Abstract
Glycosylation is an effective way to improve the water solubility of natural products. In this work, a novel glycosyltransferase gene (BbGT) was discovered from Beauveria bassiana ATCC 7159 and heterologously expressed in Escherichia coli. The purified enzyme was functionally characterized through in vitro enzymatic reactions as a UDP-glucosyltransferase, converting quercetin to five monoglucosylated and one diglucosylated products. The optimal pH and temperature for BbGT are 35 ℃ and 8.0, respectively. The activity of BbGT was stimulated by Ca2+, Mg2+, and Mn2+, but inhibited by Zn2+. BbGT enzyme is flexible and can glycosylate a variety of substrates such as curcumin, resveratrol, and zearalenone. The enzyme was also expressed in other microbial hosts including Saccharomyces cerevisiae, Pseudomonas putida, and Pichia pastoris. Interestingly, the major glycosylation product of quercetin in E. coli, P. putida, and P. pastoris was quercetin-7-O-β-d-glucoside, while the enzyme dominantly produced quercetin-3-O-β-d-glucoside in S. cerevisiae. The BbGT-harboring E. coli and S. cerevisiae strains were used as whole-cell biocatalysts to specifically produce the two valuable quercetin glucosides, respectively. The titer of quercetin-7-O-β-d-glucosides was 0.34 ± 0.02 mM from 0.83 mM quercetin in 24 h by BbGT-harboring E. coli. The yield of quercetin-3-O-β-d-glucoside was 0.22 ± 0.02 mM from 0.41 mM quercetin in 12 h by BbGT-harboring S. cerevisiae. This work thus provides an efficient way to produce two valuable quercetin glucosides through the expression of a versatile glucosyltransferase in different hosts.
Key points
• A highly versatile glucosyltransferase was identified from B. bassiana ATCC 7159.
• BbGT converts quercetin to five mono- and one di-glucosylated derivatives in vitro.
• Different quercetin glucosides were produced by BbGT in E. coli and S. cerevisiae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glycosylation is a common reaction that plays a vital role in the formation of natural glucosides with various applications (Vogt and Jones 2000). The reaction is catalyzed by dedicated glycosyltransferases (GTs). GTs transfer specific sugar moieties from activated donor molecules, mainly nucleoside diphosphate sugars (NDP-sugars) (Lairson et al. 2008), to various acceptor molecules, including many plant secondary metabolites (Yonekura‐Sakakibara and Hanada 2011). Sugar moieties play an irreplaceable role in many life processes such as cell differentiation, development, immunity, aging, carcinogenesis, and information transmission (Jiang et al. 2008). Moreover, the bioavailability, water solubility, and stability of natural products can be enhanced through glycosylation, which may further modulate their biological activities (Dou et al. 2019; Mfonku et al. 2020; Yang et al. 2018). For example, the intracellular anti-oxidative activity of apigenin was improved by C-glycosylation (Wen et al. 2017); the stability of quercetin-3-O-rutinoside (rutin) in aqueous solution at 100 °C is higher than its aglycon quercetin and its degradation rate in phosphate buffer with Fe2+ and Cu2+ is slower (Ikeda and Taguchi 2010; Makris and Rossiter 2000). Therefore, glycosylation is a commonly used tool for developing new therapeutic agents.
Many natural products such as quercetin, curcumin, and resveratrol are commonly used as dietary supplements due to their health benefits. However, these plant polyphenols often have low water solubility and poor bioavailability. For example, quercetin is a representative antioxidant flavonoid that exists in green tea, fruits, and leaf vegetables. It exerts promising pharmacological properties, such as anticancer, anti-inflammatory, antiviral, and antihypertensive properties (Cai et al. 2013; Ma et al. 2013). However, poor water solubility hinders its health benefits on humans (Li et al. 2004). Therefore, it is imperative for researchers to discover an effective approach to improve its aqueous solubility. Quercimeritrin (quercetin-7-O-β-d-glucoside) is a natural quercetin glucoside reportedly having in vitro anti-inflammatory activity (Anuradha and Sukumar 2013). It possesses strong antioxidant activity, with an oxygen radical absorbance capacity (ORAC) value of 18 ± 4 μmol Trolox® equivalents (TE)/μmol (Legault et al. 2011). Another glucoside of quercetin, isoquercitrin (quercetin-3-O-β-d-glucoside), has significant pharmacological activities against cancer, oxidative stress, cardiovascular disorders, diabetes, and allergic reactions (Chen et al. 2015; Valentová et al. 2014). Isoquercitrin showed higher inhibitory effect than quercetin in an ex vivo angiogenesis assay (Matsubara et al. 2004). Paulke et al. found that isoquercitrin has better bioavailability than quercetin (Paulke et al. 2012). Specifically, isoquercitrin gavage provides higher quercetin metabolite levels in both tissues (double to five-fold) and plasma (double to three-fold), compared to the quercetin gavage (Day et al. 2001).
While chemical synthesis of glycosides often requires extensive group protection and deprotection and use of toxic reagents, biological glycosylation through GTs represents a green and selective way to prepare desired glycosides (Cheng et al. 2019). Many researchers have worked on exploring various GTs from microorganisms or plants to increase the water solubility and bioavailability of bioactive compounds (Méndez and Salas 2001). Ko et al. found a GT named BcGT-1 from Bacillus cereus which can glycosylate different flavonoids such as quercetin, apigenin, and genistein (Hyung Ko et al. 2006). Zhao et al. isolated a flavonoid GT CsUGT73A20 from a tea plant Camellia sinensis, which exhibits a broad substrate tolerance toward multiple flavonoids (Zhao et al. 2017). Therefore, nature provides a variety of GTs that can be used to prepare glycosides of natural products. However, like other enzymes, GTs usually have high selectivity and strict substrate specificity that often only function on a particular substrate and its analogs, which limit their use as versatile biocatalysts in enzymatic preparation of desired glycosides. As such, discovery of flexible GTs would add new tools for the synthesis of various glycosides.
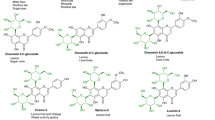
Beauveria bassiana ATCC 7159 is a filamentous fungus that has been extensively used for biotransformation of bioactive molecules (Grogan and Holland 2000). Different reactions can be catalyzed by this strain, such as oxidation, reduction, hydrolysis, methylation, hydroxylation, and glycosylation. We have previously used B. bassiana ATCC 7159 to prepare various glucosides from diverse molecules such as anthraquinones and curcuminoids (Zeng et al. 2010; Zhan and Gunatilaka 2006; Zhan and Leslie Gunatilaka 2006). The strain can selectively introduce a glucose or 4′-O-methylglucose moiety to a phenolic hydroxyl group in the substrates. In this work, we cloned the dedicated GT (BbGT) from B. bassiana ATCC 7159 and expressed it in four heterologous hosts. BbGT was functionally characterized through in vitro reactions. It is highly versatile and can convert quercetin into five monoglucosylated and one diglucosylated products (Fig. 1). Furthermore, it can accept a variety of substrates, thus representing a useful enzymatic tool for glycosylation. Interestingly, when expressed in different hosts, the enzyme generated different major glycosylation products of quercetin, which provides a convenient way to prepare different quercetin glucosides using BbGT.
Materıals and methods
General equipment and experimental materials
Products were analyzed and purified on an Agilent 1200 HPLC instrument with an Agilent Eclipse Plus-C18 column (5 μm, 250 mm × 4.6 mm). The samples were eluted with methanol–water (20:80 to 80:20, v/v, containing 0.1% formic acid) at a flow rate of 1 mL/min for 30 min. ESI–MS spectra were obtained on an Agilent 6130 single quadrupole LC–MS in the negative mode. 1D and 2D NMR spectra were recorded in deuterated dimethyl sulfoxide (DMSO-d6) on a Bruker Avance III HD Ascend-500 NMR instrument (500 MHz for 1H NMR and 125 MHz for 13C NMR) in the Department of Chemistry and Biochemistry, Utah State University. The chemical shift (δ) values are described in parts per million (ppm) and coupling constants (J values) are reported in hertz (Hz). MicroPulser™ electroporation apparatus (Bio-Rad, USA) was used for electroporation.
Phusion® High-Fidelity DNA polymerase, restriction enzymes, and T4 DNA ligase were purchased from New England Biolabs. Resibufogenin, tetracycline, quercetin, curcumin, resveratrol, and zearalenone were purchased from Sigma-Aldrich (USA). Indigoidine was produced and purified in our lab (Xu et al. 2015; Yu et al. 2013). HisPur™ Ni–NTA resin and Luria–Bertani (LB) medium were purchased from Thermo Fisher Scientific (Rockford, IL, USA). Bradford assay solution was purchased from TCI America (Portland, OR, USA). Solvents and all other chemicals were purchased from Fisher Scientific. Milli-Q water was used throughout this study.
