Abstract
Strains of the yeast genus Blastobotrys (subphylum Saccharomycotina) represent a valuable biotechnological resource for basic biochemistry research, single-cell protein, and heterologous protein production processes. Species of this genus are dimorphic, non-pathogenic, thermotolerant, and can assimilate a variety of hydrophilic and hydrophobic substrates. These can constitute a single-cell oil platform in an emerging bio-based economy as oleaginous traits have been discovered recently. However, the regulatory network of lipogenesis in these yeasts is poorly understood. To keep pace with the growing market demands for lipid-derived products, it is critical to understand the lipid biosynthesis in these unconventional yeasts to pinpoint what governs the preferential channelling of carbon flux into lipids instead of the competing pathways. This review summarizes information relevant to the regulation of lipid metabolic pathways and prospects of metabolic engineering in Blastobotrys yeasts for their application in food, feed, and beyond, particularly for fatty acid-based fuels and oleochemicals.
Key points
• The production of biolipids by heterotrophic yeasts is reviewed.
• Summary of information concerning lipid metabolism regulation is highlighted.
• Special focus on the importance of diacylglycerol acyltransferases encoding genes in improving lipid production is made.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Budding ascomycetous yeasts (e.g., Saccharomyces sp., Pichia sp., etc.) belonging to Saccharomycotina subphylum are well known for their applications in basic research, food, healthcare, and agriculture. Oleaginous yeasts (such as Yarrowia lipolytica, Lipomyces sp., Rhodotorula sp.) are explored for more than a decade for the promising role in replacing oleochemicals derived from fossil resources and food-grade oil plants. This is arising from their lipid production ability coupled to high-density growth on a wide range of low-cost substrates with unique physiological characteristics and availability of constantly evolving genetic tools (Juanssilfero et al. 2018; Ng 2020). Blastobotrys adeninivorans is another non-conventional oleaginous yeast belonging to the subphylum Saccharomycotina. The yeast is found in a diverse range of natural habitats. The first strain was isolated from soil and initially been named Trichosporon adeninovorans CBS8244T (Middelhoven et al. 1984). Further strains have been obtained from wood hydrolysates, chopped maize silage, humus-rich soil, and fermented foods such as Pu-erh tea (Wartmann et al. 1995; Abe et al. 2008; Kunze et al. 2014). Initially described as Trichosporon adeninivorans by yeast taxonomic monograph (Middlehoven et al. 1984), and later as Arxula adeninivorans (Van der Walt 1990), this dimorphic yeast was finally reclassified in the genus Blastobotrys by Kurtzman and Robnett (2007), based on a detailed phylogenetic comparison with other related species.
Among the species described in the genus Blastobotrys, B. adeninivorans (e.g., LS3, type strain CBS 8244T and CBS 7766) has been well described (Kurtzman 2007; Kurtzmann and Robnett 2007). Recently, the LS3 strain of B. adeninivorans was reassigned to B. raffinosifermentans (Thomas et al. 2019). The LS3 strain has been exploited as a microbial biocatalyst and biosensor to detect biomarkers in medicine or pollutants in aquatic samples (Malak et al. 2016; Pham et al. 2015, Theron et al. 2014; Kasprzak et al. 2016b). It exhibits unusual metabolic and physiological characteristics, e.g., the presence of n-butanol degradation pathway; thermo-, halo-, and osmo-tolerance features; and the ability to secrete high-quality proteins including intracellular and extracellular phytases, glucoamylase, cutinases, invertase, xylosidase, cellobiases, and proteases (Bischoff et al. 2015; Wartmann and Kunze 2000; Olstorpe et al. 2009; Sano et al. 1999). Post-translational modifications like O-glycosylation and N-glycosylation are reported in this yeast (Wartmann et al. 2002). Blastobotrys species share nitrate assimilation and xerotolerance as common traits (Van der Walt et al. 1990). Oleaginous traits have been described for B. adeninivorans CBS 8244T (Sanya et al. 2020) and recently for the LS3, CBS 7377, and CBS 7766 strains of the yeast (Thomas et al. 2019). When nitrogen-limitation was imposed, lipid accumulation to above 30% was observed in B. adeninivorans (Thomas et al. 2019).
The LS3 strain of B. adeninivorans emerged as an attractive host for the expression of recombinant proteins with medical or industrial interest. An increasing number of published articles have reported the LS3 strain for producing recombinant proteins, analyzing its uric acid-reducing gene, adenine-reducing gene, guanine-reducing gene, and in improving the production of polyhydroxyalkanoates (PHAs) (polyhydroxybutyrate, PHB-V) or enantiomerically pure 1-(R)-phenyl ethanol (Jankowska et al. 2013b,2013a,2015; Litwińska et al. 2019; Trautwein-Schult et al. 2013, 2014; Rauter et al. 2015; Biernacki et al. 2017; Zhang et al. 2019). Lapeña et al. (2020) confirmed the ability of the LS3 strain to assimilate a wide range of nitrogen-containing carbon sources (e.g., proteins, purines) alongside its capacity to grow well on an enzymatically saccharified sulfite-pulped spruce wood and hydrolysates of by-products from chicken. They also reported the similarity of its amino acid profile to fishmeal and soymeal, especially for essential amino acids, hence suitable for single-cell protein production. B. adeninivorans CBS 8244T described earlier for its amines, purines, and adenines assimilation ability (Middelhoven et al. 1984) has now been recognized for its capacity to hydrolyze maltose, maltose-like substrates (maltulose, melezitose), and sucrose (Visnapuu et al. 2019). Kurtzman et al. (2015) highlighted the growth of both B. adeninivorans and B. raffinosifermentans on high glucose concentration (50%, 60%, and 70%) media. The dimorphic yeast described in this review alongside the CBS 8244T strain also showed promising innate potential in the bioconversion of xylose and glucose as renewable feedstock to various molecules, including lipid, acetic, and citric acid (Thomas et al. 2019; Borelli et al. 2019).
Apart from its oleagenicity and prospects in biofuel applications, the Blastobotrys yeasts have been studied for various other biotechnological applications, listed as follows: Katongole et al. (2017) assessed the effectiveness of solid-state fermentation with B. raffinosifermentans species to improve the hygienic quality and digestibility of banana peels by mono-gastric animals. B. adeninivorans was employed for single-cell protein (SCP) production from biogas substrate. SCP derived from microbial biomass is usually used as animal feed, particularly in aquaculture (Goldberg 1985). The yeast was suitable for SCP production and its protein content displayed essential amino acids that were similar to those of soymeal and fishmeal (Lapeña et al. 2020). The yeast has also been considered a suitable host for heterologous gene expression (Wartmann and Kunze 2000).
Microbial lipases are interesting due to their function in lipid digestion for nutrient acquisition. Stehr et al. (2003) suggested a potential benefit from these enzymes in utilizing carbohydrate-restricted environments or environments where lipids are the sole carbon source. Kumari et al. (2015) applied B. raffinosifermentans as a host system in shake flask cultures to express the lipase YlLip11 from Y. lipolytica and revealed the production of 2654 U/L lipase. The extracellular lipase Alip1p, encoded by B. raffinosifermentans ALIP1, is a triacylglycerol acylhydrolase that catalyzes the hydrolysis of all ester bonds in between glycerol and medium-chain fatty acids. The genetic elements related to ALIP1 are described in Böer et al. (2005b). The ALIP1 gene regulated by carbon source or lipophilic compounds harbored an open reading frame of 1347 bp and its expressed protein (Alip1p) containing the consensus pentapeptide motif (-Gly-X-Ser-X-Gly-) for lipid recognition exhibited a dimeric structure of 100 kDa and an optimum activity at pH 7.5 and temperature 30 °C (Böer et al. 2005b).
There are a growing number of excellent reviews on genetic engineering tools for B. raffinosifermentans, much like the oleaginous yeast species of the genera Yarrowia, Rhodotorula, and Lipomyces (Malak et al. 2016; Wang et al. 2020; Park et al. 2018; Sutanto et al. 2018). Nonetheless, owing to recent studies with new approaches and technologies, we have begun to unveil the mechanisms underlying lipid biogenesis processes on a molecular level in this yeast. Additionally, Blastobotrys genus has recently attracted interest as a potential source for biofuels. In the current review, we address the recent applications made with this novel non-conventional oleaginous yeast, review genome engineering strategies, and discuss substrate constraints on lipid production and the state-of-the-art gene targeting tools available for this yeast. Most data in the literature used the name Arxula adeninivorans. In this review, however, we have used Blastobotrys adeninivorans as it remains the current taxonomically valid name.
Taxonomy and genome description
The taxonomic position of Blastobotrys species is summarized in Fig. 1. Generally, non-conventional yeasts under the Saccharomycotina subphylum include species such as Scheffersomyces (Pichia) stipitis, Komagataella phaffii (Pichia pastoris), Ogataea (Hansenula) polymorpha, Kluyveromyces lactis, Y. lipolytica, Kluyveromyces marxianus, Ogataea thermomethanolica (Cai et al. 2019), or Trichosporon cutaneum (Reiser et al. 1996). Some of them are ubiquitous to all sorts of niches, showing extreme stress tolerance phenotypes. For instance, Trichosporon cutaneum easily accommodates high concentrations of compounds like formic acid, acetic acid, levulinic acid, 5-hydroxymethylfurfural, and vanillin syringaldehyde, and Pichia kudriavzevii can sustain high furan aldehyde concentrations (Kwon et al. 2011). Phylogenetic tree reconstruction placed B. adeninivorans closest to the well-described non-conventional oleaginous yeast Y. lipolytica and coalescence-based phylogenies supported this dimorphic yeast as sister to a clade containing Saprochaete clavata and Geotrichum candidum isolate CLIB 918 (for an overview, see Shen et al. 2016). Yeast species are routinely identified using the internal transcribed spacer (ITS) region and the variable domain of the large subunit (26S) ribosomal DNA or the complete small subunit, D1/D2 domain, coupled with the gene sequences from the nearly complete large subunit (LSU) rRNA gene, mitochondrial small-subunit rRNA gene (MtSm), and cytochrome oxidase II (COXII) (Kurtzman and Robnett 2007).
B. adeninivorans LS3 is an attractive and amendable microorganism with a small genome (11.8 Mb) organized into four chromosomes—Arad1A, Arad1B, Arad1C, and Arad1D of 1659397, 2016785, 3827910, and 4300524 nucleotides, respectively. The genome size is smaller than that of S. cerevisiae S288c (12.1 Mb) and Debaryomyces hansenii CBS767 (12.2 Mb). The LS3 strain of this species is asexual with a mating-type (MAT) locus containing the transcription factor Matα1 (ARAD1D19294g, MTAL1), also observed in Y. lipolytica. However, no Matα2 (MATL2) has been uncovered in contrast to Y. lipolytica and S. cerevisiae. A genome GC content of around 48.1% resembling that of Y. lipolytica E150 (49.0%), but significantly higher than that of S. cerevisiae S288c (38.3%) and Debaryomyces hansenii CBS767 (36.3%), has been elucidated for this strain. The complete genome sequencing has been performed by Kunze and co-workers and revealed around 6000 genes in just 11.8 Mb. A single transposable element of Ty3/gypsy-type long terminal repeat superfamily of retrotransposons, Taa3 (ARAD1B13860t), has been recognized within the genome alongside genes encoding for enzymes involved in β-oxidation and the glyoxylate cycle (Kunze et al. 2014).
Morphology and growth conditions
B. adeninivorans is usually cultured at 28–37 °C (Knoll et al. 2007; Kunze et al. 2014; Tsakraklides et al. 2015; Thomas et al. 2019; Sanya et al. 2020). The yeast exhibits temperature-dependent morphology characterized by the formation of budding cells below 42 °C, the appearance of pseudo-hyphae at 42 °C, and hyphae at above 42 °C, as well as the ability to grow at temperatures up to 48 °C without prior adaptation (Choudhary et al. 2016) and a few hours of survival at 55 °C (Wartmann et al. 1995). Its dimorphism is reversible with growth conditions as budding cells are re-established below 42 °C.
