Abstract
To directly produce β-alanine from glucose by microbial fermentation, a recombinant Corynebacterium glutamicum strain with high efficiency of β-alanine production was constructed in this study. To do this, the biosynthetic pathway of β-alanine in an L-lysine-producing strain XQ-5 was modified by enhancing carbon flux in biosynthetic pathway and limiting carbon flux in competitive pathway. This study showed that replacement of L-aspartate kinase (AK) with wild-type AK and disruption of lactate dehydrogenase and alanine/valine aminotransferases increase β-alanine production because of decreasing the by-products accumulation. Moreover, L-aspartate-α-decarboxylase (ADC) from Bacillus subtilis was designed as the best enzyme for increasing β-alanine production, and its variant (BsADCE56S/I88M) showed the highest activity for catalyzing L-aspartate to generate β-alanine. To further increase β-alanine production, expression level of BsADCE56S/I88M was controlled by optimizing promoter and RBS, indicating that Pgro plus ThirRBS is the best combination for BsADCE56S/I88M expression and β-alanine production. The resultant strain XQ-5.5 produced 30.7 ± 2.3 g/L of β-alanine with a low accumulation of lactate (from 5.2 ± 0.14 to 0.2 ± 0.09 g/L) and L-alanine (from 7.6 ± 0.22 to 3.8 ± 0. 32 g/L) in shake-flask fermentation and produced 56.5 ± 3.2 g/L of β-alanine with a productivity of 0.79 g/(L·h) and the glucose conversion efficiency (α) of 39.5% in feed-batch fermentation. This is the first report of genetically modifying the biosynthetic pathway of β-alanine that improves the efficiency of β-alanine production in an L-lysine-producing strain, and these results give us a new insight for constructing the other valuable biochemical.
Key points
• Optimization and overexpression of the key enzyme BsADC increased the accumulation of β-alanine.
• The AK was replaced with wild-type AK to increase the conversion of aspartic acid to β-alanine.
• A 56.5-g/L β-alanine production in fed-batch fermentation was achieved.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
β-Alanine, also known as 3-aminopropionic acid, is the only natural β-amino acids and non-protein amino acids. As a kind of nutritional factor, β-alanine is wildly used as sports nutritional supplement or animal feed additive (Lei et al. 2020). β-alanine is an important L-aspartate derivative and a precursor for the synthesis of pantothenic acid and CoA; thus, it is mostly used in food, medicine, chemistry, feed, and other industries and also be used as an important platform chemical to produce other macromolecular substances (White 2001). At present, β-alanine is mainly produced by chemical synthesis and enzymatic synthesis in industry (White 2001). Based on the biosynthetic pathway of β-alanine (Fig. 1), L-aspartate is direct precursor of β-alanine, whereas fumaric acid is used as the precursor for L-aspartate production catalyzed by aspartase (Pei et al. 2017). Therefore, β-alanine could be produced by two-step enzymatic synthesis from fumaric acid (Song et al. 2015; Mingliang et al. 2018). In order to further increase β-alanine production, many researches focused on genetically modifying the key enzymes in two enzyme-catalyzed reactions (Gao et al. 2017; Mingliang et al. 2018; Lei et al. 2020). As the main substrate, however, the fumaric acid is made from petroleum that results in the high production cost. In addition, climate and environmental issues are becoming increasingly serious, and the use of petroleum and its derivatives are not conducive to the long-term development of society. Therefore, green and sustainable microbial fermentation urgently needed to be applied in β-alanine production in industry.
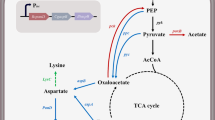
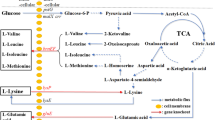
The metabolic pathway designed for the production of β-alanine and key metabolic engineering strategies. The main metabolic pathways are shown. The red cross indicates deletion of the corresponding genes. The genes, which were reformed to increase the production of β-alanine in the genome, are shown in purple. The red words indicate the main products. Enzymes encoded by the genes shown are avtA, alanine-valine aminotransferase; alaT, alanine aminotransferase; ldh, lactate dehydrogenase; lysC, Aspartate kinase; panD, L-aspartate-α-decarboxylase; aspB, aspartase; and aspC, L-aspartate aminotransferase. L-Asp L-aspartate, β-Ala β-alanine, L-Ala L-alanine, L-Lys L-lysine, Fum fumaric acid, TCA tricarboxylic acid cycle, PEP phosphoenolpyruvate, OAA oxaloacetate
Besides whole-cell catalysis for β-alanine production, microbial fermentation and metabolic modifications have been investigated to produce β-alanine (Li et al. 2018). And some researches indicated that it has great potential for the production of β-alanine by fermentation as compared to the purified enzymes and whole-cell method (Ziert 2014; Piao et al. 2019). To make the fermentation production of β-alanine from glucose with metabolic modifications, the primary work is developing an optimized metabolic pathway to maximize the yield of β-alanine. In the biosynthetic pathway of β-alanine, L-aspartate-α-decarboxylase (ADC, encoded by panD) has become the focal point for β-alanine production, which catalyzes the decarboxylation of L-aspartate (Williamson 1985; Poelje and Snell 1990; Chao et al. 2000; Leuchtenberger et al. 2005). Thus, selecting the best ADC from different sources is a novelty way to increase the production of β-alanine. Recently, the ADC from Escherichia coli (E. coli), Bacillus tequilensis, Corynebacterium glutamicum (C. glutamicum), Mycobacterium tuberculosis (M. tuberculosis), and Tribolium castaneum was introduced to improve the conversion of L-aspartate to β-alanine and indicated that ADC from Bacillus subtilis (B. subtilis) showed a higher enzyme activity and β-alanine productivity in whole-cell biocatalyst (Dusch et al. 1999; Gopalan et al. 2010; Feng et al. 2019; Liu et al. 2019; Zou et al. 2020). In addition, site-directed mutations were adopted to improve the enzyme activity and catalyze stability of ADC (Pei et al. 2017; Zhang et al. 2018). Besides the modification of ADC, the compete pathway also attracted some researchers to construct β-alanine high-producing strain. For example, Liang et al. inactivated the key enzyme-coding gene in by-products synthesis pathway and catabolic pathway of β-alanine to increase β-alanine by E. coli (Ziert 2014). However, the production of β-alanine by the modification of biosynthetic pathway and compete pathway is also too low to realize industrialized production at present. Therefore, how to construct a strain with high efficiency of β-alanine production is the top priority for increasing β-alanine production by microbial fermentation.