Strains, vectors, media, and culture condition
B. bassiana ATCC 7159, Saccharomyces cerevisiae BJ5464, Pseudomonas putida KT2440, and Pichia pastoris GS115 were obtained from the American Type Culture Collection (ATCC). E. coli XL1-Blue and BL21(DE3) were purchased from Agilent for gene cloning and protein expression, respectively. The pJET1.2 (Thermo Fisher Scientific, USA) together with pET28a( +) (Millipore Sigma, USA), pPICZB (Thermo Fisher Scientific, USA), YEpADH2p-URA3 (provided by Yi Tang at the University of California, Los Angeles), and pMiS1-mva-ges (provided by Jens Schrader and Josef Altenbuchner at Dechema-Forschungsinstitut and University of Stuttgart) vectors were, respectively, used for gene cloning and expression of the Bbgt gene. All primers were synthesized by Thermo Fisher Scientific. Carbenicillin (50 μg/mL), kanamycin (50 μg/mL), and zeocin (50 μg/mL) were supplemented into the culture media for selection of correct clones when appropriate. Wild type B. bassiana was grown in YM medium (BD Biosciences, USA) at 28 °C for 3 days in order to extract the genomic DNA. The E. coli strains were routinely grown in LB medium (Fisher Scientific, USA) at 37 °C. Synthetic complete (SC) (uracil- dropout 0.77 g/L, yeast nitrogen base 6.7 g/L and glucose 20 g/L) and YP (1% yeast extract and 2% peptone) medium were used to grow S. cerevisiae BJ5464. P. pastoris was grown in YPDS medium (1% yeast extract, 2% peptone, 2% filter-sterilized glucose, and 1 M sorbitol). MGY medium (glycerol 10 g/L, yeast nitrogen base 6.7 g/L, and biotin 0.4 mg/L) and MM medium (yeast nitrogen base 13.4 g/L and biotin 0.4 mg/L) were used for P. pastoris protein expression.
Phylogenetic analysis
For the phylogenetic analysis, the full-length amino acid sequence of BbGT was aligned with 19 GTs from other species such as plants, bacteria, and fungi using Clustal Omega. The resulting alignment was used to construct an unrooted phylogenetic tree using the neighbor-joining method.
Cloning of the Bbgt gene from B. bassiana and plasmid construction
The genomic DNA was extracted from B. bassiana using the Quick-DNA™ Fungal/Bacterial Microprep Kit (Zymo Research, USA) according to the manufacturer’s instructions. To amplify the Bbgt gene (1,386 bp), the primers (1 μM), genomic DNA (0.2 μL), dNTP mix (200 μM), 5 × buffer (4 μL), DMSO (0.4 μL), Phusion® High-Fidelity DNA Polymerase (0.2 μL at 2 U/μL), and nuclease-free water were mixed to bring the volume to 20 μL. The PCR was performed by an initial denaturation at 98 °C for 5 min, followed by 20 cycles of touchdown program (98 °C for 30 s, annealing at 65 °C for 40 s, decreasing 0.5 °C per cycle, and extension at 72 °C for 90 s), and then 20 cycles of regular program (98 °C for 30 s, annealing at 55 °C for 40 s, and extension at 72 °C for 90 s), and finally, 72 °C for 10 min of extension.
The PCR product was purified using a 0.8% agarose gel with a GeneJET Gel Extraction Kit (Thermo Fisher Scientific, USA) and ligated into the pJET1.2 cloning vector to yield pWZ7 (pJET1.2-Bbgt). The ligation product was transferred into E. coli XL1-Blue competent cells through chemical transformation, and the transformants were selected on an LB agar plate with 50 μg/mL carbenicillin. After digestion check and sequencing, the gene was then excised from pWZ7 and ligated to the pET28a expression vector between the NdeI and EcoRI restriction enzyme sites to yield expression plasmid pWZ8 (pET28a-Bbgt), and the YEpADH2p-URA3 expression vector between NdeI and PmlI to yield pWZ9 (YEpADH2p-URA3-Bbgt).
The same gene was also PCR amplified from pWZ8 using different primers (Table 1) and ligated into pJET1.2 cloning vector, yielding pJR10 (pJET1.2-Bbgt). The Bbgt gene was excised from pJR10 and ligated into the pPICZB expression vector between the EcoRI and NotI restriction enzyme sites and pMIS1-mva-ges expression vector between the PmeI and HindIII sites to yield expression plasmids pJR14 (pPICZB -Bbgt) and pJR16 (pMiS1-Bbgt), respectively (Table 1). Sequences were confirmed using the Sanger method.
Heterologous expression of BbGT and in vivo biotransformation of quercetin in E. coli BL21(DE3)
The expression plasmid pWZ8 was introduced into E. coli BL21(DE3) through chemical transformation. A colony of E. coli BL21(DE3)/pWZ8 was picked into 5 mL of LB medium containing kanamycin (50 μg/mL), which was incubated at 37 °C with shaking (250 rpm) for about 12 h. Then 500 μL of the seed culture was inoculated into 50 mL of LB broth containing kanamycin (50 μg/mL) with shaking at 250 rpm and 37 °C until an OD600 between 0.4 and 0.6 was reached. Protein expression was induced with 200 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Gold Biotechnology, USA) at 28 °C for an additional 16 h with shaking at 250 rpm. After protein expression, 0.4 mM quercetin and 0.11 M glucose were added for product analysis. The culture was maintained at 28 °C and 250 rpm for an additional 2 days. The culture was then extracted three times with 50 mL of ethyl acetate and subjected to analysis on the LC–MS at 350 nm.
Purification of BbGT from E. coil BL21(DE3)/pWZ8
E. coil BL21(DE3)/pWZ8 cells were harvested by centrifugation at 3,000 × g and 4 °C for 10 min, and subsequently washed twice with distilled water and resuspended in lysis buffer including 20 mM Tris–HCl buffer (pH 7.9) and 0.5 M NaCl with 1 mM dithiothreitol. The harvested cells were disrupted by ultrasonication (Misonix Sonicator 3000, Misonix Inc., USA) on ice. The cell lysates were centrifuged at 10,000 × g and 4 °C for 10 min to collect the soluble fraction containing target protein and analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) as soluble fraction. SDS-PAGE analysis used a premixed protein marker (EZ-Run™ Pre-Stained Rec Protein Ladder, molecular range: 11–170 kDa, Fisher BioReagents) as the reference. After electrophoresis, the gel was stained with 0.1% Coomassie Brilliant Blue R250, and destained in 20% (v/v) acetic acid–water.
To purify the enzymes, the supernatant from the cell lysates was loaded onto a HisPur™ Ni–NTA affinity column (Thermo Scientific, Rockford, USA) according to the manufacturer’s protocol. After eluting the column with cold (4 °C) washing buffers (50 mM Tris–HCl, 2 mM EDTA, 10 mM and 25 mM imidazole, pH 7.9), the recombinant BbGT was finally eluted with elution buffer (50 mM Tris–HCl, 2 mM EDTA, pH 7.9) containing 250 mM imidazole. The purified His6-tagged protein was concentrated and then desalted by buffer A (50 mM Tris–HCl, 2 mM EDTA, pH 7.9) using the 30 K Macrosep Advance Centrifugal Device (Pall Corporation, New York, USA). This enzyme was stored in 50% (v/v) glycerol at − 20 ℃. The protein concentration was determined using the Bradford assay. All purification steps were carried out at 4 °C.
Functional characterization of BbGT through in vitro enzymatic reactions
BbGT catalyzes the reaction of UDP-glucose with quercetin. Enzymatic assays were performed in a 100-μL reaction system, containing 20 mM Tris–HCl (pH 8.0), 2.5 mM substrate, 1 mM MgCl2, 2 mM sugar donor (uridine diphosphate glucose or UDP-glucose), and 12.5 μg of purified recombinant BbGT protein. The mixtures were thoroughly mixed and incubated at 30 ℃ for 6 h, and then terminated by the addition of 200 μL of HPLC-grade methanol. The reaction mixtures were centrifuged at 13,000 × g for 10 min, and supernatants were collected to analyze the products by LC–MS as described before under 350 nm.
To investigate the substrate specificity of BbGT, several sugar-acceptor substrates were tested, including curcumin, resveratrol, tetracycline, zearalenone, resibufogenin, and indigoidine. Besides UDP-glucose, another structurally similar sugar-donor substrate, UDP-glucuronic acid, was also reacted with quercetin. Reaction products were analyzed by LC–MS.
Homology modeling of BbGT
A homology model for BbGT was generated with the Swiss-Model server using the GT structure from a macrolide glycosyltransferase (PDB ID 2IYA) as a template. The homology model was aligned to UDP-glucose:flavonoid 3-O-glycosyltransferase (VvGT1), which glycosylates quercetin. Structures of VvGT1 with quercetin bound (PDB ID 2C9Z) and UDP-2-deoxy-2-fluoro glucose bound (PDB ID 2C1Z) were used to position the two molecules in the homology model.
Determination of the optimal in vitro reaction conditions
The standard curves were established using isolated quercetin glucosides to quantify product formation and conversion rate by HPLC. To determine the optimum reaction temperature, the reaction mixtures with quercetin as the substrate were incubated at varying temperatures (10, 20, 25, 30, 35, 40, 45, and 50 °C) for 3 h. Glucosylation reactions were performed as described before. To find out the optimum pH for BbGT, this enzyme was reacted with quercetin at 35 °C in 200 mM phosphate buffer with different pH values (pH 4.5, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, and 8.5). For metal ion testing, CaCl2, MgCl2, MnCl2, and ZnCl2 were tested. Control group did not contain any added metal ions. The assays were performed with individually divalent metal ion (10 mM, final concentration) using UDP-glucose as sugar donor and quercetin as sugar acceptor. All reactions were performed in triplicate and values were expressed as the mean ± standard deviation.