Cultivation mode such as shake flask (Thomas et al. 2019, Sanya et al. 2020) and fed-batch cultivation (Bernacki et al. 2017) alongside culture media encompassing selective media (Litwińska et al. 2019; Theron et al. 2014; Friedlander et al. 2016; Sanya et al. 2020) and nonselective media (Samsonova et al. 1996; Bernacki et al. 2017) are usually applied to monitor the growth of these yeasts, with the main focus on nitrogen (NH4 Cl) and carbon sources (glucose) (Sanya et al. 2020).
Oleaginous characteristics
Compared to the conventional procedures of vegetable oil production, microbial lipid production takes advantage of a short production cycle, industrial scalability of bioproducts, zero competition with food crops, independence of arable land, sustainability, and tailored processes. As such, they might circumvent the limitations posed by lipid production from oil crops and plants. For instance, Blomqvist et al. demonstrated the possibility to replace vegetable oil in the feed for Arctic char (Salvelinus alpinus) with microbial oils derived from Lipomyces starkeyi cultivated on lignocellulose hydrolysate from wheat straw (Blomqvist et al. 2018). In 2011, nearly 1500 species of ascomycete and basidiomycete yeasts belonging to 149 genera were listed and described (Kurtzman et al. 2011). Less than 30 species were characterized for their oleaginous traits (Qin et al. 2017). These oleaginous yeast species were categorized under the following classes—Saccharomycetes, Cystobasidiomycetes, Microbotryomycetes, Tremellomycetes, and Ustilaginomycetes (Sitepu et al. 2014).
As previously reported, lipid accumulation in oleaginous yeast occurs when there is depletion of nutrients in the culture medium, channeling the carbon flux towards cellular lipogenesis (Aggelis and Sourdis 1997). In the literature, two criteria are mainly described to characterize a yeast as an oleaginous microorganism. Firstly is the ability to store more than 20% lipid in their dry cell weight (DCW) predominantly in the form of triacylglycerol or TAG (Sitepu et al. 2014; Fakas 2017). For instance, the oleaginous red yeast Rhodotorula glutinis produced 70% lipid in their dry weight. Another oleaginous yeast, Lipomyces starkeyi, was reported to have lipid as high as 85.1% DCW (Li et al. 2013; Juanssilfero et al. 2018). Secondly is the presence of key enzymes, i.e., ATP citrate Lyse (ACL) and/or malic enzyme (MAE) responsible for the oleaginous trait (Ratledge 2014; Kamineni and Shaw 2020). The role of ACL and MAE enzymes as a supplier of acetyl-CoA and NADPH, respectively, appeared to be species-dependent for fatty acid (FA) synthesis. For instance, Dulermo et al. highlighted that the inactivation of the ACL1 gene in Y. lipolytica decreases FA synthesis by 60 to 80%, confirming its essential role in FA synthesis. In contrast, inactivation of the MAE1 gene has no effects on FA synthesis, except in a FA over-accumulating strain where it improves FA synthesis by 35% (Dulermo et al. 2015).
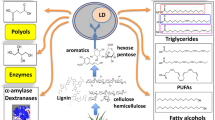
Although cultivation conditions for efficient B. raffinosifermentans-based processes (Stöckmann et al. 2009) and lipases activity were elucidated a decade ago, it is only recently that the lipid production ability and lipid-related genes have been uncovered for the yeast (Thomas et al. 2019; Sanya et al. 2020). The metabolic engineering strategies for improving its lipid production are essentially described in the works of Sanya et al. (2020). The lipid content of the unmodified wild-type strain (LS3) of B. adeninivorans was 30% DCW on glucose (Thomas et al. 2019). The lipid profile of B. adeninivorans (e.g., strains CBS 8244 and LS3), at cultivation temperatures ranging from 15 to 37 °C, contained C18:1 (n − 7), C18:1 (n − 5), C17:1, C18:3 (n − 3), and C16 and C18 FAs (Olstorpe et al. 2014; Thomas et al., 2019). Interestingly, Thomas et al. demonstrated variation in FA composition of the yeast at elevated temperatures. For instance, increasing temperature from 28 to 45 °C showed an increase in C18:0 and C18:2 in the CBS 8244T and LS3, while the relative proportions of C16:0 and C18:1 decreased (Thomas et al. 2019). In the LS3, CBS 7377, CBS 7766, and CBS 8244T strains, the dominant FA species in lipid profile, oleic acid (18:1), ranged from 43 to 52%, depending on the strain and medium (Thomas et al. 2019). These FA species are good candidates for biodiesel quality as palmitic (16:0), stearic (18:0), oleic (18:1), linoleic (18:2), and linolenic (18:3) acids are elucidated to be suitable FAs for biodiesel production (Knothe 2009). Of note, Thomas et al. (2019) demonstrated the capacity of this yeast to grow efficiently on both hydrophobic (oleic acid) and hydrophilic (glucose, xylose) substrates generating value-added bioproducts such as organic acids (citric acid and acetic acid). A cumulative concentration ranging from 3.4 to 10.1 g/L has been disclosed for both citric and acetic acid (Thomas et al. 2019). The anamorphic hemiascomycete yeast of this report contained fatty acid species retrieved from other oleaginous plants or yeasts and therefore can be exploited as renewable resources for industrial raw materials (Supplementary Table S1). It holds excellent promise as non-food feedstocks for the generation of bio-commodities. Some of the key findings regarding Blastobotrys genus are summarized in Fig. 2. In Blastobotrys species, total intracellular lipids were quantified by gas chromatography after lyophilizing the harvested wet cell pellets and transesterifying cellular fatty acid contents to produce fatty acids methyl esters (Thomas et al. 2019; Sanya et al. 2020). We compared the lipogenesis ability of B. raffinosifermentans (LS3) on some carbon substrates to that of other well-documented oleaginous yeasts in Table 1. The commonly used strains in research are summarized in Table 2.
Tools and techniques employed to engineer Blastobotrys yeasts
Genetic elements and vector systems developed in Blastobotrys genus
Most of the vector systems have been developed in Blastobotrys genus to integrate a copy of the gene of interest in its genome, for example, integration in an atrp1 (tryptophan) auxotrophic host strain of B. raffinosifermentans. Those elements consist of the ATRP1 selection marker fused to truncated ALEU2 promoter of 53 bp or TEF1 (translation-elongation factor 1α) promoter, terminator, expression vector encoding a heterologous enzyme with biotechnological interest. For instance, a high copy number (eight copies) of an amyA expression vector encoding heterologous alpha-amylase from Bacillus amyloliquefaciens instead of a single copy resulted in strains with superior productivity for a secreted recombinant alpha-amylase (Steinborn et al. 2007). The use of green fluorescent protein (GFP)-coding genes from the jellyfish Aequorea victoria as a reporter gene to assess heterologous gene expression was also reported in this yeast (Wartmann et al. 2002, Terentiev et al. 2004; Steinborn et al. 2006).
The functionality of several endogenous constitutive and inducible promoters (Supplementary Table S2), selection markers, and terminator elements (PHO5 terminator of S. cerevisiae, Kasprzak et al. 2016a; Steinborn et al. 2007) have been verified in B. raffinosifermentans. For example, the B. raffinosifermentans AHSB4 gene promoter, encoding histone H4, was uncovered to be strongly and constitutively expressed under salt-free and salt-stress conditions (Wartman et al. 2003). On the other hand, a constitutive promoter of TEF1 gene (encoding translation-elongation factor 1α) from B. raffinosifermentans that enables strong constitutive expression in various yeasts alongside genetic elements related to this gene was reported (Rösel and Kunze 1995; Steinborn et al. 2006; Terentiev et al. 2004). The Liu Y et al. (2016) demonstrated an intron in promoter’s presence as relevant for improving targeted expression in R. toruloides. Indeed, the native DGA1 gene of R. toruloides was overexpressed using two strong promoters (LDPin and GPD1) and resulted in increased lipid content by 21% in R. toruloides with the LDP intron promoter (LDPin) than with GPD promoter. This is also 55% higher than that reported in the R. toruloides wild-type strain.
The other genetic element employed in the yeast system is the transcription terminator. Eukaryotic terminators are genetically encoded elements post-transcriptionally upregulated by factors associated with cis-elements (e.g., GUUCG/U) within the 3′ untranslated region (UTR) (Ito et al. 2016). Both endogenous and heterologous terminators were either verified or reported functional in B. raffinosifermentans. For example, endogenous terminators ATRP1 have been applied to construct a selective marker module incorporated in the vector/expression system Xplor®2 or multicopy integration experiments facilitating the insertion of a yeast rDNA integrative expression cassette (YRC) or a yeast integrative expression cassette (YIC) into the genome. It is worth noting that transformants are hardly recovered when transforming B. raffinosifermentans by circular YRC and YIC. Simultaneously, a linearized form of YIC generated even lower transformants than a linearized form of YRC (Böer et al. 2009a, 2009d).
Like promoters and terminators, selection markers are also valuable in genome engineering. Some publications characterized the suitability of individual genes of B. raffinosifermentans as auxotrophic markers. For instance, ILV1, from the LS3 strain, comprising 1653 bp and encoding 550 amino acids of the threonine deaminase, has been shown useful as an auxotrophic selection marker for the transformation of plasmids into this yeast (Wartmann et al. 1995, 1998). Moreover, B. raffinosifermentans ALEU2, encoding L-isopropylmalate dehydrogenase and harboring an open reading frame of 1086 bp, was successfully employed as an auxotrophic marker (Wartmann et al. 2003). In the experiment validating its suitability to complement the auxotrophy of a B. raffinosifermentans aleu2 host, the plasmid pAL-ALEU2m containing ALEU2 as a selection marker and the 25S rDNA was used alongside the GFP gene, encoding an intracellular green fluorescent protein, and the HSA gene, encoding the secreted human serum albumin (Wartmann et al. 2003). The ATRP1 encoding a phosphoribosyl anthranilate isomerase in this yeast was also used as an auxotrophic marker in experiments focused on recombinant protein production (Steinborn et al. 2006). Antibiotic resistance markers like the Escherichia coli-derived hph gene, encoding hygromycin B phosphotransferase have been used in several studies (Wartmann et al. 2003; Rösel and Kunze 1998).
Transformation methods of Blastobotrys yeast
Preparation of competent cells of B. raffinosifermentans LS3 employed sorbitol, bicine, NaOH, and PEG-1000. Indeed, after harvesting cells cultured in rich media and washing them with water, a suspension on bicine buffer (containing 1 M sorbitol, 10 mM bicine-NaOH/pH 8.35, 3% PEG 1000, 5% DMSO), followed by centrifugation and resuspension in the same buffer generate competent cells (Kunze and Kunze 1996). After that, the linearized plasmid DNA of interest is commonly used to transform B. raffinosifermentans LS3 competent cells.
Genome editing toolbox for Blastobotrys yeast
Among several gene-editing methods that are used currently, very few have been reported in Blastobotrys species, especially B. raffinosifermentans, on the contrary to the most thoroughly studied model yeasts, Y. lipolytica (Madzak 2015) and S. cerevisiae (Pretorius 2017; Macías et al. 2019), that have plenty of genetic engineering tools. For instance, Tsakraklides and co-workers successfully increased gene targeting in B. raffinosifermentans by 3–8 fold depending on the target locus through hydroxyurea treatment (Tsakraklides et al. 2015; Galli and Schiest 1996). The development of genetic tools allowing several genetic manipulations in B. raffinosifermentans was mainly achieved by the Gotthard Kunze team (IPK, Gatersleben, Germany, Böer et al. 2009d). B. raffinosifermentans host-vector expression system was constructed for the first time by transforming auxotrophic host strain with a plasmid pAL-ATRP1-amyA containing the ATRP1, encoding a phosphoribosyl anthranilate isomerase isolated from the yeast B. raffinosifermentans, as selection marker and the 25S rDNA for targeting and equipped with an expression cassette consisting of the Bacillus amyloliquefaciens-derived amyA gene fused to the constitutive B. raffinosifermentans-derived TEF1 promoter and S. cerevisiae-derived PHO5 terminator (Steinborn et al. 2007). The sole example of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) element implicating the yeast of this report is the expression of cas9D147YP411T nuclease variant under B. raffinosifermentans-derived TEF1 promoter (Juergens et al. 2018).
