Abstract
Microbial production of fats and oils is being developed as a means of converting biomass to biofuels. Here we investigate enhancing expression of enzymes involved in the production of fatty acids and triglycerides as a means to increase production of these compounds in Aspergillus oryzae. Examination of the A. oryzae genome demonstrates that it contains two fatty acid synthases and several other genes that are predicted to be part of this biosynthetic pathway. We enhanced the expression of fatty acid synthesis-related genes by replacing their promoters with the promoter from the constitutively highly expressed gene tef1. We demonstrate that by simply increasing the expression of the fatty acid synthase genes we successfully increased the production of fatty acids and triglycerides by more than two-fold. Enhancement of expression of the fatty acid pathway genes ATP-citrate lyase and palmitoyl-ACP thioesterase increased productivity to a lesser extent. Increasing expression of acetyl-CoA carboxylase caused no detectable change in fatty acid levels. Increases in message level for each gene were monitored using quantitative real-time reverse transcription polymerase chain reaction. Our data demonstrate that a simple increase in the abundance of fatty acid synthase genes can increase the detectable amount of fatty acids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fossil fuels supply energy and raw materials to most processes in our daily life. However, increasing consumption has been an important factor in global climate change and has motivated the quest for alternative energetic compounds. Biofuels are an important alternative to petroleum-derived fuels and are considered to be carbon-neutral. To date, bioethanol is the most widely consumed liquid fuel produced using a microbial process. Ethanol is largely produced using corn starch or molasses feedstocks which are fermented by the common baker’s yeast Saccharomyces cerevisiae. Biodiesel, another liquid fuel derived from biomass, has been produced from a variety of sources by chemical conversion of triglycerides into fatty acyl methyl esters.
Some oil feedstocks used to make biodiesel include “used” cooking oils which are normally oxidized during the cooking process. This leads to high diversity of fatty acyl side chains in the triglyceride making the quality of biodiesel variable. Similar oxidation issues make the harvesting of biomass specifically for use as diesel fuel problematic as well. If triglycerides whose fatty acyl side chains are unsuitable for biodiesel are used to make biodiesel, the products exhibit undesirable characteristics such as solidification, generation of sludge during combustion, and fuel tank corrosion.
Oleaginous fungi accumulate fatty acids and triglycerides in the form of lipid droplets in their cells. The qualities of fatty acids and triglycerides synthesized by the fungi are considered stable in terms of the component varieties and constituent ratios. Therefore, if fungi were used for microbial production of biodiesel, the quality of the fuel would be more consistent. Furthermore, using genetic engineering, varieties of fatty acyl side chains could be modified for improved fuel characteristics. Genetic engineering could also be used to develop systems for fatty acid and triglyceride secretion to aid in separation of the material during processing. If genetic engineering can alter fungi to not only produce fatty acids and triglycerides more efficiently but also vary their energetic characteristics and localization, a very high quality biodiesel could be produced.
Aspergillus oryzae is a fungus that has been used industrially to make Japanese alcohol (sake), soy paste, and soy sauce. It is also the oleaginous fungus that is reported to contain high amount of lipid intracellularly (Meng et al. 2009). In addition, the whole-genome DNA sequence of the parental A. oryzae wild-type strain RIB40 is available (Machida et al. 2005). For these reasons, we believe that A. oryzae is an excellent system in which we engineer increased production of fatty acids and triglycerides genetically.
During fatty acid production in filamentous fungi, illustrated in Fig. 1, glucose is first metabolized to pyruvate by the glycolysis pathway, and the resulting pyruvate is subsequently transported into the mitochondria. The tricarboxylic acid cycle in the mitochondria converts the pyruvate to citrate which is subsequently transported into the cytosol where it stimulates the synthesis of fatty acids and triglycerides. There are four enzymes involved in the process of synthesizing palmitic acid from citrate (Fig. 1). These include ATP-citrate lyase, acetyl-CoA carboxylase, fatty acid synthase, and palmitoyl-ACP thioesterase.
Illustration of the synthesis of fatty acid and triglyceride in a filamentous fungal cell. ATP-citrate lyase (ACL), acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), and palmitoyl-ACP thioesterase (PAT) are involved in synthesis of the 16 carbon molecule palmitic acid. Fatty acid molecules composed of more than 18 carbon atoms and triglycerides are synthesized from palmitic acid. Reactions catalyzed by the enzymes in this study are represented with thick arrows
In this study, we examined the A. oryzae genome sequence to determine a set of genes that play a role in fatty acid biosynthesis. Based on predicted protein sequence similarity, we targeted four genes for promoter replacement and increased expression. Simply increasing expression of fatty acid synthase genes could significantly increase the production of these compounds. Our results also indicated which components in the pathway might be rate-limiting steps, an essential understanding needed when taking steps toward engineering an organism to produce fatty acids and triglycerides.