As the work-horse for producing amino acids, C. glutamicum has been also modified to produce β-alanine through increasing the precursor supply and decreasing the by-products accumulation (Ziert 2014). C. glutamicum XQ-5 is an L-lysine-producing strain, which was derived from the wild-type strain C. glutamicum ATCC13032 after multiple rounds of random mutagenesis and was resistant to S-2-aminoethyl-L-cysteine (AECr), 2-thiazolealanine (2-TAr), and monfluoroacetate (MFr) as well as was sensitive to L-methionine (Mets) (Wang et al. 2020a, b). Since L-aspartate is the important precursor for producing L-lysine and β-alanine, C. glutamicum XQ-5 has huge potential as chassis cells to produce β-alanine. In this study, this L-lysine-producing strain C. glutamicum XQ-5 was used as chassis cell to construct the β-alanine high-producing strain from glucose. Firstly, we restored the feedback inhibition of aspartate kinase (AK) to block L-lysine production in strain XQ-5. Then, the best ADC was screened from fifteen ADC in different branch of ADC phylogenetic tree indicating that BsADC is the best for β-alanine production in C. glutamicum. After that, the expression level of ADC was optimized by modifying of the promoter, RBS and ORF of BsADC-coding gene to further enhance the carbon flux in β-alanine biosynthetic pathway. Finally, the precursor of β-alanine was increased by overexpression of aspartase and disruption of the compete pathway. The resultant strain C. glutamicum XQ-5.5 produced 56.5 ± 3.2 g/L of β-alanine with a productivity of 0.79 g/(L·h) and the glucose conversion efficiency (α) of 39.5% in feed-batch fermentation. The present work provides a valuable strategy for producing other value-added chemicals that precursor of the target product high-producing strain can be used as the effective chassis cell to develop the high-yielding strain.
Material and methods
Microbial strains and plasmids
The E. coli JM109 was used for plasmid construction, and E. coli BL21 was be used for plasmid expression. C. glutamicum XQ-5 was used as the host strain for β-alanine production (Wang et al. 2020a, b). lysC gene was amplified from C. glutamicum 13032 and ligated to plasmid pK18mobSacB by homologous recombination. panD gene was ligated to pEC-XK99E plasmid by EcoRI/PstI. For exogenous gene integration, the relative exogenous genes were firstly inserted into the appropriate endonuclease site downstream of the Ptrc promoter in pEC-XK99E. The resultant plasmids were named as pEC-panD. The accession numbers of relevant nucleotide sequences are listed in Online Resource Table S1. The promoters Peftu, PlacM, Pgro, GroRBS, MaxRBS, SecRBS, ThirRBS, FourRBS, and FifRBS were combined on the plasmid pEC-BspanD by homologous recombination. aspB gene was amplified from Bacillus thermophilus YM55-1. The alaT gene was amplified from XQ-5 strain by using T7 terminator as primer, and then homologous recombination was performed on pK18 plasmid. Restriction endonucleases (Takara) and the DNA Ligase Kit Ver. 2.0 (Takara) were used to construct plasmids. For strain construction, plasmids were transformed into C. glutamicum by electroporation. All constructed plasmids including chromosomal deletions and integrations in the engineered strains were verified by DNA sequencing.
In this study, the plasmid pEC-XK99E was used for gene overexpression in C. glutamicum. The suicide plasmid pK18mobsacB was used for gene knockout and replace in C. glutamicum. Firstly, the constructed plasmid was electroporated into C. glutamicum, and then the positive transformants were screened with a 25 μg/mL kanamycin solution in LBH medium. The final positive transformants were obtained by eliminating the plasmids according to the sucrose lethal principle. The changes in the chromosome were verified by PCR analysis. The strains and plasmids constructed in this study are listed in Table 1. Promoters and the RBS sequences are listed in Table 2. In addition, the primers are listed in Online Resource Table S2.
Medium and culture conditions
For gene manipulation and plasmid construction, E. coli cells were grown at 37 °C in Luria–Brentani (LB) liquid broth or on LB agar plate (1.5% agar, w/v) containing (per L) tryptone 10 g, yeast extract 5 g, and NaCl 10 g. The expanded culture of C. glutamicum were performed at 30 °C in LBG medium containing (per L) tryptone 10 g, yeast extract 5 g, glucose 5 g, and NaCl 10 g. EPO medium and LB-Brain Heart Infusion-Sorbitol (LBHIS) medium were used to construct the recombinant bacteria (Rest et al. 1999). The expression of proteins was performed in TB medium containing (per L) tryptone 12 g, yeast extract 24 g, KH2PO4 2.31 g, K2HPO4 16.42 g, and glycerin 4 mL. When needed, 50 μg/mL kanamycin was added to the medium.
The single colony was inoculated in LBG liquid medium and incubated at 30 °C for 12 h with rotation speed 100 r/min. Next, 5 mL of the seed culture was transferred to 50 mL of the fermentation medium in a standard 500-mL shake flask and was cultured for 72 h at 30 °C with rotation speed 100 r/min. The fermentation medium in shake flask contained (per liter) 100 g glucose, 10 g beet molasses, 8 g corn steep powder, 40 g (NH4)2SO4, 0.02 g FeSO4, 0.02 g MnSO4, 450 μg thiamine, 8 mg niacinamide, 850 μg biotin, 0.6 mg ZnSO4, 0.53 g KCl, 1 g KH2PO4, 1 g K2HPO4, 4 g MgSO4·7H20, 50 mg betaine, and 40 g CaCO3. The fermentation medium in 5-L jar fermenter (BLBio-5GJ-2-H, Bailun Bi-Technology Co. Ltd., Shanghai, China) contained (per liter) 80 g glucose, 50 g beet molasses, 40 g (NH4)2SO4, 20 g corn steep powder, 2 g KH2SO4, 1.5 g MgSO4.7H20, 0.03 g FeSO4, 0.02 g MnSO4, 0.03 g glycine betaine, 600 μg biotin, 300 μg thiamine·HCl, and 2 mL antifoam. The temperature, pH, and the relative dissolved oxygen were set according to the previous reports (Xu et al. 2019), which were controlled by inherent equipment in fermenter OD600, and glucose concentration and β-alanine concentration were determined every 4 h during fermentation. To maintain the glucose concentration at about 5 g/L, the feed solution prepared according to our previous reports was used by adjusting the feeding rate (Xu et al. 2019).