Lipid biogenesis in oleaginous yeasts
The lipogenesis mechanism in oleaginous yeasts is primarily based on the supply of acetyl-CoA and reduced nicotinamide adenine dinucleotide phosphate (NADPH), a provider of the reducing power indispensable for fatty acid synthesis. The role of the pentose phosphate pathway (PPP) has been demonstrated in lipogenesis. Glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (PGD) in the PPP provide NADPH for lipogenesis in oleaginous microbes (Chen et al. 2015a). The other known NADPH supplier for fatty acid synthesis in oleaginous microbe is the cytosolic MAE (Ratledge 2014).
Furthermore, oleaginous yeasts exhibit different mechanisms for the conversion of glucose and acetate during lipogenesis. In the case of glucose as a substrate, depleted elements from the medium such as nitrogen (Bandhu et al. 2019), phosphate (Wang et al. 2018a), or sulfur (Wu et al. 2011) usually trigger lipogenesis. Silverman et al. (2016) summarized the biosynthesis of fatty acids in oleaginous yeasts. Nutrient depletion activates the adenosine monophosphate (AMP) deaminase and NADPH supply through the oxidative pentose phosphate pathway (oxPPP) or MAE. Increased AMP deaminase activity reduces the level of mitochondrial AMP, which reduces the activity of AMP-dependent isocitrate dehydrogenase in the TCA cycle, resulting in citrate accumulation. The excess mitochondrial citrate is transported into the cytosol to generate acetyl-CoA, which is the carbon skeleton required for fatty acid biosynthesis leading to the pool of different acyl-CoAs. De novo biosynthesis of TAGs is initiated via the Kennedy pathway, where the transfer of a fatty acyl group from acyl-CoA to glycerol-sn-3-phosphate (G3P) is catalyzed by glycerol-3-phosphate acyltransferase (GPAT) to form 1-acylglycerol-sn-3-phosphate (lysophosphatidate or LPA). This step is followed by the acylation of LPA to form 1,2-diacylglycerol-sn-3-phosphate (PA), dephosphorylation of PA to form diacylglycerol (DAG). In the final step, transfer of an acyl group on DAG under diacylglycerol acyltransferase (DGAT) activity constitutes TAG. These TAGs are generally stored in lipid bodies, which are cytoplasmic lipid-enriched organelles that originate and are closely related to the endoplasmic reticulum (ER), formed by the channeling of lipids and proteins from the ER toward newly formed lipid droplets (Jackson 2019).
In the case of acetate as substrate, direct uptake through symport at higher pH (Casal et al. 1996) or passive diffusion at lower pH (Augstein et al. 2003) have been observed in yeasts. After entering the cell, dissociated acetate is converted to acetyl-CoA by the acetyl-CoA synthetase (ACS2), and the acetyl-CoA from the acetyl-CoA pool follows different pathways. It is used (1) for fatty acid synthesis through acetyl-CoA carboxylase (2) employed as a precursor for citrate formation in the mitochondrion and oxidized to carbon dioxide in the tricarboxylic acid (TCA) cycle to make energy (ATP) for the cell or (3) is converted in the glyoxylate cycle to succinate (de la Peña Mattozzi et al. 2010). Further, the ex novo pathway routinely activated in oleaginous yeasts in response to hydrophobic substrates should be noticed. It consisted of (a) the breakdown of hydrophobic compounds into free fatty acids outside the cell-mediated by lipase-catalyzed hydrolysis, (b) the transfer of free fatty acids inside the cell via transporters, and (c) the conversion of free fatty acids and their derivatives into short-chain of acyl-CoAs and acetyl-CoAs by acyl-CoA oxidases through β-oxidation (Beopoulos et al. 2009).
A communication network between organelles has been noticed during lipid biogenesis. Valm et al. (2017) indicated that lipid metabolism is dispatched between the ER for lipid synthesis, LDs for storage and transport, mitochondria and peroxisomes for β-oxidation, and lysosomes for lipid hydrolysis and recycling. Emerging evidence revealed beyond organelles’ autonomous functions, a direct physical relationship between distinct organelles in eukaryotic cells. For example, the connection between LDs and peroxisomes is described by Joshi et al. (2018). They revealed that the biogenesis of LDs and peroxisome occurred at the same Pex30p subdomain. Wang and colleagues (2018b) also showed that the budding of both LDs and peroxisomes solicited the cooperation between Pex30 and the seipin complex or is inhibited otherwise. The connection between plasma membrane (PM) and ER can be seen through the yeast lipid transfer proteins (LTPs), Osh6p and Osh7p, that channel phosphatidylserine (PS) from the ER to PM via PS/phosphatidylinositol-4-phosphate (PI4P) exchange cycles. Giacomello and Pelligrini (2016) highlighted the motion of ions and lipid between mitochondria and the ER under the mediation of MERCs (Mitochondria-ER contacts). Other contact sites identified to date between cellular organelles are summarized in Fig. 3.
A schematic diagram showing the names of identified inter-organelle contact sites. Junction names are written in green. Single protein or protein complex involved in the junction that tethers two organelles in yeast is written in blue. PM (plasma membrane), ER (endoplasmic reticulum), LD (lipid droplet). vCOuPLE (vacuole-plasma membrane contact), pCLIP (for plasma membrane contact with lipid droplets), PerPECs (for peroxisome plasma membrane contact site), PerVale (for peroxisome vacuole contact), NVJ (nuclear-vacuole junction), MAM (mitochondria-associated ER membranes), vCLAMP (vacuole and mitochondria patch), PerMit (peroxisome-mitochondria contact site), PAM (ER-PM contact), LiDER (lipid droplet-ER contact site), vCLIP (vacuole-lipid droplet contact site), mCLIP (mitochondria-lipid droplet contact site), PerLiN (peroxisome-lipid droplet contact site), and EPCON (ER-peroxisome contact site) are showed in red. The contact sites positioned on the figure are inspired by Giacomello and Pelligrini (2016), Shai et al. (2018), Joshi et al. (2018), Kakimoto et al. (2018), and Stehlik et al. (2020).
Overview of lipid biogenesis genes in Blastobotrys
The understanding and exploration of lipid metabolism in Blastobotrys species are in their infancy. The lack of knowledge on the regulation of lipid accumulation in these oleaginous yeasts hinders their development as an industrial oil platform. To date, only a few reports explored the role of the significant proteins/enzymes in the lipid biosynthetic pathway of Blastobotrys species, unlike the case of the well-established eukaryotic model for lipid production such as Y. lipolytica and Lipomyces starkeyi. Some of the genes described for the dimorphic B. adeninivorans are summarized in Fig. 4 due to the critical role in their expressed membrane-bound enzymes catalyzing the last step of TAG synthesis. Thomas et al. (2019) conferred the oleaginous nature of this yeast by revealing lipid body formation along with the presence of two genes for ATP citrate lyase (ACL), suggesting their promising ability for the production of biofuel and lipids-derived compounds (Thomas 2019). Lipid bodies are essentially neutral lipid or TAG storage organelles having numerous physiological functions of the cell - including energy homeostasis, lipid metabolism, cellular toxins-trapping, and drug resistance (Chang et al. 2015). So far, the lipogenic multienzyme complex consisting of ACL (Kamineni and Shaw 2020), glycerol-3-phosphate acyltransferase (SCT1) (Tsakraklides et al. 2018), malate dehydrogenase (MDH), a key enzyme in the tricarboxylic acid (TCA) cycle (Kabran et al. 2012), Dga1p encoded by DGA1 (ARAD1C08250g), and Dga2p encoded by DGA2 (ARAD1D42460g) have been elucidated in LS3 strain (Sanya et al. 2020). These enzymes have been positioned on Fig. 5, which highlights the key steps in FA and TAG biosynthesis pathways. The genome of B. adeninivorans supposedly contains two different genes for DGAT (Thomas et al. 2019, Sanya et al. 2020). For further insights into the acyltransferases involved in lipid metabolism, we have proposed 3D structures of both Dga1p and Dga2p of B. adeninivorans based on local homology-based structural modeling (Supplementary Fig. 1A and 1B). Dga1p shared 13.74% sequence identity with 1-acyl-sn-glycerol-3-phosphate acyltransferase (PDB id 5kym.1) of Thermotoga maritima, which has been used as a template for Swiss-Model. The modeled structure highlighted the presence of α-helices in Dga1p (Supplementary Fig. 1A). For Dga2p, Swiss-Model predicted an α-helix (Supplementary Fig. 1B) sharing 33.82% sequence identity with human diacylglycerol O-acyltransferase 1 (PDB id 6vz1.1) that has been used as a template.
Schematic diagram of lipogenesis with emphasis on acyl-CoA and TAG synthesis in the oleaginous yeast Blastobotrys raffinosifermentans. Abbreviations and red color are for enzymes identified so far in the literature. ACL ATP-citrate lyase, Dga1p/Dga2p diacylglycerol acyltransferase, SCT1 glycerol-3-phosphate acyltransferase, MDH malate dehydrogenase.
B. raffinosifermentans SCT1 encodes the glycerol-3-phosphate acyl-transferase (GPAT) enzyme, which is known to attach the first fatty acid onto the glycerol backbone to produce LPA in the Kennedy pathway. SCT1 also maintains a high level of oleate in the cellular lipid composition in this yeast. When heterologously expressed in Y. lipolytica, it increased oleate level (Tsakraklides 2018). On the other hand, the inhibition of NAD+-dependent isocitrate dehydrogenase linked to the increased level of α-ketoglutaric acid, an intermediate in the TCA cycle, was demonstrated in B. adeninivorans VKM Y-2677 cells under thiamine limitation (Komzolova and Morgunov 2016). Sanya et al. (2020) studied the diacylglycerol acyltransferases type I and type II of the membrane protein acyl-CoA: diacylglycerol acyltransferases (DGAT) in Blastobotrys species and identified them as essential factors in the last steps of TAG biosynthesis. Four types of diacylglycerol acyltransferases (DGATs) have been described hitherto in eukaryotes: the type 1 (DGAT1) and type 2 (DGAT2) belonging to the membrane-bound O-acyltransferase (MBOAT) family (Cases et al. 1998) and monoacylgycerol acyltransferase (MGAT) family, respectively, and the dual-specificity DGAT having both wax ester synthase (WS) and DGAT activities (Kalscheuer and Steinbuchel 2003) and a soluble cytosolic DGAT (DGAT3) (Saha et al. 2006). Of note, the Dga1p enzyme encoded by the DGA1 gene belonging to the DGAT2 family plays a significant role in TAG biosynthesis in Y. lipolytica (Haïli et al. 2016). The study of the lipid metabolism pathway of B. raffinosifermentans (LS3 strain) highlighted the prominent role of the DGAT1 family member gene (DGA2). In contrast, the DGAT2 family member gene (DGA1) had little influence on the TAG synthesis (Sanya et al. 2020). To understand the phylogenetic relationships among the DGA1 and DGA2 encoded proteins expressed in B. raffinosifermentans and the other well-known DGAT proteins described in the literature, DGAT proteins employed by Chen et al. (2015b) alongside other DGATs retrieved from the NCBI platform (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) including those from Rhodotorula toruloides and Lipomyces starkeyi have been used to construct a phylogenetic tree. This analysis revealed that the targeted diacylglycerol acyltransferases of B. raffinosifermentans are clustered next to Y. lipolytica and away from Lipomyces starkeyi and Rhotorula turoloides (Fig. 6).