Materials and methods
Fungal strains and culture media
The A. oryzae NS4DLP (RIB40 ΔligD::ptrA niaD− sC− ΔpyrG) strain, which is a derivative of strain RIB40, was used to make all enhanced expression strains and also used as the parental strain control in this study (Marui et al. 2010; Mizutani et al. 2008; Yamada et al. 1997). The pyrG gene was disrupted in the strain RIB40 ΔligD::ptrA niaD− sC− by deleting a 200-bp-long region which included the start codon, bearing the strain NS4DLP (Marui et al. 2010). Control strains were made by complementing the pyrG and/or sC genes obtained from Aspergillus nidulans to the strain NS4DLP. The NS4DLDP strain (RIB40 ΔligD::ptrA niaD− ΔpyrG::sC of A. nidulans) that was made by disrupting pyrG gene of the strain RIB40 ΔligD::ptrA niaD− sC− with insertion of sC gene obtained from A. nidulans was used for the gene disruption experiments (Marui et al. 2010). The Czapek–Dox (CD) medium (2 % glucose, 0.3 % NaNO3, 0.2 % KCl, 0.1 % KH2PO4, 0.1 % MgSO4·7H2O, 1.5 × 10−4 % FeSO4·7H2O, 6.0 × 10−6 % Na2B4O7·10H2O, 6.0 × 10−5 % CuSO4·5H2O, 1.2 × 10−4 % MnSO4·4H2O, 1.2 × 10−4 % Na2MoO4·2H2O, 1.2 × 10−3 % ZnSO4·7H2O, pH 6.0) was used as a regular minimal medium (Tamano et al. 2007). Both NS4DLP and NS4DLDP strains were maintained on the CD minimal agar supplemented with 5 mM uridine, 10 mM uracil, 10 mM sodium glutamate, and/or 30 μg/ml methionine. The modified CD medium is the medium containing increased glucose concentration of 10 % and was used to cultivate hyphae for the fatty acid and triglyceride productivities assays. The A. nidulans A851 strain was obtained from the Fungal Genetics Stock Center, USA, and its chromosomal DNA was used as a template to amplify the A. nidulans pyrG, sC genes, and the promoter of the translation elongation factor 1a (tef1) gene by PCR.
Disruption of the candidate genes of fatty acid synthase
Disruption constructs for the genes predicted to code for the alpha (AO090124000083) and beta (AO090124000084) subunits of the fatty acid synthase of A. oryzae were generated by fusion PCR (Fig. 2a). All primers used in this study are listed in Table S1. The complementing pyrG gene was obtained from A. nidulans by amplification of a 1.8-kb fragment using the PU_AnpyrG and PL_AnpyrG primers. Two 1-kb-long DNA fragments of upstream and downstream of the coding sequences of candidate genes were generated using LUd/LLd and RUd/RLd primer pairs, respectively. Primer sequences in lowercase designate the tails for fusion to respective adjacent DNA fragments during fusion PCR (Table S1). Each DNA fragment was amplified using KOD-PLUS DNA polymerase (Toyobo, Japan) which is distributed as KOD HOT START DNA polymerase in the USA. The three DNA fragments were purified by agarose gel extraction and fused with primers LUd and RLd using the KOD-PLUS polymerase (Fig. 2a). The program of fusion PCR reaction was as follows: denaturation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 15 s, 66 °C for 30 s, and 68 °C for 4 min, with a final extension at 68 °C for 4 min. The resulting 3.8-kb-long DNA disruption constructs were purified by gel extraction and introduced into strain NS4DLDP. Transformations were performed following a previously described method (Tamano et al. 2007). Transformants were selected on CD agar supplemented with 1.2 M sorbitol, 0.03 % (w/v) palmitic acid, and 1 % (w/v) Tween 40 (polyoxyethylene sorbitan monopalmitate) and were confirmed by PCR using primers cUd and cLd.
Construction of DNA fragments for genetic engineering for enhanced expression. a DNA fragments for disruption and reversion of the fatty acid synthase candidate genes in NS4DLDP. b DNA fragment for constructing the enhanced expression strain of ATP-citrate lyase or fatty acid synthase. c DNA fragment for constructing the enhanced expression strain of acetyl-CoA carboxylase or palmitoyl-ACP thioesterase. d, e DNA fragments for generating the parental strain control complemented with only pyrG gene and both pyrG and sC genes of A. nidulans from NS4DLP, respectively. The box denoted by a dotted line in d shows the deleted region of pyrG in NS4DLP. The asterisk shown in e indicates the point mutation resulting in inactivation of sC
A construct for complementation of the fatty acid synthase beta-subunit candidate gene was generated by amplification of a 7.3-kb-long DNA fragment containing 1 kb of the upstream sequence and 6.3 kb of the coding sequence using LUr and LLr primers. A 1-kb fragment of the partial pyrG gene from A. nidulans was generated using RUr and RLr primers. These two DNA fragments were fused by PCR with primers LUr and RLr and an extension time of 8 min 30 s (Fig. 2a) using EX-Taq DNA polymerase (Takara, Japan). The fused DNA fragment was introduced into the mutant strain of fatty acid synthase beta-subunit. The transformants were selected on CD agar without palmitic acid and were subjected to clone check by PCR with cUr and cLr primers.