Enzyme activity analysis
The expression conditions were as follows: 37 °C, 100 rpm, 50 μg/mL kanamycin in 10-mL LB flask for 12 h, then transferred to TB with 5% inoculum. When the culture was shaken to OD600 between 0.5 and 0.6 at 37 °C, inducer IPTG (0.5–1 mmol/L) was added. Then it was cultured at 16 °C overnight. The bacteria were collected and washed with PBS buffer, and then the supernatant was collected after sonication for 10 min. L-Aspartate was catalyzed into β-alanine by supernatant, and the yield of β-alanine was determined by high-performance liquid chromatography (HPLC). The reaction system consists of 100 μL 100 g/L L-aspartate, 100 μL crude enzyme, and 300 μL PBS buffer. One unit of ADC activity was defined as the amount of enzyme that catalyzes the reaction to produce 1 μmol of β-alanine per minute under the described conditions.
Product detection conditions
The derivatization of β-alanine was performed by o-phthal-dialdehyde (OPA). Samples were analyzed by a high-performance liquid chromatography (HPLC) system (Agilent 1290 series; Agilent, Palo Alto, CA, USA) equipped with a C18 column (250 mm × 4.6 mm, 5 μm, Waters, Milford, MA, USA). The column was maintained at 30 °C. The compounds were detected at a wavelength of 360 nm with a UV detector. The mobile phase (supplied at 1 mL/min) consisted of a solution of (A) pH 6.2, 200 mM NaAc buffer and (B) acetonitrile ramping (A: B) from 95:5 to 60:40 over 25 min (Song et al. 2015; Feng et al. 2019).
Results
Restore the feedback inhibition of aspartate kinase (AK) to limit the carbon flux in L-lysine biosynthetic pathway
In microbial metabolism, L-aspartate was used as co-precursor to form L-lysine and β-alanine. However, the synthesis of L-lysine is inhibited in wild-type strains because of the feedback inhibition of AK (Xunyan et al. 2016). Thus, decreasing the carbon flux in L-lysine biosynthetic pathway increased the β-alanine yield from L-aspartate (Liang et al. 2017). In this study, the L-lysine high-producing strain C. glutamicum XQ-5 was used as β-alanine producing strain due to the fact that L-lysine and β-alanine have the same precursor L-aspartate (Hou et al. 2012). The nucleotide sequence analyses indicated that the AK-coding gene in strain XQ-5 was mutated at loci 932 to relieve the feedback inhibition of L-lysine (Fig. 2a) (Ohnishi et al. 2002). In order to restore the feedback inhibition of AK in strain XQ-5, this mutated AK was replaced by the wild-type AK from strain ATCC13032, resulting in strain XQ-5.1. As expected, the yield of L-lysine in strain XQ-5.1 was dramatically decreased as compared with original strain XQ-5 (from 48.5 ± 1.8 g/L to 2.2 ± 0.2 g/L), whereas the final β-alanine titer of strain XQ-5.1 was increased more than ten times than strain XQ-5 (0.5 ± 0.1 g/L vs 5.6 ± 0.6 g/L) (Fig. 2b). It should be noted that the β-alanine was obviously produced after mid-log phase until fermentation at 60 h (Fig. 2c). Interestingly, strain XQ-5.1 showed the better cell growth than original strain XQ-5, indicating that more carbon source was used to cell growth rather than to β-alanine biosynthesis.
The variation of strains XQ-5 and XQ-5.1. a Comparison of aspartate kinase sequence between strains XQ-5 and XQ-5.1. The 932 base was changed from T base to C base in XQ-5.1 that represents that the restoring the feedback inhibition of aspartate kinase. b The yield of L-lysine and β-alanine in strains XQ-5 and XQ-5.1. c Variation of OD600 and β-alanine concentration during fermentation in strain XQ-5 and XQ-5.1. These data represent average values and standard deviations achieved from three independent fermentation experiments
Mining a best L-aspartate-α-decarboxylase to reconstruct the biosynthetic pathway of β-alanine
In order to increase β-alanine production, carbon source should be redirected to produce β-alanine biosynthesis from cell growth. It is well-known that L-aspartate was catalyzed to form β-alanine only by ADC. Thus, the ADC with high activity and high stability is very important for β-alanine production (Leuchtenberger et al. 2005). Therefore, it is reasonable to presume that the low β-alanine production in strain XQ-5.1 may due to the low conversion between L-aspartate and β-alanine. To do this, the best ADC should be optimized at first. In this study, fifteen ADCs from different microbial hosts (i.e., E. coli, M. tuberculosis, B. subtilis, C. glutamicum, Pseudomonas aeruginosa, Muricauda ruestringensis, Ruminiclostridium termitidis, Staphylococcus aureus, Salmonella enterica, Bordetella pertussis, Enterobacter hormaechei, Sulfobacillus acidophilus, Aequorivita sublithincola, Frankia sp. CeD, and Bacteroidetes bacterium) were selected based on the phylogenetic tree of ADCs (Fig. 3a). To analyze enzymatic properties, theses ADC-coding genes were overexpressed in E. coli BL21 using E. coli-C. glutamicum shuttle expression plasmid pEC-XK99E. SDS-PAGE data indicated that these ADCs were successfully expressed and the molecular mass of π-protein was evident, which was equal to the calculated molecular weights about 11.1–11.7 KDa (Fig. 3b). However, subunits were too small to see (Fig. 3b). In addition, all ADCs showed the enzyme activity of ADC, but the enzyme activity differences among these ADCs are huge (Fig. 3c). Among these ADCs, BsADC showed the highest enzyme activity (i.e., 7.5 ± 0.16 U/(mg protein)), whereas RtADC showed the lowest (i.e., 1.7 ± 0.18 U/(mg protein)) (Fig. 3c). These results indicated that the ADCs from different strains can be expressed and showed the enzyme activity under the genetic background of pEC-XK99E.