Diacylglycerol acyltransferases phylogenetic relationships. Phylogenetic tree was based on 35 diacylglycerol acyltransferases from organisms described by Chen et al. (2015b) and retrieved from NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Phylogenetic trees were constructed with amino acid sequences exploiting the neighbor-joining (NJ) method employing the MEGA 7.0 software (http://www.megasoftware.net/) after a pairwise distances computation. Multiple sequence alignments were performed using the MUSCLE algorithm from MEGA 7.0. Values at the nodes represent 500 bootstrap replications (right). NCBI accession numbers except for DGA1 and DGA2 of Y. lipolytica and B. raffinosifermentans are left in the parentheses. Species are introduced in the bracket. Highlighted in blue color Blastobotrys raffinosifermentans diacylglycerol acyltransferases of this review
Furthermore, Stoltenburg et al. (1999) identified the gene ARFC3, encoding one subunit of the replication factor C (RF-C) complex, on chromosome 1 of the four B. raffinosifermentans chromosomes. B. raffinosifermentans appeared not to be an obligate aerobic oleaginous yeast as Sędzielewska et al. identified and characterized the gene encoding cytosolic fumarate reductase, the enzyme mediating the maintenance of redox balance through the regeneration of reduced cofactors during hypoxic growth conditions, and its promoter and terminator elements (Sędzielewska et al. 2012).
Strategies for improving lipid production in oleaginous yeasts
Several strategies, including low-cost substrate utilization combined with genetic engineering and mutagenesis, have been exploited to reach a higher yield or titer of lipid production in several species of oleaginous yeasts. The ability to metabolize pentose (xylose) or/and hexose (glucose) sugars derived from lignocellulosic biomass has been widely investigated (Poontawee et al. 2017; Meesters and Eggink 1996; Yamada et al. 2017; Chattopadhyay and Maiti 2020). Other examples include Cryptococcus terricola with a lipid content of 61.96% DCW on medium with 5% starch (Tanimura et al. 2014), Apiotrichum porosum DSM27194 with 44.3% lipid content on volatile fatty acids from waste organics (Qian et al. 2020), and Cystobasidium oligophagum with 41.54% lipid on starchy materials without the use of amylolytic enzymes (Vyas and Chhabra, 2017). The natural ability of specific yeasts to grow and accumulate lipid utilizing hydrophobic substrates has been of research interest, such as in Cryptococcus curvatus with 69.14% DCW of lipids (Patel and Matsakas, 2019). Studies also reported specific oleaginous yeasts’ capacity to simultaneously use several carbon sources (Tanimura et al. 2016; Anschau et al. 2014; Juanssilfero et al. 2018; Yamada et al. 2017), alongside the flocculation capacity (Sitepu et al. 2014). A yeast species’ ability to consume sugars withstanding the inhibitory effect of chemicals is also of utmost importance as microbial behavior changes accordingly. For instance, the oleaginous yeast L. starkeyi reaches 68.24% of its DCW in lipids while supporting a mixture of the following growth inhibitors: 5-hydroxymethylfurfural, furfural, acetic acid, formic acid, acid levulin, vanillin, and syringaldehyde (Juanssilfero et al. 2018).
Researchers have designed constructs mostly to overexpress certain enzymes of the pathway to engineer further the strains for a higher amount of metabolite production or maneuver the carbon flux towards value-added molecule production. Overexpression is commonly carried out by integrating an additional gene copy under a strong promoter (Delic et al. 2014). Kamineni and Shaw (2020) suggested three fundamental strategies to increase TAG biosynthesis in yeast: increase the metabolic flux rate from sugar to TAG, re-design lipid pathway stoichiometry for more efficient conversion of sugar’s carbon atoms and electrons into TAG, and eliminate competing by-products. Over the years, researchers have documented that the metabolic engineering for lipid production involve overexpression of enzymes of the fatty acid biosynthesis pathway/TAG biosynthesis pathway, regulation of the pathways associated with the TAG biosynthesis, TAG degradation, lipid droplet formation and lipid droplet degradation, partial blockage of competing for metabolic pathways, and the “multigene” transgenic approach to achieve these objectives (Xie et al. 2015; Liu Y et al. 2016; Ledesma-Amaro et al. 2016; Wang et al. 2020).
As previously mentioned, the DGAT enzyme plays a crucial role in TAG biosynthesis. Hence, the molecular understanding of the genes’ regulatory mechanism from the DGAT family appeared to be critical for their genetic manipulation to increase lipid production in oleaginous yeasts (Haïli et al. 2016; Xue et al. 2013). Changes in gene expression from the growth phase to the lipid biosynthesis phase in oleaginous yeasts and gene expression analysis of mutants with increased lipid productivity have revealed the expression of genes important for high lipid production. DGAT overexpression has already been performed recently to get insight into the lipid metabolism of B. adeninivorans and B. raffinosifermentans species (Sanya et al. 2020). Overexpression of native DGAT1 in B. raffinosifermentans increased lipid production (26.5% DCW) about 1.6-fold relative to the wild-type strain (LS3, 16.9% DCW) when cultivated on glucose (Sanya et al. 2020). Overexpression or deletions of acetyl-CoA carboxylase, another key enzyme of the fatty acid biosynthetic pathway, has also been attempted (Tamano et al. 2013), along with overexpression of glycerol-3-phosphate dehydrogenase (Beopoulos et al. 2008). Knowing that palmitoleic acid and oleic acid resulted from Δ9 fatty acid desaturases activities in oleaginous yeasts and they have been disclosed in the lipid profile of the yeasts of Blastobotrys genus, overexpression of this desaturase can be envisioned to enhance the cellular lipid content as it was reported from Rhodosporidium toruloides (Tsai et al. 2019). Promising approaches to improving intracellular lipid content may also exploit the repertoire of well-established genes involved in glycolysis, β-oxidation cycle, central metabolism, and lipid metabolism that is routinely reported in oleaginous yeasts (e.g., Yarrowia lipolytica, Rhodosporidium toruloides, Beopoulos et al. 2008; Zhu et al. 2012). For example, to elucidate the relevance of the metabolic circuits associated with the enhancement of lipids in Rhodosporidium toruloides, Castañeda and colleagues performed the deletion of the genes expressing proteins involved in the central nitrogen metabolism (Glutamate dehydrogenase), the tricarboxylic acid (TCA) cycle (Isocitrate dehydrogenase), and glycolysis (Glucose-6-phosphate isomerase) (Castañeda et al. 2019). On the other hand, Yuzbasheva et al. (2017) co-expressed the gene encoding NADP+-dependent glucose-6-phosphate dehydrogenase (ZWF1) and that encoding acyl-CoA binding protein (ACBP) to increase the lipid content in Y. lipolytica, while Wang et al. (2018c) improved lipid content in Y. lipolytica by inactivating MHY1 gene encoding Mhy1p that exerts regulatory functions on lipid biosynthesis, amino acid, and nitrogen metabolism and cell cycle. The inactivation of SCT1 as a potent way to boost fatty acyl pool can be envisioned in Blastobotrys genus. A similar experiment in Y. lipolytica contributes to the enhancement of the fatty acid ethyl esters pool identified as a promising renewable alternative for biodiesel (Ng et al. 2020). Breeding using mutagen has been used efficiently for improving lipid production in oleaginous yeasts (Liu et al. 2015, Yamada et al. 2017). For example, Takaku et al. (2021) applied ultraviolet treatment to generate Lipomyces starkeyi mutant capable of producing higher lipids compared to the wild-type. They also identified the increased expression of acyl-CoA synthesis- and Kennedy pathway-related genes during lipogenesis. On the other hand, Yamazaki et al. (2019) demonstrated that Percoll density gradient centrifugation is an important method recommended for the identification of high-accumulating TAG mutant strains.
Future directions and conclusion
System-level metabolic engineering that highlights the key signaling pathways regulating lipid biosynthesis is necessary to harvest the potential of Blastobotrys strains for developing these yeasts as an industrial platform for lipid production. Albeit the genetic parts uncovered in this review, more sophisticated genome editing methods should be designed and evaluated. Further detailed analyses of the deletion or knockout strategies to validate the role of the genes involved in circuits interfering with the lipid metabolism pathway will provide a novel insight into each gene’s relative importance in the lipid biosynthesis pathway. The study of the biochemical role of lipid mediators associated with fluxomics, metabolomics, proteomics, and computational methods can help design a suitable strategy for improving the intracellular lipid content. These strategies could further pave new directions for genetic improvement tailoring fatty acid synthesis with immense food, healthcare, and biofuel industries. A co-cultivation system involving the yeast species of this genus with other oleaginous yeasts can also be envisioned.