Heterologous expression of BbGT and in vivo biotransformation of quercetin in S. cerevisiae BJ5464
The expression plasmid pWZ9 was introduced into S. cerevisiae BJ5464 by LiAc transformation as described in the literature (Yu et al. 2017). The correct transformant was selected by autotrophy of uracil (Ura). Briefly, 10 μg of plasmid was mixed with 100 μL of S. cerevisiae BJ5464 competent cells in 700 μL of LiAc (100 mM Lithium Acetate, pH 7.5)/40% PEG 3350 (120 mM PEG-3350)/TE (10 mM Tris–HCl, pH 7.5 and 1 mM EDTA) solution. The mixture was incubated at 28 °C for 30 min with shaking. 88 μL of DMSO was then added and mixed evenly, followed by 42 °C heat shock for 7 min. After centrifugation for 10 s at 13,000 × g, the supernatant was discarded and cells were resuspended in 1 mL of TE buffer. The last step was repeated once and the cells were finally resuspended in 100 μL of TE buffer and spread on SC plates, which were incubated at 28 °C for 3 days. Yeast strains harboring plasmid pWZ9 were cultured in 5 mL of SC medium with shaking at 250 rpm and 28 °C overnight. Then, 500 μL of seed culture was inoculated into 50 mL of SC medium in a 250-mL Erlenmeyer flask, which was incubated at 28 °C and 250 rpm overnight. After the OD600 value reached 1.0, 50 mL of YP medium was added and incubated overnight. Finally, 0.4 mM quercetin was added the next day and the cultures were maintained under the same conditions for an additional 2 days.
Heterologous expression of BbGT and in vivo biotransformation of quercetin in P. putida KT2440
The expression plasmid pJR16 was introduced into P. putida KT2440 by electroporation as described in literature (Fidan and Zhan 2019). The cells were plated on LB agar plates containing 50 μg/mL kanamycin and incubated at 28 °C overnight. The transformants were picked and grown in 50 mL LB with 50 μg/mL kanamycin at 28 °C and 250 rpm. When the OD600 reached 0.35, 100 mg L-rhamnose was added for induction at 28 °C and 250 rpm. After 16 h, 0.4 mM quercetin and 0.11 M glucose were added into the culture for product analysis, which was incubated at 28 °C and 250 rpm for an additional 2 days.
Heterologous expression of BbGT and in vivo biotransformation of quercetin in P. pastoris GS115
The expression plasmid pJR14 was linearized by Pme I and introduced into P. pastoris GS115 by electroporation as described in the literature (Cregg and Russell 1998; Lin-Cereghino et al. 2005). After electroporation, 1 mL of ice-cold 1 M sorbitol was added to the cuvette immediately. The mixture was then transferred to a sterile 15-mL tube and incubated at 30 °C without shaking for 2 h. Two hundred microliters of the mixture was spread on YPDS agar plates containing various concentrations of Zeocin™ (10, 25, 50, 100, 200 μg/mL). The plates were incubated at 30 °C for 3 days.
Colonies only appeared on the YPDS plate with 10 μg/mL Zeocin™. A single colony of P. pastoris GS115/pJR14 was inoculated into MGY medium as seed culture and cultivated at 30 °C and 250 rpm for 24 h. Then, 1 mL of seed culture was inoculated into 50 mL of MGY medium, which was cultivated at 30 °C and 250 rpm for an additional 18 h. The culture was centrifuged at 3,000 × g for 5 min, washed with MM medium twice and suspended in MM medium to be OD600 1.0. Afterwards, 50 mL of broth was transferred into a 500-mL shake flask and cultivated at 30 °C and 250 rpm. Methanol was fed every 24 h to keep its concentration around 1%. After protein expression, 0.4 mM quercetin and 0.11 M glucose were added, and the culture was maintained at 28 °C and 250 rpm for an additional 3 days.
Extraction, analysis, and purification of compounds
The cultures of E. coli BL21(DE3)/pWZ8 and S. cerevisiae BJ5464/pWZ9 were scaled up to 400 mL as described above, respectively. The biotransformation broths were maintained at 28 °C and 250 rpm for two days. Both cultures were extracted with 400 mL of ethyl acetate three times. The extracts were dried under reduced pressure and purified by silica gel 60 column chromatography. Compounds were successively eluted with acetone-hexane (2:1, v/v), acetone, and methanol. The fractions containing the target products were further purified by HPLC using the same method mentioned above, yielding 5.6 mg of quercetin-7-O-β-d-glucoside (3) and 2.5 mg of quercetin-3-O-β-d-glucoside (4). The purified products were dissolved in DMSO-d6 for NMR spectra.
Whole-cell bioconversion of quercetin to quercetin glucosides by E. coli BL21(DE3)/pWZ8 and S. cerevisiae BJ5464/pWZ9
E. coli BL21(DE3)/pWZ8 and S. cerevisiae BJ5464/pWZ9 were grown as described above. After 16 h, the cells were collected by centrifugation for whole-cell bioconversion. Cells were re-suspended in the reaction buffer, and cell density (OD600 value) was determined using a UV–Vis spectrophotometer (Thermo Scientific, Rockford, USA). The whole-cell bioconversion conditions were investigated by testing different pH values (pH 6.0, 6.5, 7.0, 7.5, 8.0, and 8.5), temperatures (25, 30, 35, 40, and 45 °C), cell densities (OD600 2, 4, 6, 8, 10, and 12), reaction times (1, 3, 5, and 7 h), and substrate concentration (0.4, 0.8, 1.6, 3.2, and 5.0 mM). After the optimal conditions were determined, the whole-cell biotransformation experiment was performed in a 1-L reaction system.
Results
Amplification and sequence analysis of a putative GT gene from B. bassiana
The genome of B. bassiana ATCC 7159 has been previously sequenced by our group. Searching the genome sequence revealed the presence of a glucosyltransferase (GT) gene that is putatively involved in the commonly observed glucosylation and 4′-O-methylglucosylation by this strain. We named this gene as Bbgt (GenBank accession number MW736592). It was predicted that this 1,386-bp gene contains no introns, thus can be directly amplified from the genome of B. bassiana ATCC 7159. BLAST analysis indicated that the deduced protein of this gene was homologous to a variety of GTs. A phylogenetic tree was established based on the sequences of BbGT and 19 GTs from other organisms (Fig. 2). The phylogenetic tree mainly divided those 20 GTs into three classes: sterol GTs, flavonoid GTs, and terpene GTs. BbGT is more closely related to sterol GTs and flavonoid GTs than terpene GTs, suggesting that this enzyme is likely a flexible GT which can add the sugar moiety to different sugar-acceptor substrates.
Phylogenetic analysis of BbGT with other GTs. The numbers following GTs are the numbers of combined guide-tree/HMM iterations. The analyzed GTs include sterol GTs, namely BsUGT489 (Bacillus subtilis, GenBank accession no. WP_003220489, similarity: 18.0%) (Chang et al. 2018), BsUGT398 (B. subtilis, GenBank accession no. WP_003225398, similarity: 22.9%) (Chang et al. 2018), BsYjiC (B. subtilis, GenBank accession no. NP_389104.1, similarity: 19.6%) (Dai et al. 2018), UGT109A1 (B. subtilis, GenBank accession no. ASY97769.1, similarity: 18.8%) (Liang et al. 2017), BsGT1 (B. subtilis, GenBank accession no. ANP92054.1, similarity: 19.6%) (Wang et al. 2018), AtSGT (Arabidopsis thaliana, GenBank accession no. CAB06082.1, similarity: 19.5%) (Warnecke et al. 1997), SGTL1 (Withania somnifera, GenBank accession no. ABC96116.1, similarity: 22.4%) (Sharma et al. 2007), UGT51C1 (Candida albicans, GenBank accession no. AAD29571.1, similarity: 20.5%) (Warnecke et al. 1999); flavonoid GTs, namely TwUGT2 (Tripterygium wilfordii, GenBank accession no. MK035745.1, similarity: 13.6%) (Lu et al. 2020), CsUGT73A20 (Camellia sinensis, GenBank accession no. KP682358.1, similarity: 21.1%) (Zhao et al. 2017), Am4CGT (Antirrhinum majus, GenBank accession no. BAE48239.1, similarity: 22.3%) (Ono et al. 2006), PlUGT1 (Pueraria lobata, GenBank accession no. AGZ84545.1, similarity: 21.7%) (Li et al. 2014), UBGT (Scutellaria baicalensis, GenBank accession no. BAA83484.1, similarity: 22.1%) (Hirotani et al. 2000), FaGT7 (Fragaria ananassa, GenBank accession no. Q2V6J9, similarity: 20.9%) (Griesser et al. 2008); terpenoid GTs, namely, CsUGT1 (Citrus sinensis, GenBank accession no. GQ221686.1, similarity: 21.5%) (Fan et al. 2010), UGT84B1 (Arabidopsis thaliana, GenBank accession no. At2g23260, similarity: 20.9%) (Caputi et al. 2008), UGT84A4 (A. thaliana, GenBank accession no. At4g15500, similarity: 22.2%) (Caputi et al. 2008), CsUGT2 (C. sinensis, GenBank accession no. ACS87991.1, similarity: 21.5%) (Fan et al. 2010), UGT74M1 (Gypsophila vaccaria, GenBank accession no. ABK76266.1, similarity: 20.5%) (Meesapyodsuk et al. 2007)
Heterologous expression and purification of BbGT from E. coli BL21(DE3)
The gene was amplified from the genomic DNA of B. bassiana, and it was ligated to pET28a to yield the corresponding expression plasmid, pWZ8 (pET28a-Bbgt). The plasmid was expressed in E. coli BL21(DE3) with IPTG induction. The N-His6-tagged BbGT was purified through Ni–NTA column chromatography to homogeneity. The purified enzyme was analyzed by SDS-PAGE. As shown in Fig. 3a, BbGT (~ 50.4 kDa) was successfully expressed and purified from E. coli BL21(DE3)/pWZ8. The isolation yield of BbGT was 13.6 mg/L.