Construction of enhanced expression strains
Enhanced expression of fatty acid synthesis-related genes of A. oryzae was performed by replacing their original promoters with strong constitutive promoters from the tef1 genes obtained from A. oryzae or A. nidulans. Primers used to make these constructs using fusion PCR are listed in Table S1. Schematic drawings of the constructs are provided in Fig. 2b, c. The ATP-citrate lyase and fatty acid synthase are each composed of two subunits, and their two genes are situated next to each other in reverse orientation and therefore share a common promoter. In order to generate similar overexpression constructs, the original promoters were replaced with the tef1 promoters of A. oryzae (named “Aotef1p”) and A. nidulans (named “Antef1p”) (Fig. 2b). The tef1 promoter of A. oryzae (“Aotef1p”) was also used for enhanced expression of genes not requiring a bidirectional promoter (Fig. 2c). Fusion PCR was performed by the same reaction composition and program as the case of gene disruption except extension time being set at 5 min 30 s.
Southern hybridization was performed as previously described (Tamano et al. 2007). Briefly, biotin labeled probes were hybridized to the DNA fragments bound to Hybond N + nylon membrane (GE Healthcare, Piscataway, NJ, USA) and detected via chemiluminescence arisen from streptavidin–horseradish peroxidase conjugate using the chemiluminescent nucleic acid detection module (Pierce, Rockford, IL, USA). Probes were prepared by PCR from genomic DNA and labeled with biotin by the North2South biotin random prime labeling kit (Pierce), followed by purification with the QIAquick nucleotide removal kit (Qiagen, Hilden, Germany).
Intracellular fatty acid assay
Cultures were prepared with 2.5 × 107 spores of each A. oryzae strain inoculated into 50 ml of the modified CD medium supplemented with 10 mM sodium glutamate and 30 μg/ml methionine in a 250-ml flask. These were incubated at 30 °C, 200 rpm for 120 h. Hyphae were collected by filtration through Miracloth (EMD Chemicals, San Diego, CA, USA), followed by the wash with 100 ml MiliQ water twice. Excess water was removed from the filtered hyphae by squeezing between paper towels. The hyphae were immediately frozen in liquid nitrogen and lyophilized overnight. Fifty milligrams of lyophilized hyphae was mixed with 300 mg of 0.5 mm glass beads in a 2-ml screw-cap tube (BioSpec Products, Bartlesville, OK, USA) containing 720 μl MiliQ water and 720 μl chloroform containing 1 % Triton X-100. The hyphae were then homogenized with a “Mini Beadbeater-8” (BioSpec Products) at the maximal output for 1 min. After disruption, the hyphae were centrifuged at 14,000×g at 25 °C for 10 min. The bottom of the screw-cap tube was punctured with a needle, and 90 μl of the lower chloroform layer was transferred to a 1.5-ml microcentrifuge tube. The chloroform was evaporated by overnight incubation at 50 °C in a fume hood. The pellet, which was a mixture of fatty acid and Triton X-100, was further dried by lyophilization for 30 min. The pellets were then dissolved by pipetting up and down after sequential addition of 1 μl ethanol, 14 μl MiliQ water, and 735 μl of a solution containing 6 % ethanol and 6 % Triton X-100. Five microliters of the sample was then assayed for intracellular fatty acid using a commercial fatty acid assay kit (Free fatty acids, Half-micro test kit; Roche Applied Science, Mannheim, Germany).
Intracellular triglyceride assay
For determining intracellular triglyceride concentrations, flask culturing and hyphal lyophilization were performed using the same protocols as used for the intracellular fatty acid assay described above. Subsamples of lyophilized hyphae (50 mg) were mixed with 300 mg of 0.5 mm glass beads (BioSpec Products) and 800 μl MiliQ water in 2-ml screw-cap tubes. The hyphae were then homogenized as described for the intracellular fatty acid assay. To each homogenized sample, 800 μl of 10 % (w/v) IGEPAL CA-630 was added and mixed well, followed by incubation at 80 °C for 10 min. The extract was then centrifuged at 14,000×g at 25 °C for 10 min, and a 100-μl aliquot of the upper clear supernatant layer was transferred to a 1.5-ml microcentrifuge tube. A 10-fold dilution of each supernatant was made with MiliQ water, and 10 μl of each dilution was assayed for intracellular triglyceride using a triglyceride assay kit (Cayman Chemical Company, Ann Arbor, MI, USA).