References
Abe M, Naohiro T, Yoshito I, Chihiro T, Takuji I, Kiyohiko N (2008) Characteristic fungi observed in the fermentation process for Puer tea. Int J Food Microbiol 124(2):199–203. https://doi.org/10.1016/j.ijfoodmicro.2008.03.008
Aggelis G, Sourdis J (1997) Prediction of lipid accumulation-degradation in oleaginous micro-organisms growing on vegetable oils. Antonie Van Leeuwenhoek 72(2):159–165. https://doi.org/10.1023/a:1000364402110
Álvaro-Benito M, Fernández-Lobato M, Baronian K, Kunze G (2013) Assessment of Schwanniomyces occidentalis as a host for protein production using the wide-range Xplor®2 expression platform. Appl Microbiol Biotechnol 97(10):4443–4456
Anschau A, Xavier Michelle CA, Saartje H, Franco T (2014) Effect of feeding strategies on lipid production by Lipomyces starkeyi. Bioresour Technol 157:214–222. https://doi.org/10.1016/j.biortech.2014.01.104
Augstein A, Kathrin B, Marcus G, Sepp KD, Gerold B (2003) Characterization, localization and functional analysis of Gpr1p, a protein affecting sensitivity to acetic acid in the yeast Yarrowia lipolytica. Microbiology (Reading) 149(Pt 3):589–600. https://doi.org/10.1099/mic.0.25917-0
Awad D, Frank B, Norbert M, Thomas B (2019) Multi-factorial-guided media optimization for enhanced biomass and lipid formation by the oleaginous yeast Cutaneotrichosporon oleaginosus. Front Bioeng Biotechnol 7:54. https://doi.org/10.3389/fbioe.2019.00054
Bandhu S, Neha B, Diptarka D, Vivek J, Arushdeep S, Gazal K, Saugata H, Debashish G (2019) Overproduction of single cell oil from xylose rich sugarcane bagasse hydrolysate by an engineered oleaginous yeast Rhodotorula mucilaginosa IIPL32. Fuel 254:115653. https://doi.org/10.1016/j.fuel.2019.115653
Bansal N, Diptarka D, Saugata H, Thallada B, Anjan R, Debashish G (2020) Effect of utilization of crude glycerol as substrate on fatty acid composition of an oleaginous yeast Rhodotorula mucilagenosa IIPL32: Assessment of nutritional indices. Bioresour Technol 309:123330. https://doi.org/10.1016/j.biortech.2020.123330
Beopoulos A, Thierry C, Jean-Marc N (2009) Yarrowia lipolytica: a model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 91(6):692–696. https://doi.org/10.1016/j.biochi.2009.02.004
Beopoulos A, Zuzana M, France T, Marie-Thérèse LD, Ivan H, Seraphim P, Thierry C, Jean-Marc N (2008) Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol 74(24):7779–7789. https://doi.org/10.1128/aem.01412-08
Biernacki M, Marek M, Thomas R, Reinhard P, Kim B, Rüdiger B, Gotthard K (2017) Enhancement of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) accumulation in Arxula adeninivorans by stabilization of production. Microb Cell Factories 16(1):144. https://doi.org/10.1186/s12934-017-0751-4
Bischoff F, Katarzyna L, Arno C, Keith B, Rüdiger B, Frieder S, Gothhard K (2015) Three new cutinases from the yeast Arxula adeninivorans that are suitable for biotechnological applications. Appl Environ Microbiol 81(16):5497–5510. https://doi.org/10.1128/aem.00894-15
Blomqvist J, Jana P, Khalili TS, Sabine S, Nils M, Jude B, Mats S, Volkmar P (2018) Oleaginous yeast as a component in fish feed. Sci Rep 8(1):15945. https://doi.org/10.1038/s41598-018-34232-x
Böer E, Thomas W, Bianka L, Renate M, Rüdiger B, Gerd G, Gotthard K (2004a) Characterization of the AINV gene and the encoded invertase from the dimorphic yeast Arxula adeninivorans. Antonie Van Leeuwenhoek 86(2):121–134. https://doi.org/10.1023/B:ANTO.0000036135.69810.df
Böer E, Thomas W, Karen D, Gerd G, Gotthard K (2004b) Characterization of the Arxula adeninivorans AHOG1 gene and the encoded mitogen-activated protein kinase. Curr Genet 46(5):269–276. https://doi.org/10.1007/s00294-004-0535-3
Böer E, Thomas W, Silvia S, Rüdiger B, Gerd G, Gotthard K (2005a) Characterization of the AXDH gene and the encoded xylitol dehydrogenase from the dimorphic yeast Arxula adeninivorans. Antonie Van Leeuwenhoek 87(3):233–243. https://doi.org/10.1007/s10482-004-3832-4
Böer E, Peter MH, Rüdiger B, Gerd G, Gotthard K (2005b) An extracellular lipase from the dimorphic yeast Arxula adeninivorans: molecular cloning of the ALIP1 gene and characterization of the purified recombinant enzyme. Yeast 22(7):523–535. https://doi.org/10.1002/yea.1230
Böer E, Steinborn G, Florschütz K, Körner M, Gellissen G, Kunze G (2008) Arxula adeninivorans (Blastobotrys adeninivorans) — A dimorphic yeast of great biotechnological potential. In: Satyanarayana T, Kunze G (eds) Yeast biotechnology: diversity and applications. Springer, Dordrecht. https://doi.org/10.1007/978-1-4020-8292-4_27
Böer E, Schröter A, Rüdiger B, Michael P, Gotthard K (2009a) Characterization and expression analysis of a gene cluster for nitrate assimilation from the yeast Arxula adeninivorans. Yeast 26(2):83–93. https://doi.org/10.1002/yea.1653
Böer E, Gamal EM, Ayman EF, Peggy B, Thomas W, Michael P, Gotthard K (2009b) The MAPk ASTE11 is involved in the maintenance of cell wall integrity and in filamentation in Arxula adeninivorans, but not in adaptation to hypertonic stress. FEMS Yeast Res 9(3):468–477. https://doi.org/10.1111/j.1567-1364.2009.00496.x
Böer E, Rüdiger B, Hans-Peter M, Michael P, Gotthard K (2009c) Atan1p-an extracellular tannase from the dimorphic yeast Arxula adeninivorans: molecular cloning of the ATAN1 gene and characterization of the recombinant enzyme. Yeast 26(6):323–337. https://doi.org/10.1002/yea.1669
Böer E, Michael P, Gotthard K (2009d) Xplor 2—an optimized transformation/expression system for recombinant protein production in the yeast Arxula adeninivorans. Appl Microbiol Biotechnol 84(3):583–594. https://doi.org/10.1007/s00253-009-2167-5
Borelli G, Bernabe FM, Vieira DSL, Falsarella CM, Guimarães PGA, Juliana J (2019) Positive selection evidence in xylose-related genes suggests methylglyoxal reductase as a target for the improvement of yeasts’ fermentation in industry. Genome Biol Evol 11(7):1923–1938. https://doi.org/10.1093/gbe/evz036
Bui Duc M, Irene K, Horstmann C, Schmidt T, Breunig Karin D, Gotthard K (1996) Expression of the Arxula adeninivorans glucoamylase gene in Kluyveromyces lactis. Appl Microbiol Biotechnol 45(1-2):102–106. https://doi.org/10.1007/s002530050655
Cai P, Jiaoqi G, Yongjin Z (2019) CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb Cell Factories 18(1):63. https://doi.org/10.1186/s12934-019-1112-2
Casal M, Helena C, Cecilia L (1996) Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology (Reading) 142(Pt 6):1385–1390. https://doi.org/10.1099/13500872-142-6-1385
Cases S, Smith Steven J, Yao-Wu Z, Myers Heather M, Lear Steven R, Eric S, Sabine N, Colin C, Welch Carrie B, Lusis Aldons J, Erickson Sandra K, Farese Robert V Jr (1998) Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci U S A 95(22):13018–13023. https://doi.org/10.1073/pnas.95.22.13018
Castañeda MT, Sebastián N, Claudio V, De BH (2019) In silico optimization of lipid production in Rhodosporidium toruloides by gene knockout strategies*. This work was supported by funding projects from Agencia Na-cional de Promocion Científica y Tecnologica (PICT 2014-2394, 2016-2258), CONICET (PIP 2015-0837) and Universidad Nacional de La Plata (I216, PPID-I008) - Argentina. IFAC-PapersOnLine 52(1):94–99. https://doi.org/10.1016/j.ifacol.2019.06.043
Chang W, Ming Z, Sha Z, Ying L, Xiaobin L, Wei L, Gang L, Zhaomin L, Zhiyu X, Zuntian Z, Hongxiang L (2015) Trapping toxins within lipid droplets is a resistance mechanism in fungi. Sci Rep 5:15133. https://doi.org/10.1038/srep15133
Chattopadhyay A, Maiti MK (2020) Efficient xylose utilization leads to highest lipid productivity in Candida tropicalis SY005 among six yeast strains grown in mixed sugar medium. Appl Microbiol Biotechnol 104(7):3133–3144. https://doi.org/10.1007/s00253-020-10443-z
Chen H, Guangfei H, Lei W, Hongchao W, Zhennan G, Liming L, Hao Z, Wei C, Chen Yong Q (2015a) Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci Rep 5:11247. https://doi.org/10.1038/srep11247
Chen C-X, Zheng S, Hai-Sheng C, Feng-Li F, Long-Ling O, Zhi-Gang Z (2015b) Identification and characterization of three genes encoding acyl-CoA: diacylglycerol acyltransferase (DGAT) from the microalga Myrmecia incisa Reisigl. Algal Res 12:280–288. https://doi.org/10.1016/j.algal.2015.09.007
Choudhary J, Singh S, Nain L (2016) Thermotolerant fermenting yeasts for simultaneous saccharification fermentation of lignocellulosic biomass. Electron J Biotechnol 21:82–92. https://doi.org/10.1016/j.ejbt.2016.02.007
de la Peña MM, Yisheng K, Keasling Jay D (2010) Feast: Choking on Acetyl-CoA, the Glyoxylate Shunt, and Acetyl-CoA-Driven Metabolism. In: Timmis (ed) Handbook of hydrocarbon and lipid microbiology. Springer Berlin Heidelberg, Berlin, pp 1649–1660
Delic M, Graf Alexandra B, Gunda K, Christina H-T, Stephan H, Diethard M, Brigitte G (2014) Overexpression of the transcription factor Yap1 modifies intracellular redox conditions and enhances recombinant protein secretion. Microb Cell 1(11):376–386. https://doi.org/10.15698/mic2014.11.173
Dulermo T, Zbigniew L, Rémi D, Magdalena R, Ramedane H, Jean-Marc N (2015) Analysis of ATP-citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. Biochim Biophys Acta 1851(9):1107–1117. https://doi.org/10.1016/j.bbalip.2015.04.007
El Fiki A, El MG, Carmen B, Thomas W, Rūdiger B, Gerd G, Gotthard K (2007) The Arxula adeninivorans ATAL gene encoding transaldolase-gene characterization and biotechnological exploitation. Appl Microbiol Biotechnol 74(6):1292–1299. https://doi.org/10.1007/s00253-006-0785-8
Fakas S (2017) Lipid biosynthesis in yeasts: a comparison of the lipid biosynthetic pathway between the model nonoleaginous yeast Saccharomyces cerevisiae and the model oleaginous yeast Yarrowia lipolytica. Eng Life Sci 17(3):292–302. https://doi.org/10.1002/elsc.201600040
Friedlander J, Vasiliki T, Annapuma K, Greenhagen Emily H, Consiglio Andrew L, Kyle ME, Crabtree Donald V, Jonathan A, Nugent Rebecca L, Hamilton Maureen A, Joe Shaw A, South Colin R, Gregory S, Brevnova Elena E (2016) Engineering of a high lipid producing Yarrowia lipolytica strain. Biotechnol Biofuels 9:77. https://doi.org/10.1186/s13068-016-0492-3
Galli A, Schiestl RH (1996) Hydroxyurea induces recombination in dividing but not in G1 or G2 cell cycle arrested yeast cells. Mutat Res 354(1):69–75. https://doi.org/10.1016/0027-5107(96)00037-1
Gellissen G, Kunze G, Gaillardin C, Cregg JM, Berardi E, Veenhuis M, van der Klei I (2005) New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica - a comparison. FEMS Yeast Res 5(11): 1079–1096. https://doi.org/10.1016/j.femsyr.2005.06.004
Giacomello M, Pellegrini L (2016) The coming of age of the mitochondria-ER contact: a matter of thickness. Cell Death Differ 23(9):1417–1427. https://doi.org/10.1038/cdd.2016.52