Expression of BbGT in E. coli and in vitro functional characterization. a SDS-PAGE analysis of the purified recombinant BbGT from E. coli BL21(DE3). Lane 1: Purified BbGT; Lane 2: Protein ladder. b HPLC analysis of the reaction of BbGT with quercetin in the presence of UDP-glucose at 350 nm. (i) negative control (without BbGT); (ii) quercetin + BbGT. c-h ESI–MS (-) spectra of products 1–6
In vitro functional characterization of BbGT
Since the phylogenetic analysis suggested that BbGT is more related to sterol GTs and flavonoids GTs, we first tested whether it can glycosylate the sterol resibufogenin, in the presence of UDP-glucose. However, no products were formed (Fig. S1). We next tested the flavonoid quercetin as a potential substrate. HPLC analysis (Fig. 3b) showed that six more polar products were formed. ESI–MS spectra (Fig. 3c) of 1–6 showed the corresponding quasimolecular ion [M-H]− at m/z 624.9, 462.8, 462.9, 462.9, 462.9, and 462.9, respectively. Therefore, products 2–6 have the same molecular weight of 464, which is 162 mass units larger than the substrate quercetin, indicating that a glucose moiety was added to different hydroxyl groups of quercetin. These products were deduced to be quercetin monoglucosides. Since quercetin has five free phenolic hydroxyl groups, we propose that the glucose moiety was introduced to each of these hydroxyl groups to generate the five monoglucosides (Fig. 1). The molecular weight of 1 was found to be 626, which is 324 mass units larger than the substrate or 162 units larger than 2–6, suggesting that two glucose moieties were added to quercetin to form a diglucoside. Thus, its function was characterized as a UDP-glucosyltransferase.
Broad substrate specificity of BbGT toward sugar acceptor substrates
We next examined whether BbGT can use other substrates. We first tested whether it can use UDP-glucuronic acid since its structure is highly similar to UDP-glucose. However, no products were formed from quercetin (data not shown), indicating that BbGT is a specific UDP-glucosyltransferase that only recognizes UDP-glucose as a sugar donor.
The substrate specificity of BbGT toward sugar acceptor substrates was also investigated. Several substrates, including curcumin, resveratrol, zearalenone, tetracycline, and indigoidine, were incubated with BbGT in the presence of UDP-glucose. HPLC analysis showed that compared to the negative control (trace i, Fig. 4a), BbGT generated two more polar products C1 and C2 (trace ii), at 19.8 and 23.9 min respectively, from curcumin. The two products showed the UV spectra (Figs. S2a and S2b) similar to that of the substrate. The ESI–MS spectra (Figs. S3a and S3b) of the two products showed the [M-H]− ion peaks at m/z 691.0 and 528.8, indicating that their molecular weights are 692 Da and 530 Da. These are consistent with a diglucoside and a monoglucoside of curcumin, whose molecular weight is 368 Da. This result indicated that BbGT can take curcumin as the substrate for glucosylations.
HPLC analysis of glucosylation of different sugar-acceptor substrates by BbGT. a Glucosylation of curcumin (420 nm); b Glucosylation of zearalenone (250 nm); c glucosylation of resveratrol (300 nm). (i) substrate incubated with the reaction buffer without BbGT; (ii) substrate incubated with the reaction buffer with BbGT
When the macrolactone zearalenone was used as the substrate, two more polar products Z1 and Z2 at 18.5 and 22.5 min were detected by HPLC (Fig. 4b). Similarly, two products R1 and R2 appeared at 12.2 and 13.8 min when BbGT reacted with resveratrol (Fig. 4c). All products showed the similar UV spectra (Figs. S2c–f) to the substrates. The ESI–MS spectra (Figs. S3c and S3d) of Z1 and Z2 showed a [M-H]− ion peak at m/z 479.0 and 478.9, respectively, indicating that their molecular weights are 480 Da, which is consistent with the addition of a glucose moiety to zearalenone. The ESI–MS spectra (Figs. S3e and S3f) of R1 and R2 showed a [M-H]− ion peak at m/z 389.1 and 389.0, respectively. Their molecular weights were thus determined to be 390 Da, suggesting that they are monoglucosylated products of resveratrol. For tetracycline and indigoidine, no products were found (data not shown), indicating that these two substrates were not taken by BbGT as the sugar acceptor substrate.
The basis for the broad substrate specificity of BbGT was also investigated using homology modeling. The model (Fig. 5a) suggests the BbGT adopts a GT-B fold, with the C-terminal domain binding the sugar donor and the N-terminal domain binding the quercetin acceptor (Chang et al. 2011; Erb et al. 2009; Schmid et al. 2016). Residues important for binding UDP-glucose (Thr280, D374) and the active site histidine base (His20) in the flavonoid glucosyltransferase from Vitis vinifera (which uses UDP-glucose donor and quercetin acceptor molecules) (Offen et al. 2006) are conserved in the homology model of BbGT (Thr300, D390, and His31, respectively). Hydrophobic residues in BbGT are positioned on one face of the acceptor binding pocket (Fig. 5b), which is also peppered with side chains capable of interacting with the hydroxyl groups of quercetin (Fig. 5c–e). The acceptor pocket is wide and composed of numerous loops, the movement of which could help to explain the promiscuity observed in substrate selection by BbGT. Figure 5c–e show that the quercetin molecule can be positioned in the pocket in multiple conformations, each placing a different hydroxyl group of quercetin in line for attack of the UDP glucose donor, presumably using the network of available hydrogen bond donors/acceptors in the active site to reposition the acceptor molecule. The open active site (Dai et al. 2021; Offen et al. 2006), the presence of numerous loops (Schmid et al. 2016), strategic location of hydrogen-bonding acceptors/donors in a relatively hydrophobic pocket (Gatti-Lafranconi and Hollfelder 2013), and predicted conformational changes upon binding of the substrates (Hu et al. 2003; Offen et al. 2006; Schmid et al. 2016) likely all contribute to the observed promiscuity in product specificity.
Homology modeling of the BbGT. a The structure of BbGT was predicted using homology modeling. The model shows a GT-B fold where the C-terminal domain encompasses the sugar donor binding site (gray) and the N-terminal domain houses the acceptor binding site (teal) with the active site formulated at the interface. A quercetin molecule (magenta) and UDP-2-deoxy-2-fluoro glucose (green) are shown based on their positions in the 2C9Z and 2C1Z structures, respectively. b The hydrophobic character of the quercetin binding pocket is shown as red (high hydrophobicity) and white (low hydrophobicity). In c-e, the quercetin molecule (cyan with oxygen atoms colored yellow) was placed into the active site to position three of the possible nucleophilic hydroxyl groups within 4–5 Å of C-1 of the donor sugar. Potential hydrogen bond donors/acceptors (purple) are present throughout the mostly hydrophobic pocket
Determination of the optimal in vitro reaction conditions of BbGT
Quantification of quercetin conversion showed that BbGT has the highest glucosylation activity at 35 °C (Fig. 6a). The glucosylation activity of BbGT increased significantly by 31.50% when the temperature increased from 10 to 35 °C, but gradually decreased when the reaction temperatures were above 35 °C. Thus, the optimum reaction temperature for BbGT was determined to be 35 °C. The effect of pH on the glucosylation activity of BbGT was then determined. The enzyme was reacted with quercetin at 35 °C but different pH values. The conversion rate of quercetin by BbGT notably increased from 15.6 to 49.9% with the increase of pH from 4.5 to 8.0, and then decreased to 38.9% at pH 8.5 (Fig. 6b). Therefore, the optimum pH for BbGT is 8.0.
Determination of the optimum in vitro reaction conditions for BbGT. a Effect of reaction temperature on the BbGT glucosylation activity. b Effect of reaction pH on the BbGT glucosylation activity. c Effect of metal ions on the BbGT glucosylation activity. Data are presented as the mean ± SD from three independent experiments
In addition, the effect of various metal ions on the activity of BbGT was also tested. We found that BbGT activity was stimulated by Ca2+, Mg2+, and Mn2+ at the tested concentrations (Fig. 6c). In the presence of these three metal ions, the enzyme activity was 1.38-, 1.52-, and 1.48-fold higher than that of the control group, respectively. Among them, Mg2+ exhibited the strongest stimulating effect, and the conversion rate of quercetin increased by nearly 30% compared to the control. By contrast, when Zn2+ was added into the reaction system, the activity of BbGT was inhibited. The conversion rate decreased from 56 to 35.3% (Fig. 6c). Based on these results, the optimal in vitro reaction conditions for BbGT are at 35 °C, pH 8.0 with 10 mM of Mg2+.
Comparison of the in vivo glucosylation of quercetin by BbGT in different heterologous expression systems
Quercetin was first incubated with IPTG-induced E. coli BL21(DE3)/pWZ8. HPLC analysis showed that E. coli BL21(DE3)/pWZ8 was able to convert the substrate (trace i, Fig. 7a) to a major product (trace ii) that corresponds to product 3 in the in vitro enzymatic reaction. Besides the retention time, the UV and MS spectra also confirmed that this compound is identical to product 3. Several other product peaks were also detected, but were very minor.
