https://doi.org/10.1007/s10695-016-0266-7
Schmitz M, Ziv T, Admon A, Baekelandt S, Mandiki SNM, L’Hoir M, Kestemont P (2017) Salinity stress, enhancing basal and induced immune responses in striped catfish Pangasianodon hypophthalmus (Sauvage). J Proteomics 167:12–24. https://doi.org/10.1016/j.jprot.2017.08.005
Scott GR, Baker DW, Schulte PM, Wood CM (2008) Physiological and molecular mechanisms of osmoregulatory plasticity in killifish after seawater transfer. J Exp Biol 211:2450–2459. https://doi.org/10.1242/jeb.017947
Seck EH, Senghor B, Merhej V, Bachar D, Cadoret F, Robert C, Azhar EI, Yasir M, Bibi F, Jiman-Fatani AA, Konate DS, Musso D, Doumbo O, Sokhna C, Levasseur A, Lagier JC, Khelaifia S, Million M, Raoult D (2019) Salt in stools is associated with obesity, gut halophilic microbiota and Akkermansia muciniphila depletion in humans. Int J Obes 43:862–871. https://doi.org/10.1038/s41366-018-0201-3
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60
Sutriana A, Hashim R, Akter MN, Nor SAM (2018) Galactooligosaccharide and a combination of yeast and β-glucan supplements enhance growth and improve intestinal condition in striped catfish Pangasianodon hypophthalmus fed soybean meal diets. Fish Sci 84:523–533. https://doi.org/10.1007/s12562-018-1195-4
Takei Y (2021) The digestive tract as an essential organ for water acquisition in marine teleosts: lessons from euryhaline eels. Zool Lett 7:10. https://doi.org/10.1186/s40851-021-00175-x
Takei Y, Hwang P-P (2016) Homeostatic responses to osmotic stress. In: Schreck CB, Tort L, Farrell AP, Brauner CJ (eds) Fish Physiology. Elsevier, pp 207–249
Team RC (2013) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: http://www.R-project.org/
Teschler JK, Cheng AT, Yildiz FH (2017) The two-component signal transduction system VxrAB positively regulates Vibrio cholerae biofilm formation. J Bacteriol 199:e00139-e217
Tine M, Bonhomme F, McKenzie DJ, Durand J-D (2010) Differential expression of the heat shock protein Hsp70 in natural populations of the tilapia, Sarotherodon melanotheron, acclimatised to a range of environmental salinities. BMC Ecol 10:11. https://doi.org/10.1186/1472-6785-10-11
Tlaskalova-Hogenova H, Tuckova L, Mestecky J, Kolinska J, Rossmann P, Stepankova R, Kozakova H, Hudcovic T, Hrncir T, Frolova L, Kverka M (2005) Interaction of mucosal microbiota with the innate immune system. Scand J Immunol 62:106–113. https://doi.org/10.1111/j.1365-3083.2005.01618.x
Tyagi I, Tyagi K, Bhutiani R, Chandra K, Kumar V (2021) Bacterial diversity assessment of world’s largest sewage-fed fish farms with special reference to water quality: a Ramsar site. Environ Sci Pollut Res 28:42372–42386. https://doi.org/10.1007/s11356-021-13756-2
Usher ML, Talbot C, Eddy FB (1990) Effects of transfer to seawater on digestion and gut function in Atlantic salmon smolts (Salmo salar L.). Aquaculture 90:85–96. https://doi.org/10.1016/0044-8486(90)90285-U
Wexler HM (2007) Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20:593–621. https://doi.org/10.1128/CMR.00008-07
Whittamore JM (2012) Osmoregulation and epithelial water transport: lessons from the intestine of marine teleost fish. J Comp Physiol B 182:1–39. https://doi.org/10.1007/s00360-011-0601-3
Winklhofer KF, Tatzelt J, Haass C (2008) The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J 27:336–349. https://doi.org/10.1038/sj.emboj.7601930
Wong S, Rawls JF (2012) Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol Ecol 21:3100–3102. https://doi.org/10.1111/j.1365-294X.2012.05646.x
Wu J, Shi Y-H, Zhang X-H, Li C-H, Li M-Y, Chen J (2015) Molecular characterization of an IL-1β gene from the large yellow croaker (Larimichthys crocea) and its effect on fish defense against Vibrio alginolyticus infection. Dong wu xue yan jiu = Zool Res 36:133–141
Xu T, Wu Y, Lin Z, Bertram R, Götz F, Zhang Y, Qu D (2017) Identification of genes controlled by the essential YycFG two-component system reveals a role for biofilm modulation in Staphylococcus epidermidis. Front Microbiol 8:724
Xu Y, Wang N, Tan H-Y, Li S, Zhang C, Feng Y (2020) Function of Akkermansia muciniphila in obesity: interactions with lipid metabolism, immune response and gut systems. Front Microbiol 11:219
Yang W-K, Hseu J-R, Tang C-H, Chung M-J, Wu S-M, Lee T-H (2009) Na+/K+-ATPase expression in gills of the euryhaline sailfin molly, Poecilia latipinna, is altered in response to salinity challenge. J Exp Mar Bio Ecol 375:41–50
Yang W-K, Hsu A-D, Kang C-K, Lai IP, Liao P-S, Lee T-H (2018) Intestinal FXYD12 and sodium-potassium ATPase: A comparative study on two euryhaline medakas in response to salinity changes. PLoS One 13:e0201252
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. https://doi.org/10.1186/1471-2105-13-134
Zhang M, Sun Y, Liu Y, Qiao F, Chen L, Liu W-T, Du Z, Li E (2016) Response of gut microbiota to salinity change in two euryhaline aquatic animals with reverse salinity preference. Aquaculture 454:72–80. https://doi.org/10.1016/j.aquaculture.2015.12.014
Zhang T, Li Q, Cheng L, Buch H, Zhang F (2019) Akkermansia muciniphila is a promising probiotic. Microb Biotechnol 12:1109–1125. https://doi.org/10.1111/1751-7915.13410
Zhao R, Symonds JE, Walker SP, Steiner K, Carter CG, Bowman JP, Nowak BF (2020) Salinity and fish age affect the gut microbiota of farmed Chinook salmon (Oncorhynchus tshawytscha). Aquaculture 528:735539. https://doi.org/10.1016/j.aquaculture.2020.735539
Acknowledgements
The authors thank to the Académie de Recherche et d’Enseignement Supérieur (ARES) and the General Directorate and Development for Cooperation (DGD) in Belgium through the Research for Development Project (RDP) “Towards a sustainable pangasius breeding, a selection approach (PANGAGEN).”
Funding
The research was supported by the Académie de Recherche et d’Enseignement Supérieur (ARES) and the General Directorate and Development for Cooperation (DGD) in Belgium.
Author information
Authors and Affiliations
Contributions
PK, FF, and NTP designed and coordinated the project. PK, DQH, and BTBH conceived of the experiment. DQH carried out the experiment and analysed the data. BTBH, DTTH, DTY, PTL, BMT, DMH, and VNS supported to carry out and sample the experiment. LJ and MMG guided and advised for bioinformatics analysis. DQH, BTBH, JL, MMG, and PK prepared the manuscript. All the authors contributed to the article and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hieu, D.Q., Hang, B.T.B., Lokesh, J. et al. Salinity significantly affects intestinal microbiota and gene expression in striped catfish juveniles. Appl Microbiol Biotechnol 106, 3245–3264 (2022). https://doi.org/10.1007/s00253-022-11895-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11895-1