Gienow U, Gotthard K, Frieder S, Rüdiger B, Hofemeister J (1990) The Yeast Genus Trichosporon spec. LS3; Molecular characterization of genomic complexity. Zentralblatt für Mikrobiologie 145(1):3–12. https://doi.org/10.1016/S0232-4393(11)80001-6
Giersberg M, Adelheld D, Rüdiger B, Michael P, Gotthard K (2012) Production of a thermostable alcohol dehydrogenase from Rhodococcus ruber in three different yeast species using the Xplor®2 transformation/expression platform. J Ind Microbiol Biotechnol 39(9):1385–1396. https://doi.org/10.1007/s10295-012-1134-9
Guerfali M, Ines A, Nadia M, Wajdi A, Hafedh B, Rosário BM, Ribeiro Maria HL, Ali G (2019) Triacylglycerols accumulation and glycolipids secretion by the oleaginous yeast Rhodotorula babjevae Y-SL7: Structural identification and biotechnological applications. Bioresour Technol 273:326–334. https://doi.org/10.1016/j.biortech.2018.11.036
Haïli N, Julien L, Michel C, Franjo J, Christelle L-M, Thierry C, Pierre B (2016) Expression of soluble forms of yeast diacylglycerol acyltransferase 2 that integrate a broad range of saturated fatty acids in triacylglycerols. PLoS One 11(10):e0165431. https://doi.org/10.1371/journal.pone.0165431
Ito Y, Takao K, Mamoru Y, Satoshi K, Shingo I, Kenji I, Makoto F-S, Takashi M (2016) Enhancement of protein production via the strong DIT1 terminator and two RNA-binding proteins in Saccharomyces cerevisiae. Sci Rep 6:36997. https://doi.org/10.1038/srep36997
Jackson CL (2019) Lipid droplet biogenesis. Curr Opin Cell Biol 59:88–96. https://doi.org/10.1016/j.ceb.2019.03.018
Jankowska DA, Anke T-S, Arno C, Hoferichter P, Klein C, Rüdiger B, Keith B, Gotthard K (2013b) Arxula adeninivorans xanthine oxidoreductase and its application in the production of food with low purine content. J Appl Microbiol 115(3):796–807. https://doi.org/10.1111/jam.12284
Jankowska DA, Anke T-S, Arno C, Rüdiger B, Keith B, Gotthard K (2015) A novel enzymatic approach in the production of food with low purine content using Arxula adeninivorans endogenous and recombinant purine degradative enzymes. Bioengineered 6(1):20–25. https://doi.org/10.4161/21655979.2014.991667
Jankowska DA, Faulwasser K, Trautwein-Schult A, Arno C, Hoferichter P, Klein C, Rüdiger B, Keith B, Gotthard K (2013a) Arxula adeninivorans recombinant adenine deaminase and its application in the production of food with low purine content. J Appl Microbiol 115(5):1134–1146. https://doi.org/10.1111/jam.12317
Joshi AS, Benjamin N, Vineet C, Prasanna S-K, Levine Tim P, Andy G, Prinz William A (2018) Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat Commun 9(1):2940. https://doi.org/10.1038/s41467-018-05277-3
Juanssilfero AB, Prihardi K, Amza Rezky L, Nao M, Hiromi O, Hana M, Chie K, Ahmad T, Yopi OC, Bambang P, Akihiko K (2018) Effect of inoculum size on single-cell oil production from glucose and xylose using oleaginous yeast Lipomyces starkeyi. J Biosci Bioeng 125(6):695–702. https://doi.org/10.1016/j.jbiosc.2017.12.020
Juergens H, Varela Javier A, Gorter de Vries Arthur R, Thomas P, Gast Veronica JM, Gyurchev Nikola Y, Rajkumar Arun S, Robert M, Pronk Jack T, Morrissey John P, Daran Jean-Marc G (2018) Genome editing in Kluyveromyces and Ogataea yeasts using a broad-host-range Cas9/gRNA co-expression plasmid. FEMS Yeast Res 18(3). https://doi.org/10.1093/femsyr/foy012
Kabran P, Tristan R, Claude G, Jean-Marc N, Cécile N (2012) Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res 19(3):231–244. https://doi.org/10.1093/dnares/dss007
Kakimoto Y, Shinya T, Rieko K, Yuki M, Toshiya E, Yasushi T (2018) Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system. Sci Rep 8(1):6175. https://doi.org/10.1038/s41598-018-24466-0
Kalscheuer R, Alexander S (2003) A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem 278(10):8075–8082. https://doi.org/10.1074/jbc.M210533200
Kamineni A, Shaw J (2020) Engineering triacylglycerol production from sugars in oleaginous yeasts. Curr Opin Biotechnol 62:239–247. https://doi.org/10.1016/j.copbio.2019.12.022
Kamzolova SV, Morgunov IG (2016) Biosynthesis of pyruvic acid from glucose by Blastobotrys adeninivorans. Appl Microbiol Biotechnol 100(17):7689–7697. https://doi.org/10.1007/s00253-016-7618-1
Kasprzak J, Marion R, Sylvia D, Karin B, Kim B, Rüdiger B, Frieder S, Michael P, H-Mathias V, Gotthard K (2016a) Synthesis of ethyl (R)-mandelate using recombinant Carboxydothermus hydrogenoformans alcohol dehydrogenase produced by two yeast species. J Mol Catal B Enzym 133:176–186. https://doi.org/10.1016/j.molcatb.2016.08.012
Kasprzak J, Marion R, Jan R, Sebastian W, Kim B, Rüdiger B, Frieder S, Gotthard K (2016b) Characterization of an Arxula adeninivorans alcohol dehydrogenase involved in the metabolism of ethanol and 1-butanol. FEMS Yeast Res 16(3). https://doi.org/10.1093/femsyr/fow018
Katongole CB, Albina B, Volkmar P, Erik LJ (2017) Effect of solid-state fermentation with Arxula adeninivorans or Hypocrea jecorina (anamorph Trichoderma reesei) on hygienic quality and in-vitro digestibility of banana peels by mono-gastric animals. Livest Sci 199:14–21. https://doi.org/10.1016/j.livsci.2017.03.002
Kaur P, Anja L, Bijinder S, Erik B, Jelena P, Gerhard S, Rüdiger B, Gerd G, Tulasi S, Gotthard K (2007) APHO1 from the yeast Arxula adeninivorans encodes an acid phosphatase of broad substrate specificity. Antonie Van Leeuwenhoek 91(1):45–55. https://doi.org/10.1007/s10482-006-9094-6
Knoll A, Stefan B, Bernward H, Philip E, Kirsten S, Betina R, Christoph S, Juri S, Jochen B (2007) High cell density cultivation of recombinant yeasts and bacteria under non-pressurized and pressurized conditions in stirred tank bioreactors. J Biotechnol 132(2):167–179. https://doi.org/10.1016/j.jbiotec.2007.06.010
Knothe G (2009) Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ Sci 2(7):759–766. https://doi.org/10.1039/B903941D
Kot AM, Stanislaw B, Agnieszka K, Joanna B, Iwona G, Anna B-W, Maliszewska M, Lidia R (2017) Effect of initial pH of medium with potato wastewater and glycerol on protein, lipid and carotenoid biosynthesis by Rhodotorula glutinis. Electron J Biotechnol 27:25–31. https://doi.org/10.1016/j.ejbt.2017.01.007
Kumari A, Keith B, Gotthard K, Rani G (2015) Extracellular expression of YlLip11 with a native signal peptide from Yarrowia lipolytica MSR80 in three different yeast hosts. Protein Expr Purif 110:138–144. https://doi.org/10.1016/j.pep.2015.02.016
Kunze G, Kunze I (1996) Arxula adeninivorans. In: Nonconventional yeasts in biotechnology. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-79856-6_11
Kurtzman CP, Fell JW, Teun B (eds) (2011) The yeasts: a taxonomic study, vol 1–3, 5th edn. Elsevier, Amsterdam
Kunze G, Claude G, Malgorzata C, Pascal D, Tiphaine M, Erik B, Toni G, Cruz Jose A, Emmanuel T, Christian M, André G, Valérie B, Philippe B, Keith B, Sebastian B, Claudine B, Rüdiger B, Serge C, Laurence D, Cécile F, Martin G, Piotr GP, Urs H, Anja H, Dagmara J, Claire J, Paul J, Ingrid L, Véronique L-L, Marc L, Marina M-H, Martin M, Guillaume M, Guy-Franck R, Jan R, Christine S, Anasua S, Guilhem S, Joseph S, Sherman David J, Nils S, Marie-Laure S, Agnès T, Anke T-S, Benoit V, Eric W, Sebastian W, Bernard D, Jean-Luc S, Patrick W, Uwe S, Cécile N (2014) The complete genome of Blastobotrys (Arxula) adeninivorans LS3 - a yeast of biotechnological interest. Biotechnol Biofuels 7:66. https://doi.org/10.1186/1754-6834-7-66
Kurtzman CP (2007) Blastobotrys americana sp. nov., Blastobotrys illinoisensis sp. nov., Blastobotrys malaysiensis sp. nov., Blastobotrys muscicola sp. nov., Blastobotrys peoriensis sp. nov. and Blastobotrys raffinosifermentans sp. nov., novel anamorphic yeast species. Int J Syst Evol Microbiol 57(Pt 5):1154–1162. https://doi.org/10.1099/ijs.0.64847-0
Kurtzman CP, Quintilla MR, Anna K, Bart T, Vincent R, Teun B (2015) Advances in yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Res 15(6). https://doi.org/10.1093/femsyr/fov050
Kurtzman CP, Robnett CJ (2007) Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and 14 new species combinations. FEMS Yeast Res 7(1):141–151. https://doi.org/10.1111/j.1567-1364.2006.00157.x
Kwon Y-J, An-Zhou M, Qian L, Feng W, Guo-Qiang Z, Chun-Zhao L (2011) Effect of lignocellulosic inhibitory compounds on growth and ethanol fermentation of newly-isolated thermotolerant Issatchenkia orientalis. Bioresour Technol 102(17):8099–8104. https://doi.org/10.1016/j.biortech.2011.06.035
Lapeña D, Gergley K, Hansen Line D, Mydland Liv T, Volkmar P, Horn Svein J, Eijsink Vincent GH (2020) Production and characterization of yeasts grown on media composed of spruce-derived sugars and protein hydrolysates from chicken by-products. Microb Cell Factories 19(1):19. https://doi.org/10.1186/s12934-020-1287-6
Ledesma-Amaro R, Zbigniew L, Magdalena R, Zhongpeng G, Florian F, Coq CA-MC-L, Jean-Marc N (2016) Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab Eng 38:115–124. https://doi.org/10.1016/j.ymben.2016.07.001
Li Z, Hanxiao S, Xuemei M, Xiuying L, Bo X, Peng T (2013) Overexpression of malic enzyme (ME) of Mucor circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Appl Microbiol Biotechnol 97(11):4927–4936. https://doi.org/10.1007/s00253-012-4571-5
Litwińska K, Felix B, Falko M, Rüdiger B, Twan R, Gotthard K (2019) Characterization of recombinant laccase from Trametes versicolor synthesized by Arxula adeninivorans and its application in the degradation of pharmaceuticals. AMB Express 9(1):102. https://doi.org/10.1186/s13568-019-0832-3
Liu Y, Amy YS, John KCM, Lianghui J (2016) Developing a set of strong intronic promoters for robust metabolic engineering in oleaginous Rhodotorula (Rhodosporidium) yeast species. Microb Cell Factories 15(1):200. https://doi.org/10.1186/s12934-016-0600-x
Liu L, Anny P, Caitlin S, Nijia Z, Alper Hal S (2015) An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metab Eng 29:36–45. https://doi.org/10.1016/j.ymben.2015.02.003
Liu L-p, Min-hua Z, Yang H, Ning L, Wen-yong L, Hong W (2017) Efficient microbial oil production on crude glycerol by Lipomyces starkeyi AS 2.1560 and its kinetics. Process Biochem 58:230–238. https://doi.org/10.1016/j.procbio.2017.03.024
Macías LG, Miguel M, Christina T, Eladio B (2019) Comparative Genomics Between Saccharomyces kudriavzevii and S. cerevisiae Applied to Identify Mechanisms Involved in Adaptation. Front Genet 10:187. https://doi.org/10.3389/fgene.2019.00187
Madzak C (2015) Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl Microbiol Biotechnol 99(11):4559–4577. https://doi.org/10.1007/s00253-015-6624-z
Maina S, Chrysanthi P, Nikolaos K, Spiros P, Drosinos Eleftherios H, Seraphim P, Apostolis K (2017) Microbial oil production from various carbon sources by newly isolated oleaginous yeasts. Eng Life Sci 17(3):333–344. https://doi.org/10.1002/elsc.201500153