HPLC analysis of glucosylation of quercetin by BbGT in different heterologous expression systems. a HPLC traces (350 nm) of the standard of quercetin and biotransformation product profiles from different engineered strains. (i) commercial standard of quercetin; (ii) quercetin + E. coli BL21(DE3)/pWZ8; (iii) quercetin + P. putida KT2440/pJR16; (iv) quercetin + S. cerevisiae BJ5464/pWZ9; (v) quercetin + P. pastoris GS115/pJR14. Asterisked peaks in trace iv are the metabolites from the host. b UV spectra comparison of the substrate and product 3. c UV spectra comparison of the substrate and product 4. d Selected HMBC correlations for products 3 and 4
While E. coli is the most commonly used microbial host, other microorganisms such as S. cerevisiae BJ5464, P. putida KT2440, and P. pastoris GS115 are also utilized in the biotechnological production of medicinally important natural products (Fidan and Zhan 2015). It is interesting that BbGT expressed in E. coli showed different product profiles in vivo and in vitro. We thus wanted to test additional microbial hosts and see what major products could be formed. When we introduced the plasmid pJR16 (pMiS1-Bbgt) into P. putida KT2440, the product profile (trace iii, Fig. 7a) was almost the same as in E. coli, with 3 as the major product. However, when BbGT was expressed in S. cerevisiae BJ5464, incubation of quercetin with this strain yielded 4 as the major product and 3 was not detected at all (trace iv, Fig. 7a). We next introduced BbGT into another yeast strain, P. pastoris GS115. HPLC analysis showed that this engineered strain, like the two bacterial strains, converted quercetin to product 3, although with a much lower efficiency (trace v, Fig. 7a). Therefore, products 3 and 4 are the two major glucosylation products of quercetin in the BbGT-harboring microbial hosts. Their UV spectra are shown in Fig. 7b and c, which are similar to that of the substrate.
Characterization two major glycosylated products of quercetin in different microbial hosts
We next scaled up the biotransformation of quercetin by E. coli BL21(DE3)/pWZ8 and S. cerevisiae BJ5464/pWZ9. The two major products 3 and 4 were purified from these two strains, respectively, and subjected to NMR analysis (Figs. S4–S13). The proton and carbon signals were assigned based on the 1D and 2D NMR spectra (Table S1). The 13C NMR spectra of compounds 3 and 4 both revealed 21 carbon signals. In addition to the signals of quercetin, six additional carbon signals at δC 99.9, 77.1, 76.4, 73.1, 69.5, and 60.6 for 3 and δC 100.8, 77.6, 76.5, 74.1, 69.9, and 61.0 for 4 were found in the spectra, further suggesting that a glucose moiety was added to one of the hydroxyl groups in quercetin. As shown in the 1H NMR spectra (Figs. S4 and S9), the anomeric proton signals at δ 5.07 (J = 7.5 Hz), and 5.46 (J = 7.5 Hz) of 3 and 4, respectively, indicated the β-configuration of these two compounds for the glucopyranosyl moiety. The 13C NMR spectra showed the glucose anomeric carbon signals at δ 99.9 and 100.8 for compounds 3 and 4, respectively. HMBC spectrum of 3 revealed the correlation of H-1″ (δ 5.07, d, J = 7.5 Hz) to C-7 at δ 162.7 (Fig. 7d), which confirmed that 3 has a glucose moiety at C-7. Similarly, H-1″ (δ 5.46, d, J = 7.5 Hz) of 4 had HMBC correlation to C-3 at δ 133.3, indicating that the glucose moiety was introduced at C-3 (Fig. 7d). Therefore, products 3 and 4 were characterized as quercetin-7-O-β-d-glucoside and quercetin-3-O-β-d-glucoside, respectively. Their chemical shifts determined in this work are consistent with those reported in literature (Lee et al. 2008; Yang et al. 2008).
Optimized production of quercetin-7-O-β-d-glucoside with the engineered E. coli BL21(DE3)
We first investigated the effect of different cell concentrations ranging from OD600 2.0 to 12.0 on the formation of quercetin glucosides at 28 °C for 3 h. To ensure that the substrate is enough for the reactions, 1.3 mM quercetin was used together with 0.11 M glucose. We found that the product titer increased with higher cell concentrations (Fig. 8a). When the OD600 value was 10.0, the conversion rate of quercetin in the biotransformation system reached 43.5%, which was 24.5% higher than the titer at OD600 2.0.
Optimization of the glucosylation of quercetin by E. coli BL21(DE3)/pWZ8. a Effect of cell density on quercetin glucosylation by E. coli BL21(DE3)/pWZ8. b Effect of reaction pH on quercetin glucosylation by E. coli BL21(DE3)/pWZ8. c Effect of temperature on quercetin glucosylation by E. coli BL21(DE3)/pWZ8. d Effect of reaction time on quercetin glucosylation by E. coli BL21(DE3)/pWZ8. e Effect of substrate concentration on quercetin glucosylation by E. coli BL21(DE3)/pWZ8. Data are presented as the mean ± SD from three independent experiments
The production of quercetin-7-O-β-d-glucoside at different pH values (pH 6.0–8.5) was determined by HPLC. The highest conversion rate reached 54.0% at pH 7.0 (Fig. 8b), which is 15.6% higher than that at pH 6.0. This is also consistent with the optimum pH for in vitro reactions. In order to investigate the impact of temperature on the whole-cell conversion efficiency of quercetin to its glucosides, E. coli BL21(DE3)/pWZ8 cells (OD600 10.0) were incubated in 200 mM phosphate buffer (pH 7.0) at different temperatures ranging from 25 to 45 °C. The data showed that the increase in the reaction temperature could effectively improve the conversion of quercetin to quercetin-7-O-β-d-glucoside. The highest conversion rate reached 43.0% at 35 °C (Fig. 8c).
We next determined how the bioconversion time affects the conversion rate of quercetin in the engineered E. coli strain. To this end, we conducted a time course analysis for the conversion of 10 mM quercetin to it glucosides by BbGT-expressing E. coli BL21(DE3) strain (OD600 10.0) at 35 °C in 200 mM phosphate buffer (pH 7.0). The reaction was sampled at 1, 3, 5, and 7 h. The conversion rate of quercetin increased from 22.9 to 35.4% in the first 3 h (Fig. 8d). After that, the increase of the conversion rate slowed down. Therefore, the optimum reaction time for the conversion of quercetin to quercetin-7-O-β-d-glucoside by E. coli BL21(DE3)/pWZ8 was determined to be 3 h.
To further optimize the production of quercetin-7-O-β-d-glucoside, we next tested its production from different quercetin concentrations ranging from 0.4 to 5.0 mM. The reactions were conducted at pH 7.0, OD600 10, and 35 °C for 3 h. When the substrate concentration increased from 0.4 to 0.8 mM, the conversion of quercetin to its glucosides also increased from 71.8 to 79.8%. However, when the concentration of quercetin was further increased to 1.6 mM, the conversion rate dropped to 44.5% (Fig. 8e). Based on the productivity of quercetin-7-O-β-d-glucoside, 0.8 mM was deemed to be the optimal substrate concentration. We then scaled up the reaction to 1 L. The titer of quercetin-7-O-β-d-glucoside reached 0.34 ± 0.02 mM (equivalent to 158 ± 8 mg/L) from a total of 0.83 mM (equivalent to 250 mg/L) of quercetin in 24 h.
Optimized production of quercetin-3-O-β-d-glucoside with the engineered S. cerevisiae BJ5464
We first investigated the effect of different cell concentrations ranging from OD600 2.5 to 15.0 on the conversion of quercetin to quercetin-3-O-β-d-glucoside by S. cerevisiae BJ5464/pWZ9 cells at 35 °C for 3 h. To ensure that the substrate is enough for the reactions, 0.8 mM quercetin was used together with 0.11 M glucose. We found that the product titer increased with increasing cell concentrations (Fig. 9a). When the OD600 value was 7.5, the conversion rate of quercetin reached 53.8%, which was 24.4% higher than the titer at OD600 2.5.
Optimization of the whole-cell conversion of BbGT to quercetin-3-O-β-d-glucoside by S. cerevisiae BJ5464/pWZ9. a Effect of cell density on the production of quercetin-3-O-β-d-glucoside by S. cerevisiae BJ5464/pWZ9. b Effect of reaction pH on the production of quercetin-3-O-β-d-glucoside by S. cerevisiae BJ5464/pWZ9. c Effect of temperature on the production of quercetin-3-O-β-d-glucoside by S. cerevisiae BJ5464/pWZ9. d Effect of reaction time on the production of quercetin-3-O-β-d-glucoside by S. cerevisiae BJ5464/pWZ9. e Effect of substrate concentration on the production of quercetin-3-O-β-d-glucoside by S. cerevisiae BJ5464/pWZ9. Data are presented as the mean ± SD from three independent experiments
In order to investigate the impact of pH on the whole-cell conversion efficiency of quercetin, S. cerevisiae BJ5464/pWZ9 cells (OD600 7.5) were incubated with the substrate in 200 mM phosphate buffer (pH 6.5–8.5) at 28 °C for 12 h. At pH 6.5, the conversion rate was extremely low. With the increasing pH values, the conversion rate also increased apparently. The highest conversion rate reached 55.1% at pH 8 (Fig. 9b), which is higher than the optimal pH for E. coli BL21(DE3)/pWZ8 cells. The production of quercetin-3-O-β-d-glucoside at different temperatures (20–40 °C) was also investigated. The highest conversion rate reached 59.8% at 35 °C (Fig. 9c), which is 31.9% higher than that at 20 °C.
We then examined how the reaction time affects the conversion of quercetin in the engineered yeast. To this end, we conducted a time course analysis for the conversion of 0.8 mM quercetin to quercetin-3-O-β-d-glucoside by BbGT-expressing S. cerevisiae BJ5464 strain (OD600 7.5) at 35 °C in 200 mM phosphate buffer (pH 8.0). The reaction was sampled at 3, 6, 9, 12, and 15 h, and the product was quantified by HPLC. The production of quercetin-3-O-β-d-glucoside increased apparently from 3 to 12 h (Fig. 9d), and the conversion rate was increased from 6.3 to 45.5%. After 12 h, the increase of the conversion rate slowed down. Therefore, the optimum reaction time for this reaction using S. cerevisiae BJ5464/pWZ9 is 12 h under the tested conditions.