Selecting the best L-aspartate-α-decarboxylase for producing β-alanine. a The phylogenetic tree of ADCs from different strains. The strains marked are selected as test strains in this experiment. b SDS-PAGE of plasmids pEC-panDs. 1 blank control, 2–16 crude enzymes, M protein marker. The target molecular weight is 14 KDa which was marked by red box. c Comparison of β-alanine production in the engineered strains overexpressing ADCs. The deep bar represents overexpressing BsADC obtained the highest yield. d Enzyme activities of different ADCs. The light blue bar represents BsADC has the highest enzyme activity. These data represent average values and standard deviations achieved from three independent fermentation experiments
Subsequently, the above-mentioned E. coli-C. glutamicum shuttle expression plasmid pEC-XK99E-XpanD (“X” represents the different microbial hosts) was transferred into strain XQ-5.1 to investigate the β-alanine production. As expected, All ADCs overexpression strains were able to accumulate β-alanine, indicating that these ADCs are the functional forms of ADC (Fig. 3d). As can be seen from Fig. 3d, the recombinant strain XQ-5.1/pEC-XK99E-BspanD with overexpression of BsADC showed the highest β-alanine production (i.e., 15.3 ± 1.4 g/L), whereas the lower β-alanine production (i.e., ≤ 7 g/L) was found in other recombinant strains (i.e., XQ-5.1/pEC-XK99E-SaDpanD, XQ-5.1/pEC-XK99E-FrpanD and XQ-5.1/pEC-XK99E-AspanD). The specific values are listed in Online Resource Table S3. Based on these results, we conceived that heterogeneous expression of BsADC in strain XQ-5.1 is beneficial to construct a β-alanine producing strain of good productive performance.
Optimization the expression level of L-aspartate-α-decarboxylase to enhance the carbon flux in β-alanine biosynthetic pathway
As mentioned above, BsADC showed the best catalytic performance for producing β-alanine (Fig. 3d). However, BsADC belongs to pyridoxal phosphate-dependent enzymes, which has an inherent flaw of mechanism-based inactivation (Zhang et al. 2018). In recent years, many studies focusing on improving catalytic stability of BsADC by mutating key sites have been reported, for example, BsADCE56S variant (Zhang et al. 2018) as well as BsADCV68I and BsADCI88M variants (Pei et al. 2017). In addition, previous report indicated that the C-terminus of ADC shows the critical role in increasing the stability of the enzyme (Pei et al. 2017). To increase catalytic stability and enzyme activity of BsADC, iterative mutation was performed at position 56, 88, and 126 amino acids residues in this study. As can be seen from Fig. 4a, all BsADC mutants showed the increased enzyme activity. Among these variants, BsADCE56S/I88M exhibited the highest enzyme activity, which increased by 60%. It should be noted that three-mutational variant (i.e., BsADCE56S/I88M/I126*) based on the BsADCE56S/I88M variant did not obviously increase the enzyme activity although single-mutational variant (i.e., BsADCI126*) was beneficial to increase enzyme activity, in which enzyme activity of BsADCE56S/I88M/I126* increased by 56% while enzyme activity of BsADCI126* increased by 54% (Fig. 4a). The specific values are listed in Online Resource Table S4. Thus, this two-mutational variant BsADCE56S/I88M was used to modify to further increase the activity of ADC.
Optimizing the best L-aspartate-α-decarboxylase for producing β-alanine. a Enzyme activities of different mutation of BsADC. The light green bar represents the best mutant for producing β-alanine. b Comparison of β-alanine production in the engineered strains expressing BsADC driving by different promoter and RBS combinations. An array of combination cassettes designed to promote BsADC expression, which were named A–R. The red bar represents the best combination of Pgro promoter and ThirRBS for producing β-alanine. The data represent mean values and standard deviations obtained from three independent cultivations. c Variation of OD600 and β-alanine concentration during fermentation in strain XQ-5 and XQ-5.1. These data represent average values and standard deviations achieved from three independent fermentation experiments
To do this, promoter and RBS were optimized, resulting in different expression levels of ADC (Gupta and Srivastava 2021), to obtain the best BsADC variant with high catalytic efficiency. In this study, three strong promoters (PlacM, Peftu, Pgro (Haefner et al. 2005)) and six RBSs with different predicted expression levels were investigated, and these six RBSs were named GroRBS, MaxRBS, SecRBS, ThirRBS, FourRBS, and FifRBS, respectively. The above-mentioned promoters and RBSs were randomly coupled. Firstly, the Ptrc promoter of pEC-BsADC was replaced by three promoters, respectively. For the next study, we deleted part of sequence of gene lacIq. Then six RBS were integrated into plasmids by seamless cloning. Finally, we formed eighteen recombinants constitutive expression plasmids (Fig. 4b). These plasmids transferred into strain XQ-5.1 by electroporation method to analyze the production of β-alanine. As can be seen from Fig. 4b, these recombinant strains of optimizing promoter and RBS showed the increased β-alanine production as compared with the original strain XQ-5.1, while the cell growth was not obviously changed after genetic modification. Interestingly, the high expression level in theory (i.e., with the strong promoter and the high-level RBS) was not beneficial to increase β-alanine production (Fig. 4b). For example, Pgro plus ThirRBS was the best combination for β-alanine production, and the resultant strain XQ-5.1/pEC-Pgro-ThirRBS-BspanD produced 28.7 ± 0.2 g/L of β-alanine after 72-h cultivation in shake flask (Online Resource Table S5). In order to avoid using antibiotics to maintain the stability of recombinant expression plasmid, we tried to introduce the expression cassette of BsADCE56S/I88Mwith Pgro and ThirRBS at CgpanD gene loci in strain XQ-5.1. Finally, the resultant strain C. glutamicum XQ-5.1 CgpanD::Pgro-ThirRBS-BsADCE56S/I88M (i.e., strain XQ-5.2) produced 25.8 ± 0.7 g/L of β-alanine, increasing 4.6 times compared with strain XQ-5.1 (Fig. 4c).
Overexpression of aspartase to enhance the precursors supply for increasing β-alanine production
In microbial metabolism, glucose was converted to L-aspartate by tricarboxylic acid cycle or other metabolic pathways (Fig. 1). In order to further increase the production of β-alanine, we try to increase the L-aspartate supply to producing β-alanine, because many studies reported that increasing the carbon flux of L-aspartate is beneficial to the accumulation of β-alanine (Song et al. 2015). Aspartase (encoded by aspB) reversibly catalyzes fumarate to form L-aspartate (Veetil et al. 2010; Fibriansah et al. 2011). In this study, aspB from Bacillus thermophilus YM55-1 (i.e., BtaspB), which has high enzyme activity and amination ability (Song et al. 2015), was firstly overexpressed to enhance L-aspartate supply under the control of plasmid pEC-XK99E (Fig. 5a). But in order to improve the expression stability of aspB, the CgaspA gene loci in strain XQ-5.2 was replaced by the expression cassette of BtaspB with promoter Ptrc, resulting in strain XQ-5.3. Unexpectedly, the yield of β-alanine in strain XQ-5.3 was not significantly increased as compared with strain XQ-5.2 (from 25.8 ± 1.7 g/L to 26.9 ± 1.1 g/L), only increasing by 4.3% (Fig. 5b).