Quantitative reverse transcription polymerase chain reaction
To determine transcript levels using reverse transcription polymerase chain reaction (RT-PCR), each A. oryzae strain was cultured at 30 °C, 200 rpm for 48 h in the same modified CD medium as was used for cultivation for assay of intracellular fatty acid and triglyceride. Hyphae were harvested, immediately frozen with liquid nitrogen, and total RNA (2 μg) was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Contaminating chromosomal DNA was removed by treatment with RNase-Free DNase I (New England Biolabs, Ipswich, MA, USA). These samples were then used as templates for cDNA synthesis reaction using the high-capacity RNA-to-cDNA kit from Applied Biosystems (Foster City, CA, USA). One microliter of the resulting cDNA solutions was applied to quantitative real-time PCR with the Power SYBR Green PCR master mix (Applied Biosystems) using the Applied Biosystems 7900HT Fast Real-time PCR instrument, and the C T values were obtained. Also DNA standards of the genes subjected to RT-PCR were made by PCR with “real_fwd” and “real_rev” primers specific to each gene (Table S1). They were purified by gel extraction using QIAquick gel extraction kit (Qiagen, Germany), and the DNA concentration of each standard solution was assayed using the Quant-iT dsDNA BR assay kits (Invitrogen, Carlsbad, CA, USA) before being utilized in quantitative PCR. An absolute value standard curve was then made from serial dilutions of the DNA standard for each gene product from 4 pM to 4 nM by plotting C T values against concentrations. Absolute transcript level of each gene in parental and mutant strains was determined by measuring the cDNA concentration corresponding to the C T value using the standard curve. Previous DNA microarray results indicated conserved expression levels of the β-tubulin gene, AO090038000317, over a variety of culture conditions (Tamano et al. 2008). Thus, the template cDNA amount was normalized by the expression level of the β-tubulin gene determined by RT-PCR with primers 038-0317real_fwd and 038-0317real_rev.
Phylogenetic analysis
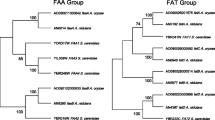
The analysis was performed referring to previous phylogenetic analysis of ketosynthase domains of polyketide synthase performed previously (Baker et al. 2012; Kroken et al. 2003). Amino acid sequences of fatty acid synthases derived from the seven fungal species whose whole genome had been sequenced were used for the analysis. The amino acid sequences of the ketoacyl synthase (KS) domain of alpha-subunit and the malonyl/palmitoyl transferase (MPT) domain of beta-subunit of fatty acid synthase were aligned using CLUSTAL X and subsequently manually adjusted. The genealogy of fatty acid synthases on the bases of the KS domain for the alpha-subunit and the MPT domain for the beta-subunit was inferred by maximum-parsimony (MP) and maximum-likelihood (ML) analysis using MEGA software package version 5 (Tamura et al. 2011). The majority rule consensus bootstrap tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. The MP trees were obtained using the close-neighbor-interchange algorithm with search level 3, in which the initial trees were obtained with the random addition of sequences (ten replicates). The trees are drawn to scale, with branch lengths calculated using the average pathway method, and are in the units of the number of changes over the whole sequence. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). In addition, a more robust approach was used to infer phylogenetic relationship among the fatty acid synthases studied by performing a randomized bootstrap ML analysis using MEGA version 5, setting the bootstrap analysis to 1,000 runs. The Dayhoff mutation data matrix was used for the analysis of the alignment. And the ML trees were obtained using the nearest-neighbor-interchange algorithm, in which the initial trees were obtained automatically.
Results
Identification of the genes of A. oryzae related to fatty acid synthesis
Using genomic information available for A. oryzae, genes predicted to encode four enzymes for fatty acid synthesis were found. Their names and roles in fatty acid synthesis are shown in Fig. 1. The ATP-citrate lyase (ACL) and fatty acid synthase (FAS) enzymes are reported to be composed of two subunits in the filamentous fungus A. nidulans (Brown et al. 1996; Hynes and Murray 2010; Schüller et al. 1992). Therefore, it seemed likely that there would be two genes encoding them in A. oryzae as well. Using sequence similarity to the orthologs of S. cerevisiae (except ACL) and A. nidulans, the genes encoding ACL (AO090023000205 for subunit 2; AO090023000206 for subunit 1), acetyl-CoA carboxylase (AO090011000838), and palmitoyl-ACP thioesterase (AO090012000721) were putatively identified. On the other hand, more than two gene pairs were predicted for fatty acid synthase subunits with significant homology. Therefore, detailed analyses including phylogenetic analysis were performed to identify the most probable orthologous gene pair working predominantly in A. oryzae.