To find out the optimal substrate concentration, we incubated the strain with different quercetin concentrations ranging from 0.2 to 3.2 mM. The reactions were conducted at pH 8.0, OD600 7.5, and 35 °C for 12 h. When the substrate concentration increased from 0.2 to 0.4 mM, the conversion rates of quercetin to quercetin-3-O-β-d-glucoside were similar and both were higher than 60%. However, when the concentration of quercetin was 0.8 mM, the conversion rate dropped to 39.9% (Fig. 9e). Based on the productivity of glucosylated quercetin, 0.4 mM quercetin was selected for scaled-up reaction. The titer of quercetin-3-O-β-d-glucoside in a 1-L reaction system was 0.22 ± 0.02 mM (equivalent to 99 ± 8 mg/L) from a total of 0.41 mM (equivalent to 125 mg/L) of quercetin in 12 h. As a result, S. cerevisiae BJ5464/pWZ9 represents a promising strain for cost-effective production of quercetin-3-O-β-d-glucoside.
Discussion
B. bassiana has attracted a lot of attention because of its ability to perform diverse enzymatic modifications (including glycosylation) of exogenous substrates (Sordon et al. 2019; Strugała et al. 2017; Zeng et al. 2010). We previously used this strain to synthesize curcumin-8′-O-4″-O-methyl-β-d-glucopyranoside from curcumin. The water solubility of this glycoside is 39,000-fold higher than the substrate (Zeng et al. 2010). In this study, we discovered and cloned a versatile GT BbGT from B. bassiana ATCC 7159, which can glycosylate the representative flavonoid quercetin in a highly flexible way. In addition, BbGT showed a flexible substrate specificity toward sugar acceptor substrates, and can glycosylate resveratrol, curcumin and zearalenone into their glucosides.
The optimum reaction temperature for BbGT was found to be 35 °C in phosphate buffer. It is similar to those reported GTs, namely, BsGT1, BsGT2, and BsGT110 (40 °C) from Bacillus subtilis (Cheng et al. 2019; Chiang et al. 2018) and UGT74AN3 from Catharanthus roseus (40 °C) (Wen et al. 2020). The optimum pH for BbGT was found to be 8.0, which is the same as the optimum pH for three recently reported UDP-glucosyltransferases, including BsGT1, BsGT2 from B. subtilis and UGT74AC1 from Siraitia grosvenorii, and three ApUFGTs from Andrographis paniculata (Cheng et al. 2019; Li et al. 2019; Mu et al. 2020). However, this value is higher than that of BsGT110 from B. subtilis (pH 7.0) (Chiang et al. 2018). Temperature and pH have significant impacts on the conversion of quercetin into quercetin glucosides, and these results indicate that UDP-glucosyltransferases are more active in mild conditions.
Metal ions play an important role in the biological function of many enzymes (Riordan 1977; Sigel and Pyle 2007). Furthermore, previous studies on UGTs showed that Mg2+, Ba2+, and Ca2+ ions exhibited a consistently positive effect on the enzymatic activity (Mu et al. 2020). Therefore, we also tested the effect of metal ions (Ca2+, Mg2+, Mn2+, and Zn2+) on the activity of BbGT. Among the four tested metal ions, Ca2+, Mg2+, and Mn2+ can stimulate the activity of BbGT. This result is in accordance with the previous report that GTs can utilize divalent metal ions such as Mn2+ and Mg2+ as enzyme reaction cofactors (Chiang et al. 2018). Li et al. also found that Ca2+ enhanced the activity of the GT from Bacillus circulans because of an additional calcium-binding site in the enzyme structure (Li et al. 2013). By contrast, Zn2+ showed inhibition to the enzymatic activity of BbGT. This is similar to the previous finding where UGT74AC1 from S. grosvenorii was strongly inhibited by Zn2+ (Mu et al. 2020). Researchers have previously found that Zn2+ is a reversible and competitive inhibitor within the active site of the glucosyltransferase at the fructose subsite (Devulapalle and Mooser 1994), which may explain its inhibition effect on BbGT enzyme reported here.
Plants and microorganisms are known to contain GTs that can catalyze the glycosylation of flavonoids. Because the 3- and 7-OH are commonly present in many flavonoids, 3-O-GT and 7-O-GT are the most widely investigated (Cao et al. 2015; Yang et al. 2018). However, most GTs have strict regional selectivity and substrate specificity (Cartwright et al. 2008; Overwin et al. 2015). BbGT identified in this work was found to be a highly flexible enzyme in terms of the glucosylation position. The homology model of BbGT (Fig. 5) showed that the cavity is large enough for quercetin to easily rotate and expose different hydroxyl groups to the sugar donor for glucosylations, which explains the production of various quercetin monoglucosides. It is also a versatile enzyme as it can accept a variety of aromatic structures and convert them into monoglucosides and even diglucosides.
There are two novel phenomena observed for BbGT. First, this fungal UGT showed different product profiles in vitro and in vivo, when quercetin was used as a substrate. The enzyme generated six quercetin glucosides in the in vitro reactions, while it produced quercetin-7-O-β-d-glucoside as the dominant product in E. coli. No diglucoside of quercetin was detected in vivo. The different product profiles between in vitro and in vivo are likely due to the availability of UDP-glucose and interference of other endogenous proteins in the host. Second, different in vivo systems also showed different product profiles. E. coli, P. putida, and P. pastoris had similar products, but S. cerevisiae produced quercetin-3-O-β-d-glucoside as the major product. The different product profiles may be indicative of host-dependent changes in the BbGT substrate specificity using host-specific post-translational modifications or oligomerization patterns and/or protein regulators. Future work on identification of the post-translational modification patterns observed in different hosts could provide some clues into this interesting phenomenon.
Whole-cell biotransformation eliminates the need of cell disruption and enzyme purification, and can provide UDP-glucose in situ for the glucosylation of quercetin, thus representing a better way to produce quercetin glucosides than in vitro enzymatic reactions. Meanwhile, process optimization is a topic of central importance in industrial production processes. The temperature and pH play critical roles in product formation during whole-cell biotransformation. The optimum temperature for whole-cell bioconversion is the same as the purified enzyme. However, the optimum pH for whole-cell bioconversion with engineered E. coli is 7, which is lower than that in vitro reactions. In the 1-L reaction system, the titer of quercetin-7-O-β-d-glucoside in engineered E. coli strain was 158 ± 8 mg/L from 250 mg/L quercetin in 24 h. With the engineered S. cerevisiae BJ5464/pWZ9 strain, the optimum temperature and pH were same as those for the purified enzyme. The titer of quercetin-3-O-β-d-glucoside in shaker flasks was 99 ± 8 mg/L from 125 mg/L quercetin in 12 h. Lim used the E. coli BL21 culture expressing UGT73B3 as the whole-cell biocatalyst to produce quercetin-3-O-β-d-glucoside, and the titer was around 100 mg/L after 20 h under fermenter-scale condition (Lim et al. 2004). Xia expressed UGT73B3 in E. coli MEC367 with 30 g/L glucose as the sole carbon source, and the production titer of quercetin-3-O-β-d-glucoside reached 3.9 g/L in 56 h in controlled bioreactors (Xia and Eiteman 2017). To improve the production of these glucosides by BbGT, the expression of BbGT may be enhanced through synthetic biology and metabolic engineering approaches such as codon optimization of Bbgt and increased production of UDP-glucose in the host. It also was reported that bioreactors can provide better oxygen transfer rates than shaker flaks (Xia and Eiteman 2017), which can further increase the production of these glucosides.
In summary, this work identified a highly flexible and versatile GT for glucosylation of a variety of natural products. The enzyme showed different product profiles from quercetin in vitro and in various microbial systems. This work also provides two engineered strains for specific and efficient production of quercetin-7-O-β-d-glucoside and quercetin-3-O-β-d-glucoside, by using BbGT in different microbial hosts and optimizing the corresponding reaction conditions.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Anuradha R, Sukumar D (2013) In vitro anti-inflammatory compound quercimeritrin isolated from tithonia diversifolia flowers by hrbc membrane stabilization. World J Pharm Res 3(1):426–431
Cai X, Fang Z, Dou J, Yu A, Zhai G (2013) Bioavailability of quercetin: problems and promises. Curr Med Chem 20(20):2572–2582. https://doi.org/10.2174/09298673113209990120
Cao H, Chen X, Jassbi AR, Xiao J (2015) Microbial biotransformation of bioactive flavonoids. Biotechnol Adv 33(1):214–223. https://doi.org/10.1016/j.biotechadv.2014.10.012
Caputi L, Lim EK, Bowles DJ (2008) Discovery of new biocatalysts for the glycosylation of terpenoid scaffolds. Chemistry 14(22):6656–6662. https://doi.org/10.1002/chem.200800548
Cartwright AM, Lim E-K, Kleanthous C, Bowles DJ (2008) A kinetic analysis of regiospecific glucosylation by two glycosyltransferases of Arabidopsis thaliana: domain swapping to introduce new activities. J Biol Chem 283(23):15724–15731. https://doi.org/10.1074/jbc.M801983200
Chang A, Singh S, Phillips GN Jr, Thorson JS (2011) Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr Opin Biotechnol 22(6):800–808. https://doi.org/10.1016/j.copbio.2011.04.013
Chang T-S, Wu J-Y, Wang T-Y, Wu K-Y, Chiang C-M (2018) Uridine diphosphate-dependent glycosyltransferases from Bacillus subtilis ATCC 6633 catalyze the 15-O-glycosylation of ganoderic acid A. Int J Mol Sci 19(11):3469. https://doi.org/10.3390/ijms19113469
Chen Q, Li P, Xu Y, Li Y, Tang B (2015) Isoquercitrin inhibits the progression of pancreatic cancer in vivo and in vitro by regulating opioid receptors and the mitogen-activated protein kinase signalling pathway. Oncol Rep 33(2):840–848. https://doi.org/10.3892/or.2014.3626
Cheng Y, Zhang J, Shao Y, Xu Y, Ge H, Yu B, Wang W (2019) Enzyme-catalyzed glycosylation of curcumin and its analogues by glycosyltransferases from Bacillus subtilis ATCC 6633. Catalysts 9(9):734. https://doi.org/10.3390/catal9090734
Chiang C-M, Wang T-Y, Yang S-Y, Wu J-Y, Chang T-S (2018) Production of new isoflavone glucosides from glycosylation of 8-hydroxydaidzein by glycosyltransferase from Bacillus subtilis ATCC 6633. Catalysts 8(9):387. https://doi.org/10.3390/catal8090387
Cregg JM, Russell KA (1998) Transformation Pichia protocols. Springer 27−39.