The variation of strains XQ-5.2 and XQ-5.3. a SDS-PAGE of plasmid pEC-aspB. 1 blank control, 2 crude enzyme, M protein marker. The target molecular weight is 52 KDa which was marked by red box. b OD600 and β-alanine concentration during fermentation in strain XQ-5.2, XQ-5.3. These data represent average values and standard deviations achieved from three independent fermentation experiments
Blocking competitive pathway to decrease by-products accumulation
Although strain XQ-5.3 did not obviously increase the β-alanine production, it accumulated large number of by-products, such as lactate and L-alanine (Fig. 6a). In order to decrease the by-products accumulation and to increase carbon source for β-alanine production, the competitive consumption pathway of pyruvate was deleted. Firstly, lactate dehydrogenase (encoded by ldh), which is the key enzyme in lactate production, was deleted to reduce lactate biosynthesis, resulting in strain XQ-5.4. Furthermore, the L-alanine biosynthetic pathway was modified to decrease L-alanine accumulation in the next experiment. Given that L-alanine is essential for cell growth, the L-alanine biosynthetic pathway should be rationally modified to maintain cell growth. To do this, we try to delete avtA gene (encoding alanine-valine aminotransferase) and add T7 (strength 243 a.u.) terminator (Chen et al. 2013) in front of alaT gene (encoding alanine aminotransferase) to reduce L-alanine biosynthesis, resulting in strain XQ-5.5 (Marienhagen and Eggeling 2008). As can be seen from Fig. 6a, the lactate accumulation in strain XQ-5.5 was dramatically decreased (from 5.6 ± 0.3 to 0.2 ± 0.1 g/L). Compared with strain XQ-5.3 and XQ-5.5, the production of L-alanine in strain XQ-5.5 was significantly decreased (Fig. 6a). The specific values are listed in Online Resource Table S6. As we expected, moreover, the yield of β-alanine in strain XQ-5.5 was further increased, reaching 30.7 ± 2.3 g/L (Fig. 6b). However, the growth of strain XQ-5.5 was obviously decreased as compared with strain XQ-5.3, in which the OD600 of strain XQ-5.5 was 28.6 ± 1.7 while the OD600 of strain XQ-5.3 was 36.3 ± 1.2 (Fig. 6b).
Variation of strains XQ-5.3 and XQ-5.5. a Variation of by-products concentration in strain XQ-5.3 and XQ-5.5. b Variation of OD600 and β-alanine concentration in strain XQ-5.3 and XQ-5.5. These data represent average values and standard deviations achieved from three independent fermentation experiments
Fed-batch fermentation
To test the ability of strain XQ-5.5 for β-alanine production, the production performance of the strain XQ-5.5 was investigated in fed-batch fermentation. Figure 7 shows the time profiles of strain XQ-5.5 by fed-batch fermentations in a 5-L jar fermenter. During the entire fermentation period, about 143±6.0 g/L of glucose was consumed (Fig. 7). Different from the fermentation in shake flask, interestingly, β-alanine was first accumulated at the logarithmic early growth (i.e., at 8 h) and then continually increased to 56.5±3.2 g/L at 72 h (Fig. 7). By-products’ concentration also increased. The specific values are listed in Online Resource Table S7. The overall β-alanine productivity was about 0.79 g/(L·h), and the glucose conversion efficiency (α) was 39.5% after 72 h. In addition, strain XQ-5.5 grew slowly, and it reached to the maximum value (i.e., OD600=43.2) after 36 h, which was consistent with that in shake flask (Figs. 6b and 7).
β-alanine fed-batch fermentations of strain XQ-5.5. The β-alanine production (squares, brown), OD600 (triangle, green), and glucose (circle, purple) of strains cultivated in 5-L fermenters. The data represent mean values and standard deviations obtained from three independent cultivations. These data represent average values and standard deviations achieved from three independent fermentation experiments
Discussion
In recent years, the demand of β-alanine has continued to increase for its wide applications in industry. With the development of genetic engineering technology, a β-alanine high-yielding strain based on metabolic engineering has become a research focus. Most previous studies mainly focus on how to increase the production of β-alanine in E. coli (Song et al. 2015; Liang et al. 2017; Li et al. 2018). So far, C. glutamicum is commonly used to produce amino acids and organic acids. The engineering attempt for the direct fermentation production of β-alanine is rarely reported and usually associated with very low production in C. glutamicum (Ziert 2014). Therefore, it is necessary to produce high-yield β-alanine in C. glutamicum. It should be noted that the biosynthesis of L-lysine and β-alanine shows the same precursor, i.e., L-aspartate (Piao et al. 2019). In the present study, we devote to reasonably modify an L-lysine high-yielding strain C. glutamicum XQ-5 to construct a β-alanine high-producing strain. To do this, the AK in L-lysine high-yielding strain XQ-5 was replaced by wild-type AK from strain ATCC13032 to redirect L-aspartate into β-alanine biosynthetic pathway rather than to L-lysine biosynthetic pathway. Then, the β-alanine biosynthetic pathway was further enhanced by selecting and optimizing ADC expression level, introducing exogenous aspB, and blocking the competitive pathway. The specific values of shake-flask fermentation yield of strains are listed in Online Resource Table S8. As a result, a β-alanine high-producing strain C. glutamicum XQ-5.5 was obtained, which produced 56.5 ± 3.2 g/L of β-alanine with productivity of about 0.79 g/(L·h) and the α of 39.5% after 72 h in fed-batch fermentation.