In the original genomic analysis and the annotation-improved analysis of A. oryzae, ten candidates were predicted to code for fatty acid synthases (Machida et al. 2005; Vongsangnak et al. 2008). Five of them encode alpha subunits, while another five encode beta-subunits. Among them, the AO090124000083 and AO090124000084 genes were considered most plausible in A. oryzae. This conclusion was drawn from the following three observations: (a) They had the highest amino acid sequence homologies to the orthologs of S. cerevisiae and A. nidulans that were essential to growth, respectively. The amino acid sequences translated from AO090124000083 and AO090124000084 are identical at 63 and 56 % to YPL231W and YKL182W of S. cerevisiae and at 88 and 89 % to AN9407 and AN9408 of A. nidulans, respectively, from the BLASTP analyses; (b) their cDNA was read most frequently among the candidate genes in the previous expressed sequence tag analysis of A. oryzae (Akao et al. 2007); (c) we made molecular phylogenetic trees of genes encoding fatty acid synthases from some fungi that have sequenced whole genomes (Figs. 3 and S1) (Andersen et al. 2011; Galagan et al. 2003, 2005; Goffeau et al. 1996; Jones et al. 2004; Machida et al. 2005; Nierman et al. 2005; Pel et al. 2007), and AO090124000083 and AO090124000084 were located nearest the orthologous genes of S. cerevisiae and A. nidulans among the genes encoding fatty acid synthase of A. oryzae. Given these data taken from genome sequence information alone, we felt it was plausible to target these two genes for gene disruption analysis.
Genealogy of fatty acid synthases, inferred by maximum-parsimony analysis of the ketoacyl synthase domain for the alpha-subunit, whereas the malonyl/palmitoyl transferase domain for the beta-subunit. Left panel shows the tree of alpha-subunit genes, while right panel shows that of beta-subunit genes. Systematic name and derivation of each gene are written at the right side of each tree. Numbers close to branches indicate percentage bootstrap support for each clade (numbers below 50 % are not shown). Two genes “jgi 206414” and “jgi 43457” marked with asterisks are from A. niger ATCC 1015 strain sequenced by the Joint Genome Institute of DOE (Andersen et al. 2011). They are orthologous to “An01g00050” and “An09g01740” genes from A. niger CBS 513.88 strain whose genome was sequenced by DSM Food Specialties (Pel et al. 2007). The reason why their sequences were used for this analysis instead of orthologs from CBS 513.88 is that the predicted sequences of CBS orthologs seem shorter than the actual lengths and do not contain malonyl/palmitoyl thioesterase domain required to the analysis, resulting in unavailability for it
Fatty acid synthesis is linked to cell membrane synthesis because fatty acids are used to make phospholipids, the major components of cell membrane. Null mutations of fatty acid synthesis-related genes in S. cerevisiae and A. nidulans are not viable on minimal media (Brown et al. 1996; Schüller et al. 1992). This indicates fatty acid synthesis is essential. Supplementation with palmitic acid or myristic acid restores viability to these mutants (Brown et al. 1996; Schüller et al. 1992). We reasoned that if we targeted the primary genes responsible for fatty acid synthesis in A. oryzae, the strains would display a similar phenotype. Therefore, we targeted the genes AO090124000083 and AO090124000084 for gene replacement and tested the mutants for viability on minimal media. The mutant of AO090124000084 could not grow in the absence of palmitic acid (Fig. 4), and it lost the ability to make spores when grown even with palmitic acid supplementation. A complementation test of the AO090124000084 mutant was performed by reintroducing the whole gene at the original locus. The resulting strain recovered both the capability to grow without palmitic acid and to form spores (Fig. 4). By comparison, attempts at gene replacement of AO090124000083 resulted only in obtaining heterokaryons that had nuclei of both parental strain and mutant. Though single spore isolation was performed three times in the presence of palmitic acid, no mutant homokaryons were obtained. This may indicate that the gene is essential and required for viability even in the presence of palmitic acid supplementation. From these data, both genes seem likely to play an important role in fatty acid synthesis in A. oryzae.
Fatty acid synthase beta-subunit disruption mutant. Growth of the disruptant and the revertant of fatty acid synthase beta-subunit gene (AO090124000084) on CD plates supplemented with (left panel) or without (right panel) 0.03 % palmitic acid and 1 % Tween 40 is shown. The disruptant strain (ΔFASβ) was inoculated with hyphae. NS4DLP complemented with both pyrG and sC of A. nidulans as a parental strain control (WT) and the revertant (ΔFASβ/FASβ) were spot-inoculated with 1 × 104 spores, respectively. All plates were incubated at 30 °C for 96 h
Generating the enhanced expression strains of genes involved in fatty acid synthesis in A. oryzae
We made enhanced expression strains of the two primary genes for fatty acid synthesis in an attempt to increase the production of fatty acid. Promoter sequence of the tef1 gene was used to replace their original promoters for enhancing expression (Kitamoto et al. 1998). Schematic drawings of the constructs for increased expression are illustrated in Fig. 2. The tef1 gene promoter of A. oryzae was reported to be useful in overexpression of genes when used to replace native promoters (Marui et al. 2010). The tef1 gene promoter of A. nidulans has not yet been reported to be similarly useful; however, its expression is reported to be constitutively high by the DNA microarray analysis (Mogensen et al. 2006). Therefore, we used both tef1 promoters for making enhanced expression strains in this study.