Dai L, Li J, Yang J, Zhu Y, Men Y, Zeng Y, Cai Y, Dong C, Dai Z, Zhang X (2018) Use of a promiscuous glycosyltransferase from Bacillus subtilis 168 for the enzymatic synthesis of novel protopanaxatriol-type ginsenosides. J Agric Food Chem 66(4):943–949. https://doi.org/10.1021/acs.jafc.7b03907
Dai L, Qin L, Hu Y, Huang J-w, Hu Z, Min J, Sun Y, Guo R-T (2021) Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity. Biochem Biophys Res Commun 534:73–78. https://doi.org/10.1016/j.bbrc.2020.11.104
Day AJ, Mellon F, Barron D, Sarrazin G, Morgan MR, Williamson G (2001) Human metabolism of dietary flavonoids: identification of plasma metabolites of quercetin. Free Radic Res 35(6):941–952. https://doi.org/10.1080/10715760100301441
Devulapalle KS, Mooser G (1994) Subsite specificity of the active site of glucosyltransferases from Streptococcus sobrinus. J Biol Chem 269(16):11967–11971
Dou F, Wang Z, Li G, Dun B (2019) Microbial transformation of flavonoids by Isaria fumosorosea ACCC 37814. Molecules 24(6):1028. https://doi.org/10.3390/molecules24061028
Erb A, Weiss H, Härle J, Bechthold A (2009) A bacterial glycosyltransferase gene toolbox: generation and applications. Phytochemistry 70(15–16):1812–1821. https://doi.org/10.1016/j.phytochem.2009.05.019
Fan J, Chen C, Yu Q, Li Z-G, Gmitter FG (2010) Characterization of three terpenoid glycosyltransferase genes in ‘Valencia’ sweet orange (Citrus sinensis L. Osbeck). Genome 53(10):816–823. https://doi.org/10.1139/G10-068
Fidan O, Zhan J (2015) Recent advances in engineering yeast for pharmaceutical protein production. RSC Adv 5(105):86665–86674. https://doi.org/10.1039/C5RA13003D
Fidan O, Zhan J (2019) Discovery and engineering of an endophytic Pseudomonas strain from Taxus chinensis for efficient production of zeaxanthin diglucoside. J Biol Eng 13:66. https://doi.org/10.1186/s13036-019-0196-x
Gatti-Lafranconi P, Hollfelder F (2013) Flexibility and reactivity in promiscuous enzymes. ChemBioChem 14(3):285–292. https://doi.org/10.1002/cbic.201200628
Griesser M, Vitzthum F, Fink B, Bellido ML, Raasch C, Munoz-Blanco J, Schwab W (2008) Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria×ananassa) achene and receptacle. J Exp Bot 59(10):2611–2625. https://doi.org/10.1093/jxb/ern117
Grogan G, Holland H (2000) The biocatalytic reactions of Beauveria spp. J Mol Catal B Enzym 9(1–3):1–32. https://doi.org/10.1016/S1381-1177(99)00080-6
Hirotani M, Kuroda R, Suzuki H, Yoshikawa T (2000) Cloning and expression of UDP-glucose: flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 210(6):1006–1013. https://doi.org/10.1007/PL00008158
Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S (2003) Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci USA 100(3):845–849. https://doi.org/10.1073/pnas.0235749100
Ikeda K, Taguchi R (2010) Highly sensitive localization analysis of gangliosides and sulfatides including structural isomers in mouse cerebellum sections by combination of laser microdissection and hydrophilic interaction liquid chromatography/electrospray ionization mass spectrometry with theoretically expanded multiple reaction monitoring. Rapid Commun Mass Spectrom 24(20):2957–2965. https://doi.org/10.1002/rcm.4716
Jiang J-r, Yuan S, Ding J-f, Zhu S-c, Xu H-d, Chen T, Cong X-d, Xu W-p, Ye H, Dai Y-j (2008) Conversion of puerarin into its 7-O-glycoside derivatives by Microbacterium oxydans (CGMCC 1788) to improve its water solubility and pharmacokinetic properties. Appl Microbiol Biotechnol 81(4):647–657. https://doi.org/10.1007/s00253-008-1683-z
Ko JH, Kim BG, Joong-Hoon A (2006) Glycosylation of flavonoids with a glycosyltransferase from Bacillus cereus. FEMS Microbiol Lett 258(2):263–268. https://doi.org/10.1111/j.1574-6968.2006.00226.x
Lairson L, Henrissat B, Davies G, Withers S (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77:521–555. https://doi.org/10.1146/annurev.biochem.76.061005.092322
Lee S, Park HS, Notsu Y, Ban HS, Kim YP, Ishihara K, Hirasawa N, Jung SH, Lee YS, Lim SS (2008) Effects of hyperin, isoquercitrin and quercetin on lipopolysaccharide-induced nitrite production in rat peritoneal macrophages. Phytother Res 22(11):1552–1556. https://doi.org/10.1002/ptr.2529
Legault J, Perron T, Mshvildadze V, Girard-Lalancette K, Perron S, Laprise C, Sirois P, Pichette A (2011) Antioxidant and anti-inflammatory activities of quercetin 7-O-β-D-glucopyranoside from the leaves of Brasenia schreberi. J Med Food 14(10):1127–1134. https://doi.org/10.1089/jmf.2010.0198
Li C, Ban X, Gu Z, Li Z (2013) Calcium ion contribution to thermostability of cyclodextrin glycosyltransferase is closely related to calcium-binding site CaIII. J Agric Food Chem 61(37):8836–8841. https://doi.org/10.1021/jf4024273
Li D, Park J-H, Park J-T, Park CS, Park K-H (2004) Biotechnological production of highly soluble daidzein glycosides using Thermotoga maritima maltosyltransferase. J Agric Food Chem 52(9):2561–2567. https://doi.org/10.1021/jf035109f
Li J, Li Z, Li C, Gou J, Zhang Y (2014) Molecular cloning and characterization of an isoflavone 7-O-glucosyltransferase from Pueraria lobata. Plant Cell Rep 33(7):1173–1185. https://doi.org/10.1007/s00299-014-1606-7
Li Y, Li X-L, Lai C-J-S, Wang R-S, Kang L-P, Ma T, Zhao Z-H, Gao W, Huang L-Q (2019) Functional characterization of three flavonoid glycosyltransferases from Andrographis paniculata. R Soc Open Sci 6(6):190150. https://doi.org/10.1098/rsos.190150
Liang H, Hu Z, Zhang T, Gong T, Chen J, Zhu P, Li Y, Yang J (2017) Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab Eng 44:60–69. https://doi.org/10.1016/j.ymben.2017.07.008
Lim E-K, Ashford DA, Hou B, Jackson RG, Bowles DJ (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotech Bioeng 87(5):623–631. https://doi.org/10.1002/bit.20154
Lin-Cereghino J, Wong WW, Xiong S, Giang W, Luong LT, Vu J, Johnson SD, Lin-Cereghino GP (2005) Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. Biotechniques 38(1):44–48. https://doi.org/10.2144/05381BM04
Lu Y, Ma B-W, Gao J, Tu L-C, Hu T-Y, Zhou J-W, Liu Y, Tu Y-H, Lin Z-S, Huang L-Q (2020) Isolation and characterization of a glycosyltransferase with specific catalytic activity towards flavonoids from Tripterygium wilfordii. J Asian Nat Prod Res 22(6):537–546. https://doi.org/10.1080/10286020.2019.1642330
Ma B, Zeng J, Shao L, Zhan J (2013) Efficient bioconversion of quercetin into a novel glycoside by Streptomyces rimosus subsp. rimosus ATCC 10970. J Biosci Bioeng 115(1):24–26. https://doi.org/10.1016/j.jbiosc.2012.07.020
Makris DP, Rossiter JT (2000) Heat-induced, metal-catalyzed oxidative degradation of quercetin and rutin (quercetin 3-O-rhamnosylglucoside) in aqueous model systems. J Agric Food Chem 48(9):3830–3838. https://doi.org/10.1021/jf0001280
Matsubara K, Ishihara K, Mizushina Y, Mori M, Nakajima N (2004) Anti-angiogenic activity of quercetin and its derivatives. Lett Drug Des Discov 1(4):329–333. https://doi.org/10.2174/1570180043398533
Meesapyodsuk D, Balsevich J, Reed DW, Covello PS (2007) Saponin biosynthesis in Saponaria vaccaria. cDNAs encoding β-amyrin synthase and a triterpene carboxylic acid glucosyltransferase. Plant Physiol 143(2):959–969. https://doi.org/10.1104/pp.106.088484
Méndez C, Salas JA (2001) Altering the glycosylation pattern of bioactive compounds. Trends Biotechnol 19(11):449–456. https://doi.org/10.1016/S0167-7799(01)01765-6