AK is the first key enzyme in the biosynthetic pathway of L-aspartate family amino acid (i.e., AFAA) in C. glutamicum, but it is inhibited by L-lysine and L-threonine (Kato et al. 2004). Thus, the feedback inhibition of AK should be relieved for breeding AFAA high-producing strain except L-aspartate producing strain (Ohnishi et al. 2002). However, our results indicated that the AK with feedback inhibition is beneficial to increase β-alanine production (Fig. 2). The similar results were also found in the previous results, in which AK-deficient strain showed the obvious increase of β-alanine production (Liang et al. 2017). This is because more L-aspartate can be used to biosynthesize β-alanine rather than other products. Interestingly, although the original strain XQ-5 is an L-lysine high-producing strain and the inherent AK in strain XQ-5 was replaced by the wild-type AK from strain ATCC13032 (i.e., strain XQ-5.1), the β-alanine production of strain XQ-5.1 was not increased to the expected value (Fig. 2). We presumed that the ADC in strain XQ-5.1 loses the high catalytic efficiency for β-alanine production because the original strain XQ-5 was obtained by multiple rounds of random mutagenesis. In previous research, different ADCs were selected and used to accumulate efficient β-alanine by whole-cell biocatalyst or fermentation (Dusch et al. 1999; Zhang et al. 2018; Li et al. 2018). These results suggested that overexpressing ADC could increase the production of β-alanine (Liang et al. 2017). In this study, BsADC, selected from different sources of ADC, is proved the best ADC for β-alanine production in C. glutamicum (Fig. 3). These results were consistent with the previous results reported by Pei et al. (2017), in which the BsADC showed higher specific activity and thermostability than CgADC and EcADC (Pei et al. 2017; Zhang et al. 2018). In addition, the mutant BsADCE56S/I88M showed the highest enzyme activity among the other mutations constructed in this study and wild type, which increased by more than 60% (Fig. 4a). Previous results indicated that BsADC variants with mutation of Glu56Ser or I88M exhibited an improved activity and significantly attenuated the mechanism-based ADC inactivation (Pei et al. 2017; Zhang et al. 2018). Interestingly, three-mutational variant (i.e., BsADCE56S/I88M/I126*) did not obviously increase the enzyme activity, although the single-mutational variant (i.e., BsADCI126*) was beneficial to increase enzyme activity (Fig. 4a). The single I126* mutation, which makes the enzyme two amino acid shorter, remarkably changed the activity and catalytic stability of the enzyme. This result suggests that the C-terminus of ADC is an important region which affects its catalysis. However, stacking different mutation points does not necessarily increase the mutation effect (Qian et al. 2018). In order to further increase the BsADC activity, the expression level of BsADCE56S/I88M-coding gene was optimized in this study. Previous results pointed out that promoters of different intensities combined with RBSs of different expression intensity would produce different expression effect at transcription level in C. glutamicum (Duan et al. 2021). And the results of this study confirmed it once more, in which Pgro plus ThirRBS was the best combination for increasing ADC activity and β-alanine production (Fig. 4b and c).
Previous reports have revealed that L-aspartate could be produced through aspartate aminotransferase (encoded by aspC)-catalyzed transamination from OAA and aspartase (encoded by aspB or aspA)-catalyzed direct amination from fumarate (Ziert 2014; Song et al. 2015; Piao et al. 2019). As suggested in our study, aspartase could be used for improving the supply of L-aspartate in XQ-5.2, and the resultant strain XQ-5.3 showed the slight increase of β-alanine from 25.8 ± 1.7 to 26.9 ± 1.1 g/L, only increasing by 4.3% as compared with strain XQ-5.2 (Fig. 5b). The similar results were also found in previous results reported by Zou et al. (2020), in which overexpression of the native aspartase from E. coli combined with overexpression of sdhCDAB operon to increase fumarate supply did not significantly increase β-alanine production because of the enzymatic properties of aspartase (Zou et al. 2020). Aspartase (also referred to as aspartate ammonia lyases) catalyzed the reversible deamination of L-aspartate to yield fumarate and ammonia, and the enzyme has a rather narrow substrate specificity (Fibriansah et al. 2011). These indicated that native aspartase is a rate-limited key enzyme in the conversion from fumarate to L-aspartate, and retinal modification of aspartase could be of great potential to improve L-aspartate supply and β-alanine production in the future. It should be noted that the strain XQ-5.3 accumulated a large number of by-products, such as lactate and L-alanine (Fig. 6a). In previous study, the gene clusters frdABCD (fumarate reductase), genes lysC (aspartate kinase), panC (pantoate–beta-alanine ligase), ptsG (phosphoglucomutase), aspA (aspartase), sdh (succinate dehydrogenase), ldh (L-lactate dehydrogenase), and alaT (alanine aminotransferase) were deleted to decrease by-products accumulation, and the resultant strain also showed the increase of β-alanine production (Ziert 2014; Liang et al. 2017; Piao et al. 2019). In this study, the genes ldh and avtA were deleted, and gene alaT was weaken to block the carbon loss from pyruvate, thus reducing the accumulation of lactate and L-alanine (Fig. 6a). Although the cell growth of strain XQ-5.5 was dramatically decreased as compared with strain XQ-5.3 (Fig. 6b), it showed the better growth performance than that of strain with double deletion of genes avtA and alaT (Marienhagen and Eggeling 2008; Ziert 2014). In C. glutamicum, alanine-valine aminotransferase (encoded by avtA) and alanine aminotransferase (encoded by alaT) are the key enzyme in L-alanine biosynthesis, and L-alanine is essential for cell growth (Marienhagen and Eggeling 2008; Hou et al. 2012), and thus, the L-alanine biosynthetic pathway should be rationally modified to maintain cell growth. For example, deletion of avtA gene combined with attenuation of alaT gene was proved to decrease L-alanine accumulation rather than to obviously decrease cell growth (Marienhagen and Eggeling 2008; Wang et al. 2020a, b). As expected, the resultant strain XQ-5.5 showed the increase of β-alanine production and the decrease of by-products accumulation (Fig. 6). These results have once again proven that the decrease of by-products redirects the carbon flux into β-alanine biosynthesis. Finally, the production performance of strain XQ-5.5 was studied in a fed-batch process. During fermentation in a 5-L fermenter with 1-L media, β-alanine production was started at the logarithmic early growth and then continually increased to 56.5 ± 3.2 g/L at 72 h (Fig. 7). This is different from the fermentation in shake flask (Figs. 6 and 7), but that may be because the component of medium is different between in shake flask and in fermenter (see “Material and methods”). Beet molasses (Mustafa et al. 2020) and corn steep powder (Amartey and Jeffries 1994) are complex nutrients and are rich in growth factor (e.g., vitamin B1 and vitamin H) and inorganic ions (e.g., Mg2+ and Mn2+), thus beneficial to cell growth.