As described above, two genes encoding distinct subunits of ATP-citrate lyase and fatty acid synthase are situated side by side in outward directions on the chromosome of A. oryzae. The promoters of both genes were replaced with tef1 promoters. In this replacement, if tef1 promoters derived from the same species were to be used, the resulting construct proved to be unstable due to proximity of the same two DNA sequences. Constructs containing inverted copies of the identical tef1 promoter sequence were attempted, but no positive clones were obtained in this study. In an attempt to construct a more stable bidirectional promoter, we used both tef1 promoters of A. oryzae and A. nidulans to make the enhanced expression strains. Only the tef1 promoter of A. oryzae was used in the construction of enhanced expression strains of the single genes encoding acetyl-CoA carboxylase and palmitoyl-ACP thioesterase.
Each enhanced expression strain obtained was tested for growth on CD agar plates supplemented with sodium glutamate and methionine. Each of the mutants was spotted with 5 × 104 spores at the center of a minimal agar plate. Another set of plates were prepared by spreading 5 × 105 spores across the surface of an entire minimal media plate. These plates were incubated at 30 °C for 6 days and growth was observed. The upper row in Fig. 5a shows the plates where spores were spotted at the center, while the lower row contains images of the plates where spores were spread for spore production. Enhanced expression of fatty acid synthase caused a decrease in colony diameter. Enhanced expression of ATP-citrate lyase also had a negative effect on overall colony growth rate while increased expression of palmitoyl-ACP thioesterase or acetyl-CoA carboxylase had little impact on growth. The parental strain control made the largest colony. The diameter of each colony was measured and represented as a graph (Fig. 5b). After 6 days of incubation, the fatty acid synthase expression strain was 27 mm in diameter, which was only 39 % of the size of the parental strain. The enhanced ATP-citrate lyase expression strain was 44 mm in diameter, which was 64 % of the parental strain.
Comparison of growth between enhanced expression strains and the parental strain control. The parental strain control (WT) is NS4DLP complemented with the pyrG of A. nidulans. The data of enhanced expression strains were indicated with abbreviations of the genes subjected to enhanced expression, respectively. Graphs show the averages and standard deviations of data from three independent clones. Experiments were performed in triplicate. a Growth phenotypes of the tested strains. The mutants and the parental strain control were spot-inoculated with 5 × 104 spores (upper row) or spread with 5 × 105 spores (lower row) onto the CD agar supplemented with 10 mM sodium glutamate and 30 μg/ml methionine, followed by incubation at 30 °C for 6 days. b Colony diameters of the mutants compared with the parental strain control. These plates were spot-inoculated as for the upper row of a and were incubated at 30 °C for 3–6 days. Colony diameters were measured. c Comparison of the efficiency of spore formation. The mutants and the parental strain control were spread-inoculated as for the lower row of a and were incubated at 30 °C for 6 days. Spores were collected and their numbers were counted with the improved Neubauer hemocytometer. d Lyophilized mutant and the parental strain control hyphae from 2 and 5 day flask cultures were weighed
With the spread-inoculated spore production plates (Fig. 5a lower row), the enhanced fatty acid synthase expression strain appeared to be making more spores than the parental strain control plates while the enhanced ATP-citrate lyase and palmitoyl-ACP thioesterase expression strains appeared to be making fewer spores. Enhanced expression of acetyl-CoA carboxylase appeared to have no effect on spore production. To quantify this, we collected spores from each plate and counted the total number of spores per plate (Fig. 5c). The enhanced fatty acid synthase expression strain had 1.8-fold more spores relative to the parental strain control per plate while the ATP-citrate lyase and palmitoyl-ACP thioesterase strains produced fewer spores and the acetyl-CoA carboxylase had no effect on spore production. These results demonstrate that enhanced expression of fatty acid synthase negatively affects radial growth rate, but does not have a negative impact on asexual sporulation.
Quantification of intracellular fatty acid and triglyceride
Growth rates in modified CD media supplemented with sodium glutamate and methionine were determined to be similar for all strains used in this study over a period of 5 days (Fig. 5d). Also they proved to be increasing cellular mass even at the passage of 5 days after inoculation (Fig. 5d). Therefore, fatty acid and triglyceride productivities were determined from hyphae cultivated for 5 days.