Malak A, Keith B, Gotthard K (2016) Blastobotrys (Arxula) adeninivorans: a promising alternative yeast for biotechnology and basic research. Yeast 33(10):535–547. https://doi.org/10.1002/yea.3180
Maza DD, Viñarta Silvana C, Ying S, Manuel GJ, Aybar Manuel J (2020) Growth and lipid production of Rhodotorula glutinis R4, in comparison to other oleaginous yeasts. J Biotechnol 310:21–31. https://doi.org/10.1016/j.jbiotec.2020.01.012
Meesters PAE, Eggink G (1996) Isolation and characterization of a delta-9 fatty acid desaturase gene from the oleaginous yeast Cryptococcus curvatus CBS 570. Yeast 12(8):723–730. https://doi.org/10.1002/(sici)1097-0061(19960630)12:8<723::aid-yea963>3.0.co;2-o
Middelhoven WJ, Hoogkamer-Te Niet MC, Kreger-Van Rij NJ (1984) Trichosporon adeninovorans sp. nov., a yeast species utilizing adenine, xanthine, uric acid, putrescine and primary n-alkylamines as the sole source of carbon, nitrogen and energy. Antonie Van Leeuwenhoek 50(4):369–378. https://doi.org/10.1007/bf00394651
Ng T-K, Ai-Qun Y, Hua L, Kurniasih PJN, Jae CW, Jan LSS, Wook CM (2020) Engineering Yarrowia lipolytica towards food waste bioremediation: Production of fatty acid ethyl esters from vegetable cooking oil. J Biosci Bioeng 129(1):31–40. https://doi.org/10.1016/j.jbiosc.2019.06.009
Olstorpe M, Schnürer J, Passoth V (2009) Screening of yeast strains for phytase activity. FEMS Yeast Res 9(3):478–488. https://doi.org/10.1111/j.1567-1364.2009.00493.x
Olstorpe M, Jana P, Anders K, Volkmar P (2014) Strain- and temperature-dependent changes of fatty acid composition in Wickerhamomyces anomalus and Blastobotrys adeninivorans. Biotechnol Appl Biochem 61(1):45–50. https://doi.org/10.1002/bab.1130
Park Y-K, Jean-Marc N, Rodrigo L-A (2018) The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol 36(3):304–317. https://doi.org/10.1016/j.tibtech.2017.10.013
Patel A, Matsakas L (2019) A comparative study on de novo and ex novo lipid fermentation by oleaginous yeast using glucose and sonicated waste cooking oil. Ultrason Sonochem 52:364–374. https://doi.org/10.1016/j.ultsonch.2018.12.010
Pham HTM, Martin G, Linda G, Karima H, Jochen T, Steffen U, Gerold H, Peter W, Kirsten S, Keith B, Gotthard K (2015) The determination of pharmaceuticals in wastewater using a recombinant Arxula adeninivorans whole cell biosensor. Sensors Actuators B Chem 211:439–448. https://doi.org/10.1016/j.snb.2015.01.107
Poontawee R, Wichien Y, Savitree L (2017) Efficient oleaginous yeasts for lipid production from lignocellulosic sugars and effects of lignocellulose degradation compounds on growth and lipid production. Process Biochem 53:44–60. https://doi.org/10.1016/j.procbio.2016.11.013
Pretorius IS (2017) Synthetic genome engineering forging new frontiers for wine yeast. Crit Rev Biotechnol 37(1):112–136. https://doi.org/10.1080/07388551.2016.1214945
Qian X, Xinhai Z, Lin C, Xiaoyu Z, Fengxue X, Weiliang D, Wenming Z, Katrin O, Min J (2020) Bioconversion of volatile fatty acids into lipids by the oleaginous yeast Apiotrichum porosum DSM27194. Fuel 119811. https://doi.org/10.1016/j.fuel.2020.119811
Qin L, Lu L, An-Ping Z, Dong W (2017) From low-cost substrates to Single Cell Oils synthesized by oleaginous yeasts. Bioresour Technol 245(Pt B):1507–1519. https://doi.org/10.1016/j.biortech.2017.05.163
Ratledge C (2014) The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: a reappraisal and unsolved problems. Biotechnol Lett 36(8):1557–1568. https://doi.org/10.1007/s10529-014-1532-3
Rauter M, Alexandra P, Jakub K, Karin B, Keith B, Rudiger B, Gothhard K, Vorbrodt Matthias H (2015) Coexpression of Lactobacillus brevis ADH with GDH or G6PDH in Arxula adeninivorans for the synthesis of 1-(R)-phenylethanol. Appl Microbiol Biotechnol 99(11):4723–4733. https://doi.org/10.1007/s00253-014-6297-z
Rauter M, Jakub K, Karin B, Jan R, Sebastian W, Anja H, Martin M, Uwe S, Keith B, Rudiger B, Frieder S, Matthias Vorbrodt H, Gotthard K (2016) Aadh2p: an Arxula adeninivorans alcohol dehydrogenase involved in the first step of the 1-butanol degradation pathway. Microb Cell Factories 15(1):175. https://doi.org/10.1186/s12934-016-0573-9
Reiser J, Ochsner UA, Kälin M, Glumoff V, Fiechter A (1996) Trichosporon. In: Wolf K (ed) Nonconventional yeasts in biotechnology: a handbook. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 581–606
Rösel H, Kunze G (1995) Cloning and characterization of a TEF gene for elongation factor 1α from the yeast Arxula adeninivorans. Curr Genet 28(4):360–366. https://doi.org/10.1007/BF00326434
Rösel H, Kunze G (1998) Integrative transformation of the dimorphic yeast Arxula adeninivorans LS3 based on hygromycin B resistance. Curr Genet 33(2):157–163. https://doi.org/10.1007/s002940050322
Saha S, Balaji E, Sona R, Ram R (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol 141(4):1533–1543. https://doi.org/10.1104/pp.106.082198
Samsonova IA, Gotthard K, Rüdiger B, Böttcher F (1996) A set of genetic markers for the chromosomes of the imperfect yeast Arxula adeninivorans. Yeast 12(12):1209–1217. https://doi.org/10.1002/(sici)1097-0061(19960930)12:12<1209::aid-yea12>3.0.co;2-w
Sano K, Fukuhara H, Nakamura Y (1999) Phytase of the yeast Arxula adeninivorans. Biotechnol Lett 21:33–38. https://doi.org/10.1023/A:1005438121763
Sanya DRA, Djamila O, Gotthard K, Cécile N, Crutz L-CA-M (2020) The native acyltransferase-coding genes DGA1 and DGA2 affect lipid accumulation in Blastobotrys raffinosifermentans differently when overexpressed. FEMS Yeast Res. https://doi.org/10.1093/femsyr/foaa060
Sara M, Kaur BS, François BJ (2016) Lipid production by Yarrowia lipolytica grown on biodiesel-derived crude glycerol: optimization of growth parameters and their effects on the fermentation efficiency. RSC Adv 6(93):90547–90558. https://doi.org/10.1039/C6RA16382C
Sędzielewska KA, Erik B, Carmen B, Thomas W, Rüdiger B, Michael M, Keith B, Gotthard K (2012) Role of the AFRD1-encoded fumarate reductase in hypoxia and osmotolerance in Arxula adeninivorans. FEMS Yeast Res 12(8):924–937. https://doi.org/10.1111/j.1567-1364.2012.00842.x
Shai N, Eden Y, van Roermund Carlo WT, Nir C, Chen B, Lodewijk I, Laetitia C, Julie M, Ramona S, Lior Z, Mari Muriel C, Reggiori Fulvio M, Hughes Adam L, Mafalda E-H, Cohen Mickael M, Waterham Hans R, Wanders Ronald JA, Maya S, Einat Z (2018) Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat Commun 9(1):1761. https://doi.org/10.1038/s41467-018-03957-8
Shapaval V, Jule B, Johanna B, Valeria T, Volkmar P, Mats S, Achim K (2019) Biochemical profiling, prediction of total lipid content and fatty acid profile in oleaginous yeasts by FTIR spectroscopy. Biotechnol Biofuels 12:140. https://doi.org/10.1186/s13068-019-1481-0
Shen X-X, Xiaofan Z, Jacek K, Kurtzman Cletus P, Todd HC, Antonis R (2016) Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda) 6(12):3927–3939. https://doi.org/10.1534/g3.116.034744
Silverman AM, Kangjian Q, Peng X, Gregory S (2016) Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 100(8):3781–3798. https://doi.org/10.1007/s00253-016-7376-0
Sitepu IR, Garay Luis A, Ryan S, David L, Block David E, Bruce GJ, Boundy-Mills Kyria L (2014) Oleaginous yeasts for biodiesel: current and future trends in biology and production. Biotechnol Adv 32(7):1336–1360. https://doi.org/10.1016/j.biotechadv.2014.08.003
Stehlik T, Marco K, Jörg K, Michael B, Johannes F (2020) Peroxisomal targeting of a protein phosphatase type 2C via mitochondrial transit. Nat Commun 11(1):2355. https://doi.org/10.1038/s41467-020-16146-3
Stehr F, Kretschmar M, Kröger C, Hube B, Schäfer W (2003) Microbial lipases as virulence factors. J Mol Catal B Enzym 22(5):347–355. https://doi.org/10.1016/S1381-1177(03)00049-3
Steinborn G, Erik B, Anja S, Kristina T, Gotthard K, Gerd G (2006) Application of a wide-range yeast vector (CoMed) system to recombinant protein production in dimorphic Arxula adeninivorans, methylotrophic Hansenula polymorpha and other yeasts. Microb Cell Factories 5:33. https://doi.org/10.1186/1475-2859-5-33
Steinborn G, Gerd G, Gotthard K (2007) A novel vector element providing multicopy vector integration in Arxula adeninivorans. FEMS Yeast Res 7(7):1197–1205. https://doi.org/10.1111/j.1567-1364.2007.00280.x
Stöckmann C, Marco S, Barbara D, Armin M, Grit H, Georg M, Doris K, Jochen B, Ah KH, Gerd G (2009) Process development in Hansenula polymorpha and Arxula adeninivorans, a re-assessment. Microb Cell Factories 8:22. https://doi.org/10.1186/1475-2859-8-22
Stoltenburg R, Lösche O, Klappach G, Kunze G (1999) Molecular cloning and expression of the ARFC3 gene, a component of the replication factor C from the salt-tolerant, dimorphic yeast Arxula adeninivorans LS3. Curr Genet 35(1):8–13. https://doi.org/10.1007/s002940050426
Sutanto S, Siti Z, Lan T-NP, Suryadi I, Yi-Hsu J (2018) Lipomyces starkeyi: its current status as a potential oil producer. Fuel Process Technol 177:39–55. https://doi.org/10.1016/j.fuproc.2018.04.012
Takaku H, Sayaka E, Kotoha K, Rikako S, Satoshi A, Haruka K, Tomohiko M, Katsuno Y, Hideo A, Yosuke S, Wataru O, Koji I, Sachiyo A, Harutake Y (2021) Isolation and characterization of Lipomyces starkeyi mutants with greatly increased lipid productivity following UV irradiation. J Biosci Bioeng. https://doi.org/10.1016/j.jbiosc.2021.01.006
Tamano K, Bruno Kenneth S, Karagiosis Sue A, Culley David E, Shuang D, Collett James R, Myco U, Hideaki K, Baker Scott E, Masayuki M (2013) Increased production of fatty acids and triglycerides in Aspergillus oryzae by enhancing expressions of fatty acid synthesis-related genes. Appl Microbiol Biotechnol 97(1):269–281. https://doi.org/10.1007/s00253-012-4193-y
Tanimura A, Takashima M, Sugita T, Endoh R, Kikukawa M, Yamaguchi S, Sakuradani E, Ogawa J, Ohkuma M, Shima J (2014) Cryptococcus terricola is a promising oleaginous yeast for biodiesel production from starch through consolidated bioprocessing. Sci Rep 4:4776. https://doi.org/10.1038/srep04776
Tanimura A, Takashima M, Sugita T, Endoh R, Ohkuma M, Kishino S, Ogawa J, Shima J (2016) Lipid production through simultaneous utilization of glucose, xylose, and L-arabinose by Pseudozyma hubeiensis: a comparative screening study. AMB Express 6(1):58. https://doi.org/10.1186/s13568-016-0236-6
Terentiev Y, Huarto PA, Erik B, Thomas W, Jens K, Uta B, Wolfgang B, Manfred S, Gerd G, Gotthard K (2004) A wide-range integrative yeast expression vector system based on Arxula adeninivorans-derived elements. J Ind Microbiol Biotechnol 31(5):223–228. https://doi.org/10.1007/s10295-004-0142-9