In conclusion, a β-alanine high-yielding strain C. glutamicum XQ-5.5 was obtained from a L-lysine high-producing strain XQ-5 by enhancing carbon flux in biosynthetic pathway and limiting carbon flux in competitive pathway. The resultant strain C. glutamicum XQ-5.5 produced 56.5 ± 3.2 g/L of β-alanine with a productivity of 0.79 g/(L·h) and a α of 39.5% in feed-batch fermentation. Although the productivity is lower than that of strain E. coli FZβA-10 reported by Zou et al. (2020), the yield is the highest value for β-alanine production, as far as we know (Table 3), demonstrating that strain XQ-5.5 is a competitive platform strain for β-alanine production. As can be seen from Fig. 6b, the cell growth was dramatically affected after modification of strain XQ-5. Thus, how to improve the cell growth of β-alanine high-yielding strain is a critical factor for increasing the β-alanine productivity in the next study. In addition, as the metabolites of β-alanine, pantothenic acid accumulation will lead to the decrease of β-alanine accumulation. Although deletion of gene panC could block β-alanine breakdown, the cell growth of this mutant was obvious disrupted (Liang et al. 2017). So, decrease of pantothenic acid through weakening the expression of key enzymes is a better way to increase the production of β-alanine (Sahm and Eggeling 1999).
Data availability
Construction of recombinant plasmids and strains; primer pairs used in this study; analyzing the expression of genes; analyzing of β-alanine production by shake-flask fermentation; the ADC genes used in the study; primer pairs used in the study.
All data generated or analyzed during this study are included in the published article.
Code availability
Not applicable
References
Amartey S, Jeffries TW (1994) Comparison of corn steep liquor with other nutrients in the fermentation of D-Xylose by Pichia stipitis CBS 6054. Biotechnol Lett 16(2):211–214. https://doi.org/10.1007/BF01021673
Chao YP, Lo TE, Luo NS (2000) Selective production of L-aspartic acid and L-phenylalanine by coupling reactions of aspartase and aminotransferase in Escherichia coli. Enzyme Microb Technol 27(1–2):19–25. https://doi.org/10.1016/S0141-0229(00)00149-6
Chen YJ, Liu P, Nielsen AK, Brophy J, Clancy K, Peterson T, Voigt CA (2013) Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods 10(7):659–664. https://doi.org/10.1038/nmeth.2515
Duan Y, Zhai W, Liu W, Zhang X, Shi JS, Zhang X, Xu Z (2021) Fine-tuning multi-gene clusters via well-characterized gene expression regulatory elements: case study of the arginine synthesis pathway in C. glutamicum. ACS Synth Biol 10(1):38–48. https://doi.org/10.1021/acssynbio.0c00405
Dusch N, Pühler A, Kalinowski J (1999) Expression of the Corynebacterium glutamicum panD gene encodingL-aspartate-alpha-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl Environ Microbiol 65(4):1530–1539. https://doi.org/10.1109/5.118982
Feng Z, Zhang J, Chen G, Ge Y, Zhu H (2019) Extracellular expression of L-aspartate-α-decarboxylase from Bacillus tequilensis and its application in the biosynthesis of β-alanine. Appl Biochem Biotechnol 189(10). https://doi.org/10.1007/s12010-019-03013-1
Fibriansah G, Veetil VP, Poelarends GJ, Thunnissen A-MWH (2011) Structural basis for the catalytic mechanism of aspartate ammonia lyase. Biochem 50(27):6053–6062. https://doi.org/10.1021/bi200497y
Gao L, Qiu J (2007) Research advances in L-aspartate decarboxylase. Ind Microbiol 37:54–59. https://doi.org/10.1016/S1872-2040(07)60059-0
Gao Y, Liu Z, Liu K, Zhou Z, Cui W (2017) Biocatalytic access to β-alanine by a two-enzyme cascade synthesis. Chin J Biotechnol 33(5):875–879. https://doi.org/10.13345/j.cjb.160416
Gopalan G, Chopra S, Ranganathan A, Swaminathan K (2010) Crystal structure of uncleaved L-aspartate-α-decarboxylase from Mycobacterium tuberculosis. Proteins 65(4):796–802. https://doi.org/10.1002/prot.21126
Gupta JK, Srivastava S (2021) The effect of promoter and RBS combination on the growth and glycogen productivity of sodium-dependent bicarbonate transporter (SbtA) overexpressing Synechococcus sp. PCC 7002 cells. Front Microbiol 12(April 2021):607411. https://doi.org/10.3389/fmicb.2021.607411
Haefner S, Klopprogge C, Kroeger B, Schroeder H, Zelder O (2005) Pgro expression units
Hou X, Chen X, Yue Z, He Q, Zhang W (2012) (L)-Valine production with minimization of by-products’ synthesis in Corynebacterium glutamicum and Brevibacterium flavum. Amino Acids 43(6):2301–2311. https://doi.org/10.1007/s00726-012-1308-9
Kato C, Kurihara T, Kobashi N, Yamane H, Nishiyama M (2004) Conversion of feedback regulation in aspartate kinase by domain exchange. Biochem Biophys Res Commun 316(3):802–808. https://doi.org/10.1016/j.bbrc.2004.02.122
Lei W, Xiaoyu P, Shumei C, Meirong H, Yong T (2020) Enhanced production of β-alanine through co-expressing two different subtypes of l-aspartate-α-decarboxylase. J Ind Microbiol Biotechnol 6–7:6–7. https://doi.org/10.1007/s10295-020-02285-5
Leuchtenberger W, Huthmacher K, Drauz K (2005) Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol 69(1):1–8. https://doi.org/10.1007/s00253-005-0155-y
Li H, Lu X, Chen K, Yang J, Zhang A, Wang X, Ouyang P (2018) β-alanine production using whole-cell biocatalysts in recombinant Escherichia coli. Mol Catal 449:93–98. https://doi.org/10.1016/j.mcat.2018.02.008
Liang SS, Li Z, Zhang B, Zhou ZM (2017) Metabolic engineering of Escherichia coli for the production of β-alanine. Food Ferment Ind