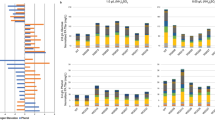
Intracellular fatty acid and triglyceride were assayed using commercially available bioassay kits as described in “Materials and methods.” The results of these assays showed that the greatest increases in both fatty acids and triglycerides were observed in the enhanced fatty acid synthase expression strain. Relative to the parental strain, in the fatty acid synthase expression strain, the fatty acids per gram of lyophilized hyphae increased 2.1-fold and triglycerides increased 2.2-fold (Fig. 6a, c). Expressed as production per liter of flask culture, fatty acids increased 2.4-fold and triglycerides increased 2.8-fold in the fatty acid synthase expression strain relative to the parental strain (Fig. 6b, d). The resulting mass productivity of fatty acid in the enhanced expression strain of fatty acid synthase was 0.047 mmol/g of lyophilized hyphae, which corresponded to 12 mg of palmitic acid-converted weight (Fig. 6a). From the point of unit culture volume, 0.26 mmol fatty acid was produced per liter, which was equivalent to 67 mg of palmitic acid-converted weight (Fig. 6b). For triglycerides, 0.25 mmol was made per gram of lyophilized hyphae, which corresponded to 221 mg triglyceride (Fig. 6c). And it was 1.39 mmol of the production per liter of culture, corresponding to 1.23 g triglyceride (Fig. 6d).
Results of assays measuring intracellular fatty acid and triglycerides. NS4DLP complemented with pyrG of A. nidulans was used as the control of parental strain phenotype. Data from enhanced expression strains are indicated by abbreviations of the genes subjected to enhanced expression. Amounts of intracellular fatty acid per gram of lyophilized hyphae and those per liter of culture are represented in panels (a, b), respectively. Amounts of intracellular triglyceride per gram of lyophilized hyphae and per liter of culture are represented at panels (c, d), respectively. Graphs show the averages and standard deviations of data for three independent clones
The second greatest increase in productivity was seen in the ATP-citrate lyase-enhanced expression strain, where a 1.7-fold increase in the productivities of both fatty acids and triglycerides per gram of lyophilized hyphae relative to the parental strain was observed (Fig. 6a, c). Expressed relative to culture volume, this amounted to 1.7- and 1.9-fold increases in fatty acid and triglyceride per liter, respectively, relative to the parental strain control (Fig. 6b, d). The palmitoyl-ACP thioesterase-enhanced expression strain also showed increased productivity of fatty acids and triglycerides (1.5- and 1.4-fold increase, respectively). These increases were similar to the enhanced ATP-citrate lyase expression strain. The acetyl-CoA carboxylase-enhanced expression strain did not show a significant increase in productivities of either fatty acids or triglycerides relative to the parental strain.
Quantitative real-time RT-PCR determination of transcriptional levels of fatty acid synthesis-related genes from enhanced expression strains
As described in “Materials and methods,” the tef1 promoters of A. oryzae and A. nidulans were used to make strains with enhanced expression of several fatty acid synthesis-related genes. The tef1 promoter of A. oryzae was reported to be effective in achieving high level expression of target genes in A. oryzae (Marui et al. 2010), whereas the effectiveness of the tef1 promoter from A. nidulans was unknown. Therefore, quantitative real-time RT-PCR was used to determine the expression levels of genes whose promoters were replaced with the tef1 promoters from A. oryzae and A. nidulans. The results shown in Table 1 showed that increased expression was obtained for all genes investigated. The greatest fold increases in mRNA levels relative to the parental strain were observed for the palmitoyl-ACP thioesterase-enhanced expression strain (5.7-fold) and for the acetyl-CoA carboxylase-enhanced expression strain, (3.8-fold). Expression levels of the ATP-citrate lyase and fatty acid synthase genes in these enhanced expression strains were also elevated, but to a lesser extent (1.2- and 1.8-fold, respectively).
It is interesting to note that the fold increases in production of fatty acids and triglycerides were inversely related to the fold increases in transcript level. And since the increases of expression levels were 1.2- to 1.8-fold, there seemed to be quantitative relationship between them and increase ratios of the productivities of fatty acid and triglyceride.
The promoter of tef1 gene has been reported to constitutively express high levels of homologous and heterologous transcripts in A. oryzae and A. nidulans (Kitamoto et al. 1998; Mogensen et al. 2006). However, our results indicated that the level of expression of homologous genes whose promoters were replaced with the tef1 promoter was quite variable. To further investigate this phenomenon, quantitative real-time RT-PCR was used to compare the expression levels of genes subjected to enhanced expression with the native tef1 gene in those strains. While expression of each gene was enhanced when the native promoter was replaced by the tef1 promoter, the results presented in Table 1 indicate that expression levels were all lower than the expression of the native tef1 gene. The smallest expressional difference from tef1 gene was observed in the ATP-citrate lyase subunit 1-enhanced expression strain with an expression level that was one third of that of tef1 gene. The largest expressional difference from tef1 gene occurred in the ATP-citrate lyase subunit 2-enhanced expression strain where the expression level was 100 times less than that of tef1 gene.
Discussion
This study utilized genetic engineering methods in A. oryzae to increase production of fatty acids and triglycerides, the source materials of biodiesel. Enhancement of expression of the genes directly involved in fatty acid synthesis was chosen as the strategy. Our results indicated that enhancing expressions of the fatty acid synthase genes yielded the greatest increases in production of both fatty acids and triglycerides.