Mfonku NA, Mbah JA, Kodjio N, Gatsing D, Zhan J (2020) Isolation and selective glycosylation of antisalmonellal anthraquinones from the stem bark of Morinda lucida Benth. (Rubiaceae). Phytochem Lett 37:80–84. https://doi.org/10.1016/j.phytol.2020.04.011
Mu S, Li J, Liu C, Zeng Y, Men Y, Cai Y, Chen N, Ma H, Sun Y (2020) Effective glycosylation of cucurbitacin mediated by UDP-glycosyltransferase UGT74AC1 and molecular dynamics exploration of its substrate binding conformations. Catalysts 10(12):1466. https://doi.org/10.3390/catal10121466
Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25(6):1396–1405. https://doi.org/10.1038/sj.emboj.7600970
Ono E, Fukuchi-Mizutani M, Nakamura N, Fukui Y, Yonekura-Sakakibara K, Yamaguchi M, Nakayama T, Tanaka T, Kusumi T, Tanaka Y (2006) Yellow flowers generated by expression of the aurone biosynthetic pathway. Proc Natl Acad Sci USA 103(29):11075–11080. https://doi.org/10.1073/pnas.0604246103
Overwin H, Wray V, Hofer B (2015) Flavonoid glucosylation by non-Leloir glycosyltransferases: formation of multiple derivatives of 3,5,7,3′,4′-pentahydroxyflavane stereoisomers. Appl Microbiol Biotechnol 99(22):9565–9576. https://doi.org/10.1007/s00253-015-6760-5
Paulke A, Eckert GP, Schubert-Zsilavecz M, Wurglics M (2012) Isoquercitrin provides better bioavailability than quercetin: comparison of quercetin metabolites in body tissue and brain sections after six days administration of isoquercitrin and quercetin. Pharmazie 67(12):991–996
Riordan JF (1977) The role of metals in enzyme activity. Ann Clin Lab Sci 7(2):119–129
Schmid J, Heider D, Wendel NJ, Sperl N, Sieber V (2016) Bacterial Glycosyltransferases: challenges and opportunities of a highly diverse enzyme class toward tailoring natural products. Front Microbiol 7:182. https://doi.org/10.3389/fmicb.2016.00182
Sharma LK, Madina BR, Chaturvedi P, Sangwan RS, Tuli R (2007) Molecular cloning and characterization of one member of 3β-hydroxy sterol glucosyltransferase gene family in Withania somnifera. Arch Biochem Biophys 460(1):48–55. https://doi.org/10.1016/j.abb.2007.01.024
Sigel RK, Pyle AM (2007) Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem Rev 107(1):97–113. https://doi.org/10.1021/cr0502605
Sordon S, Popłoński J, Tronina T, Huszcza E (2019) Regioselective O-glycosylation of flavonoids by fungi Beauveria bassiana, Absidia coerulea and Absidia glauca. Bioorg Chem 93:102750. https://doi.org/10.1016/j.bioorg.2019.01.046
Strugała P, Tronina T, Huszcza E, Gabrielska J (2017) Bioactivity in vitro of quercetin glycoside obtained in Beauveria bassiana culture and its interaction with liposome membranes. Molecules 22(9):1520. https://doi.org/10.3390/molecules22091520
Valentová K, Vrba J, Bancířová M, Ulrichová J, Křen V (2014) Isoquercitrin: pharmacology, toxicology, and metabolism. Food Chem Toxicol 68:267–282. https://doi.org/10.1016/j.fct.2014.03.018
Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5(9):380–386. https://doi.org/10.1016/S1360-1385(00)01720-9
Wang D-D, Jin Y, Wang C, Kim Y-J, Perez ZEJ, Baek NI, Mathiyalagan R, Markus J, Yang D-C (2018) Rare ginsenoside Ia synthesized from F1 by cloning and overexpression of the UDP-glycosyltransferase gene from Bacillus subtilis: Synthesis, characterization, and in vitro melanogenesis inhibition activity in BL6B16 cells. J Ginseng Res 42(1):42–49. https://doi.org/10.1016/j.jgr.2016.12.009
Warnecke D, Erdmann R, Fahl A, Hube B, Müller F, Zank T, Zähringer U, Heinz E (1999) Cloning and functional expression of UGT genes encoding sterol glucosyltransferases from Saccharomyces cerevisiae, Candida albicans, Pichia pastoris, and Dictyostelium discoideum. J Biol Chem 274(19):13048–13059. https://doi.org/10.1074/jbc.274.19.13048
Warnecke DC, Baltrusch M, Buck F, Wolter FP, Heinz E (1997) UDP-glucose:sterol glucosyltransferase: cloning and functional expression in Escherichia coli. Plant Mol Biol 35(5):597–603. https://doi.org/10.1023/A:1005806119807
Wen C, Huang W, He M-M, Deng W-L, Yu H-H (2020) Cloning and characterization of a glycosyltransferase from Catharanthus roseus for glycosylation of cardiotonic steroids and phenolic compounds. Biotechnol Lett 42(1):135–142. https://doi.org/10.1007/s10529-019-02756-5
Wen L, Zhao Y, Jiang Y, Yu L, Zeng X, Yang J, Tian M, Liu H, Yang B (2017) Identification of a flavonoid C-glycoside as potent antioxidant. Free Radic Biol Med 110:92–101. https://doi.org/10.1016/j.freeradbiomed.2017.05.027
Xia T, Eiteman MA (2017) Quercetin glucoside production by engineered Escherichia coli. Appl Biochem Biotechnol 182(4):1358–1370. https://doi.org/10.1007/s12010-017-2403-x
Xu F, Gage D, Zhan J (2015) Efficient production of indigoidine in Escherichia coli. J Ind Microbiol Biotechnol 42(8):1149–1155. https://doi.org/10.1007/s10295-015-1642-5
Yang B, Liu H, Yang J, Gupta VK, Jiang Y (2018) New insights on bioactivities and biosynthesis of flavonoid glycosides. Trends Food Sci Technol 79:116–124. https://doi.org/10.1016/j.tifs.2018.07.006
Yang Y, Wu T, Yang WX, Aisa HA, Zhang TY, Ito Y (2008) Preparative isolation and purification of four flavonoids from Flos Gossypii by high-speed countercurrent chromatography. J Liq Chromatogr Relat Technol 31(10):1523–1531. https://doi.org/10.1080/10826070802039663
Yonekura-Sakakibara K, Hanada K (2011) An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J 66(1):182–193. https://doi.org/10.1111/j.1365-313X.2011.04493.x
Yu D, Xu F, Valiente J, Wang S, Zhan J (2013) An indigoidine biosynthetic gene cluster from Streptomyces chromofuscus ATCC 49982 contains an unusual IndB homologue. J Ind Microbiol Biotechnol 40(1):159–168. https://doi.org/10.1007/s10295-012-1207-9
Yu D, Xu F, Zhang S, Zhan J (2017) Decoding and reprogramming fungal iterative nonribosomal peptide synthetases. Nat Commun 8(1):1–11. https://doi.org/10.1038/ncomms15349
Zeng J, Yang N, Li XM, Shami PJ, Zhan J (2010) 4’-O-Methylglycosylation of curcumin by Beauveria bassiana. Nat Prod Commun 5(1):77–80. https://doi.org/10.1177/1934578X1000500119
Zhan J, Gunatilaka AA (2006) Microbial transformation of amino- and hydroxyanthraquinones by Beauveria bassiana ATCC 7159. J Nat Prod 69(10):1525–1527. https://doi.org/10.1021/np060339k
Zhan J, Leslie Gunatilaka AA (2006) Selective 4’-O-methylglycosylation of the pentahydroxy-flavonoid quercetin by Beauveria bassiana ATCC 7159. Biocatal Biotransfor 24(5):396–399. https://doi.org/10.1080/10242420600792169
Zhao X, Wang P, Li M, Wang Y, Jiang X, Cui L, Qian Y, Zhuang J, Gao L, Xia T (2017) Functional characterization of a new tea (Camellia sinensis) flavonoid glycosyltransferase. J Agric Food Chem 65(10):2074–2083. https://doi.org/10.1021/acs.jafc.6b05619
Funding
This work was supported by the National Science Foundation Award CBET-2044558. The Bruker Avance III HD Ascend-500 NMR instrument used in this research was funded by the National Science Foundation Award CHE–1429195.
Author information
Authors and Affiliations
Contributions
JR, WT, and JZ conceived and designed research. JR, WT, CDB, OP, MWM, AP, BW, SEH, and LPS conducted experiments. JR, WT, JH, and JZ analyzed data. JR, WT, and JZ wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jie Ren and Wenzhu Tang contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ren, J., Tang, W., Barton, C.D. et al. A highly versatile fungal glucosyltransferase for specific production of quercetin-7-O-β-d-glucoside and quercetin-3-O-β-d-glucoside in different hosts. Appl Microbiol Biotechnol 106, 227–245 (2022). https://doi.org/10.1007/s00253-021-11716-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11716-x