Liu Z, Zheng W, Ye W, Wang C, Zhou Z (2019) Characterization of cysteine sulfinic acid decarboxylase from Tribolium castaneum and its application in the production of β-alanine. Appl Microbiol Biotechnol 103(25). https://doi.org/10.1007/s00253-019-10139-z
Marienhagen J, Eggeling L (2008) Metabolic Function of Corynebacterium glutamicum Aminotransferases AlaT and AvtA and Impact on L-Valine Production. Appl Environ Microbiol 74(24):7457–7462. https://doi.org/10.1128/AEM.01025-08
Mingliang C, Ying QI, Yanming X, Chao H, Jingya LI, Apeng L (2018) Biocatalytic synthesis of β-alanine from fumaric acid by a two-enzyme system. Bull Ferment Sci Technol
Mustafa G, Arshad M, Bano I, Abbas M (2020) Biotechnological applications of sugarcane bagasse and sugar beet molasses. Biomass Convers Biorefin (11)
Ohnishi J, Mitsuhashi S, Hayashi M, Ando S, Ikeda M (2002) A novel methodology employing Corynebacterium glutamicum genome information to generate a new L-lysine-producing mutant. Appl Microbiol Biotechnol 58(2):217–223. https://doi.org/10.1007/s00253-001-0883-6
Pei W, Zhang J, Deng S, Tigu F, Li Y, Li Q, Cai Z, Li Y (2017) Molecular engineering of L-aspartate-α-decarboxylase for improved activity and catalytic stability. Appl Microbiol Biotechnol 101(15):6015–6021. https://doi.org/10.1007/s00253-017-8337-y
Piao X, Wang L, Lin B, Chen H, Liu W (2019) Metabolic engineering of Escherichia coli for production of L-aspartate and its derivative β-alanine with high stoichiometric yield. Metab Eng. https://doi.org/10.1016/j.ymben.2019.04.012
Poelje P, Snell EE (1990) Cloning, sequencing, expression, and site-directed mutagenesis of the gene from Clostridium perfringens encoding pyruvoyl-dependent histidine decarboxylase. Biochem 29(1):132–139. https://doi.org/10.1021/bi00453a016
Qian Y, Liu J, Song W, Chen X, Luo Q, Liu L (2018) Production of β-alanine from fumaric acid using a dual-enzyme cascade. ChemCatChem 10. https://doi.org/10.1002/cctc.201801050
Rest M, Lange C, Molenaar D (1999) A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52(4):541–545. https://doi.org/10.1007/s002530051557
Sahm H, Eggeling L (1999) d-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding l-valine synthesis for d-pantothenate overproduction. Appl Environ Microbiol 65(5):1973–1979. https://doi.org/10.1016/S0027-5107(99)00041-X
Song CW, Lee J, Ko YS, Lee SY (2015) Metabolic engineering of Escherichia coli for the production of 3-aminopropionic acid. Metab Eng 30(3):121–129. https://doi.org/10.1016/j.ymben.2015.05.005
Veetil VP, Raj H, Quax WJ, Janssen DB, Poelarends GJ (2010) Site-directed mutagenesis, kinetic and inhibition studies of aspartate ammonia lyase from Bacillus sp. YM55-1. Febs J 276(11):2994–3007. https://doi.org/10.1111/j.1742-4658.2009.07015.x
Wang L, Yu H, Xu J, Ruan H, Zhang W (2020a) Deciphering the crucial roles of AraC-type transcriptional regulator Cgl2680 on NADPH metabolism and L-lysine production in Corynebacterium glutamicum. World J Microbiol Biotechnol 36(6):82. https://doi.org/10.1007/s11274-020-02861-y
Wang Y, Shi K, Chen P, Zhang F, Zhang WG (2020) Rational modification of the carbon metabolism of Corynebacterium glutamicum to enhance L-leucine production. J Ind Microbiol Biotechnol 47(5). https://doi.org/10.1007/s10295-020-02282-8
White WH (2001) Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of β-alanine production from spermine. J Biol Chem 276(14):10794–10800. https://doi.org/10.1074/jbc.M009804200
Williamson JM (1985) [75] L-Aspartate α-decarboxylase. Methods Enzymol 113:589–595
Xu JZ, Yu HB, Han M, Liu LM, Zhang WG (2019) Metabolic engineering of glucose uptake systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production. J Industrial Microbiol Biotechnol 46(7):937–949. https://doi.org/10.1007/s10295-019-02170-w
Xunyan D, Yue Z, Jianxun Z, Xiaoyuan W (2016) Characterization of aspartate kinase and homoserine dehydrogenase from Corynebacterium glutamicum IWJ001 and systematic investigation of L-isoleucine biosynthesis. J Ind Microbiol Biotechnol 43(6). https://doi.org/10.1007/s10295-016-1763-5
Zhang T, Zhang R, Xu M, Zhang X, Yang T, Liu F, Yang S, Rao Z (2018) Glu56Ser mutation improves the enzymatic activity and catalytic stability of Bacillus subtilis L-aspartate α-decarboxylase for an efficient β-alanine production. Process Biochem 70(JUL):117–123. https://doi.org/10.1016/j.procbio.2018.04.004
Ziert C (2014) Metabolic engineering of Corynebacterium glutamicum for the production of L-aspartate and its derivatives β-alanine and ectoine. publication/2691217
Zou X, Guo L, Huang L, Li M, Zhang S, Yang A, Zhang Y, Zhu L, Zhang H, Zhang J, Feng Z (2020) Pathway construction and metabolic engineering for fermentative production of beta-alanine in Escherichia coli. Appl Microbiol Biotechnol 104(6):2545–2559. https://doi.org/10.1007/s00253-020-10359-8
Funding
This work was funded by the National Key Research and Development Program of China [No. 2021YFC2100900] and the Top-Notch Academic Programs Project of Jiangsu Higher Education Institutions, the 111 project (Grant number 111–2-06).
Author information
Authors and Affiliations
Contributions
J.X. and Z.R. conceived the experiments. J.W. and W.Z. designed and performed the experiments and analyzed the data. J.W. and J.X. wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
Not applicable
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, JY., Rao, ZM., Xu, JZ. et al. Enhancing β-alanine production from glucose in genetically modified Corynebacterium glutamicum by metabolic pathway engineering. Appl Microbiol Biotechnol 105, 9153–9166 (2021). https://doi.org/10.1007/s00253-021-11696-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11696-y