Theron CW, Labuschagné M, Gudiminchi R, Albertyn J, Smit MS (2014) A broad-range yeast expression system reveals Arxula adeninivorans expressing a fungal self-sufficient cytochrome P450 monooxygenase as an excellent whole-cell biocatalyst. FEMS Yeast Res 14(4):556–566. https://doi.org/10.1111/1567-1364.12142
Thomas S, Akiola SDR, Florian F, Huu-Vang N, Gotthard K, Cécile N, Anne-Marie C-LC (2019) Blastobotrys adeninivorans and B. raffinosifermentans, two sibling yeast species which accumulate lipids at elevated temperatures and from diverse sugars. Biotechnol Biofuels 12:154. https://doi.org/10.1186/s13068-019-1492-x
Tiukova IA, Sylvain P, Jens N, Mats S, Kerkhoven Eduard J (2019) Genome-scale model of Rhodotorula toruloides metabolism. Biotechnol Bioeng 116(12):3396–3408. https://doi.org/10.1002/bit.27162
Trautwein-Schult A, Dagmara J, Arno C, Petra H, Christina K, Andrea M, Hans-Peter M, Keith B, Rüdiger B, Gotthard K (2013) Arxula adeninivorans recombinant urate oxidase and its application in the production of food with low uric acid content. J Mol Microbiol Biotechnol 23(6):418–430. https://doi.org/10.1159/000353847
Trautwein-Schult A, Dagmara J, Arno C, Petra H, Christina K, Andrea M, Hans-Peter M, Keith B, Rüdiger B, Gotthard K (2014) Arxula adeninivorans recombinant guanine deaminase and its application in the production of food with low purine content. J Mol Microbiol Biotechnol 24(2):67–81. https://doi.org/10.1159/000357674
Tsai Y-Y, Takao O, Chih-Chan W, Dolgormaa B, Ryo M, Savitree L, Kazuhito F (2019) Delta-9 fatty acid desaturase overexpression enhanced lipid production and oleic acid content in Rhodosporidium toruloides for preferable yeast lipid production. J Biosci Bioeng 127(4):430–440. https://doi.org/10.1016/j.jbiosc.2018.09.005
Tsakraklides V, Annapurna K, Consiglio Andrew L, Kyle ME, Jonathan F, Blitzblau Hannah G, Hamilton Maureen A, Crabtree Donald V, Austin S, Jonathan A, Sullivan John E, Greg LW, South Colin R, Greenhagen Emily H, Joe SA, Brevnova Elena E (2018) High-oleate yeast oil without polyunsaturated fatty acids. Biotechnol Biofuels 11:131. https://doi.org/10.1186/s13068-018-1131-y
Tsakraklides V, Elena B, Gregory S, Joe SA (2015) Improved gene targeting through cell cycle synchronization. PLoS One 10(7):e0133434. https://doi.org/10.1371/journal.pone.0133434
Valm AM, Sarah C, Legant Wesley R, Justin M, Uri H, Eric W, Cohen Andrew R, Davidson Michael W, Eric B, Jennifer L-S (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546(7656):162–167. https://doi.org/10.1038/nature22369
Van der Walt JP, Smith MT, Yamada Y (1990) Arxula gen. nov. (Candidaceae), a new anamorphic, arthroconidial yeast genus. Antonie Van Leeuwenhoek 57(1):59–61. https://doi.org/10.1007/bf00400338
Visnapuu T, Aivar M, Kristina P, Katrin V, Karin E, Tiina A (2019) Characterization of a maltase from an early-diverged non-conventional yeast Blastobotrys adeninivorans. Int J Mol Sci 21(1). https://doi.org/10.3390/ijms21010297
Vyas S, Chhabra M (2017) Isolation, identification and characterization of Cystobasidium oligophagum JRC1: A cellulase and lipase producing oleaginous yeast. Bioresour Technol 223:250–258. https://doi.org/10.1016/j.biortech.2016.10.039
Wang Y, Sufang Z, Zhiwei Z, Hongwei S, Xinping L, Xiang J, Xiang J, Kent ZZ (2018a) Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol Biofuels 11:148. https://doi.org/10.1186/s13068-018-1134-8
Wang S, Fatima-Zahra I, Martin H, Alexandra G, Ejsing Christer S, Pedro C (2018b) Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat Commun 9(1):2939. https://doi.org/10.1038/s41467-018-05278-2
Wang G, Delong L, Zhengang M, Shanshan Z, Wenxing L, Lin L (2018c) Comparative transcriptome analysis reveals multiple functions for Mhy1p in lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica. Biochim Biophys Acta Mol Cell Biol Lipids 1863(1):81–90. https://doi.org/10.1016/j.bbalip.2017.10.003
Wang J, Rodrigo L-A, Yongjun W, Boyang J, Xiao-Jun J (2020) Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica—a review. Bioresour Technol 313:123707. https://doi.org/10.1016/j.biortech.2020.123707
Wang Q, Yan C, Biswarup S, Wenmeng M, Lynn ZR, Xianhua L, Guangyi W (2017) Characterization and robust nature of newly isolated oleaginous marine yeast Rhodosporidium spp. from coastal water of Northern China. AMB Express 7(1):30–30. https://doi.org/10.1186/s13568-017-0329-x
Wartmann T, Rösel H, Kunze I, Bode R, Kunze G (1998) AILV1 gene from the yeast Arxula adeninivorans LS3-a new selective transformation marker. Yeast 14(11):1017–1025. https://doi.org/10.1002/(sici)1097-0061(199808)14:11<1017::Aid-yea314>3.0.Co;2-0
Wartmann T, Erik B, Huarto PA, Heike S, Oliver B, Gerd G, Gotthard K (2002) High-level production and secretion of recombinant proteins by the dimorphic yeast Arxula adeninivorans. FEMS Yeast Res 2(3):363–369. https://doi.org/10.1016/s1567-1356(02)00086-7
Wartmann T, Gellissen G, Kunze G (2001) Regulation of the AEFG1 gene, a mitochondrial elongation factor G from the dimorphic yeast Arxula adeninivorans LS3. Curr Genet 40(3):172–178. https://doi.org/10.1007/s002940100254
Wartmann T, Kunze I, Duc Bui M, Manteuffel R, Kunze G (1995) Comparative biochemical, genetical and immunological studies of glucoamylase producing Arxula adeninivorans yeast strains. Microbiol Res 150(2):113–120. https://doi.org/10.1016/s0944-5013(11)80045-5
Wartmann T, Kunze G (2000) Genetic transformation and biotechnological application of the yeast Arxula adeninivorans. Appl Microbiol Biotechnol 54(5):619–624
Wartmann T, Kunze G (2003) Temperature-dependent dimorphism of the non-conventional yeast Arxula adeninivorans. In: Wolf K, Breunig K, Barth G (eds) Non-conventional yeasts in genetics, biochemistry and biotechnology: practical protocols. Springer Berlin Heidelberg, Berlin, pp 3–6
Wartmann T, Regina S, Erik B, Heike S, Oliver B, Gerd G, Gotthard K (2003) The ALEU2 gene—a new component for an Arxula adeninivorans-based expression platform. FEMS Yeast Res 3(2):223–232. https://doi.org/10.1016/s1567-1356(02)00190-3
Williams J, Anke T-S, Gottard K, Kim B (2017) Environmental and metabolic parameters affecting the uric acid production of Arxula adeninivorans. Appl Microbiol Biotechnol 101(11):4725–4736. https://doi.org/10.1007/s00253-017-8199-3
Wu S, Xin Z, Hongwei S, Qian W, Zhao Zongbao K (2011) Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour Technol 102(2):1803–1807. https://doi.org/10.1016/j.biortech.2010.09.033
Xavier MCA, ALV C, Carolina DA, Teixeira FT (2017) Lipid production from hemicellulose hydrolysate and acetic acid by Lipomyces starkeyi and the ability of yeast to metabolize inhibitors. Biochem Eng J 118:11–19. https://doi.org/10.1016/j.bej.2016.11.007
Xenopoulos E, Ioannis G, Afroditi C, Apostolis K, Seraphim P (2020) Lipid production by yeasts growing on commercial xylose in submerged cultures with process water being partially replaced by olive mill wastewaters. Processes 8(7). https://doi.org/10.3390/pr8070819
Xie D, Jackson EN, Zhu Q (2015) Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: from fundamental research to commercial production. Appl Microbiol Biotechnol 99(4):1599–1610. https://doi.org/10.1007/s00253-014-6318-y
Xue Z, Sharpe Pamela L, Seung-Pyo H, Yadav Narenda S, Dongming X, Short David R, Damude Howard G, Rupert Ross A, Seip John E, Jamie W, Pollak Dana W, Bostick Michael W, Bosak Melissa D, Macool Daniel J, Hollerbach Dieter H, Hongxiang Z, Arcilla Dennis M, Bledsoe Sidney A, Kevin C, McCord Elizabeth F, Tyreus Bjorn D, Jackson Ethel N, Quinn Z (2013) Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol 31(8):734–740. https://doi.org/10.1038/nbt.2622
Yamada R, Azusa Y, Tomomi K, Hiroyasu O (2017) Evaluation of lipid production from xylose and glucose/xylose mixed sugar in various oleaginous yeasts and improvement of lipid production by UV mutagenesis. Biochem Eng J 128:76–82. https://doi.org/10.1016/j.bej.2017.09.010
Yamazaki H, Suzuka K, Sayaka E, Shiho A, Satoshi A, Yosuke S, Wataru O, Katsurou Y, Hideo A, Hiroaki T (2019) Highly selective isolation and characterization of Lipomyces starkeyi mutants with increased production of triacylglycerol. Appl Microbiol Biotechnol 103(15):6297–6308. https://doi.org/10.1007/s00253-019-09936-3
Yang X, Guojie J, Zhiwei G, Hongwei S, Fengwu B, Kent ZZ (2014) Recycling biodiesel-derived glycerol by the oleaginous yeast Rhodosporidium toruloides Y4 through the two-stage lipid production process. Biochem Eng J 91:86–91. https://doi.org/10.1016/j.bej.2014.07.015
Yuzbasheva EY, Mostova Elizaveta B, Andreeva Natalia I, Yuzbashev Tigran V, Laptev Ivan A, Sobolevskaya Tatiana I, Sineoky Sergey P (2017) Co-expression of glucose-6-phosphate dehydrogenase and acyl-CoA binding protein enhances lipid accumulation in the yeast Yarrowia lipolytica. New Biotechnol 39(Pt A):18–21. https://doi.org/10.1016/j.nbt.2017.05.008
Zhang R, Shi-Jue G, Chun-Yan Z, Yu S, Xiao-Ling L, Mou-Ming Z, Cheng-Hua W (2019) Characterization of a novel alkaline Arxula adeninivorans urate oxidase expressed in Escherichia coli and its application in reducing uric acid content of food. Food Chem 293:254–262. https://doi.org/10.1016/j.foodchem.2019.04.112
Zhu Z, Sufang Z, Hongwei L, Hongwei S, Xinping L, Fan Y, Zhou Yongjin J, Guojie J, Mingliang Y, Hanfa Z, Zhao Zongbao K (2012) A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun 3:1112. https://doi.org/10.1038/ncomms2112
Acknowledgments
Authors acknowledge Hiroko KITAMOTO, Institute for Agro-Environmental Sciences, National Agriculture and Food Research Organisation, Tsukuba, Ibaraki, Japan, and Zongbao Kent Zhao, Laboratory of Biotechnology, Dalian Institute of Chemical Physics, CAS, 457 Zhongshan Road, Dalian, 116023 People’s Republic of China and Mahesh Khot from FONDECYT (Fondo Nacional de Desarrollo Científico y Tecnológico/National Fund for Scientific and Technological Development), Government of Chile, Chile, Santiago in Postdoctoral Project Grant context (No. 3180134), for proofreading this review document.
Author information
Authors and Affiliations
Contributions
DRAS has prepared manuscript, performed literature analysis, and prepared tables and figures. DO, VP, MKM, AC, and MBK have revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants performed by any of the authors
Consent for publication
Not applicable.
Conflict of interest
All authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 173 kb)
Rights and permissions
About this article
Cite this article
Sanya, D.R.A., Onésime, D., Passoth, V. et al. Yeasts of the Blastobotrys genus are promising platform for lipid-based fuels and oleochemicals production. Appl Microbiol Biotechnol 105, 4879–4897 (2021). https://doi.org/10.1007/s00253-021-11354-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11354-3