Fatty acid synthase is formed from two subunits encoded by separate genes. However, in A. oryzae, ten genes that compose five gene pairs with homology to known fatty acid synthases were found. The gene pair that we chose to investigate was predicted to be most important in fatty acid synthesis based on (1) homology to the orthologs of S. cerevisiae and A. nidulans that were essential to their growth and (2) showed the highest level of expression in A. oryzae microarray data (Tamano et al. 2008). Consequently, the gene pair of AO090124000083 and AO090124000084 was predicted to be most important for fatty acid synthesis in A. oryzae. This conclusion was supported by the results obtained from disruption of these genes.
It should be noted that the orthologous gene AN9407.1 of A. nidulans corresponding to AO090124000083 encoding fatty acid synthase alpha-subunit was successfully disrupted in the previous study (Brown et al. 1996). Differently from A. nidulans, disruptants of A. oryzae, which possesses multiple nuclei (heterokaryon), must be purified two to three times by the single spore isolation to exclude the transformants having the intact nucleus. During this process, the cells having the intact nuclei might have been more efficiently selected due to better growth than the true disruptant without the intact nucleus (homokaryon), if any, even when palmitic acid was supplemented. As a result, we might have failed to obtain the homokaryon disruptant.
One hypothesis concerning the function of two of the four additional fatty acid synthase gene pair homologues (other than AO090124000083 and AO090124000084) is based on the observation that some polyketide synthases involved in synthesis of secondary metabolites use the fatty acid synthase product fatty acyl CoA as a substrate. In these instances, genes of both fatty acid synthase and polyketide synthase are often situated proximally in a secondary metabolite gene cluster. In A. oryzae, the polyketide synthase genes AO090026000009 and AO090010000114 are situated near the fatty acid synthase gene pairs AO09002600012/3 and AO090010000107/8, respectively. The former is incorporated in the aflatoxin biosynthesis gene cluster that is inactivated in A. oryzae by spontaneous mutations, while the latter seems to be incorporated in a secondary metabolite gene cluster of unknown function. Further support for the hypothesis that these additional fatty acid synthase genes being involved in secondary metabolite production rather than fatty acid synthesis is provided by the fact that they are phylogenetically distinct from the essential fatty acid synthase genes (Fig. 3).
The subunit genes of the two remaining fatty acid synthase gene pairs in A. oryzae (AO090010000156/71 and AO090011000040/6) are located 28 and 37 kb apart, with 12 and five genes intervening between them, respectively. In addition, these genes are not associated with identifiable polyketide gene clusters. The function of these genes is undetermined. Possibly these genes originated through duplication and translocation of the other fatty acid synthase genes at some point in the evolution of A. oryzae. In support of this hypothesis, phylogenetic analysis indicated that the AO090011000040/6 and AO090010000156/71 gene pair clusters were segregated with AO090124000083–4 and AO090010000107–8 (Fig. 3), respectively. This leads to the hypothesis that AO090011000040/6 and AO090010000156/71 are derived from AO090124000083–4 and AO090010000107–8, respectively.
In our enhanced expression strains, a correlation between colony size and increase in fatty acid and triglyceride productivities was observed (Fig. 5a, b). The enhanced fatty acid synthase expression strain that showed the greatest increase in fatty acid and triglyceride productivity made the smallest colonies among the tested mutants. The enhanced acetyl-CoA carboxylase expression strain that showed no increase in fatty acid or triglyceride production produced colonies indistinguishable from the parental strain control. The other strains showed intermediate colony size and productivity phenotypes. On the other hand, differences in hyphal growth were not obvious between the transformant and the wild-type strains at any time points during the cultivation when their spores were spread over the entire agar surface of the plate. Taking the reduced hyphal growth above when the spores are spotted at the center of the agar plate into consideration, the results suggest reduction of hyphal extension along the agar surface. Therefore, it is possible that the correlation between the enhanced expression and the hyphal growth in Fig. 5a may be related to the diversion of acetyl-CoA from sterol synthesis to fatty acid and triglyceride synthesis affecting hyphal growth characteristics. Further, transformant of the enhanced FAS expression in the flask cultivation showed morphological change of mycelia from clustered globular shape of about 2 mm in diameter to evenly dispersed thin filaments in the medium (data not shown). This supports the idea that sterol biosynthesis essential to plasma membrane might be affected by the enhancement of the fatty acid biosynthesis. Another interesting observation was that the enhanced fatty acid synthase expression strain made more spores than any of the other strains tested. This strain made 1.8 times more spores than the parental strain control, whereas the other strains made the same or fewer spores. The reason is unclear, but this characteristic should not be a drawback for further improvements or industrial use in the future.
Further enhancement of expression of these enzymatic functions may result from improvements in the expression of the genes involved. Microarray data obtained by our group indicated that expression of the tef1 gene is constitutively high (Tamano et al. 2008). In this study, we observed that, although expression was driven by the same tef1 promoter, the levels of expression of each fatty acid synthesis-related gene were less than expression of the native tef1 gene. The reason for this reduced expression is not clear; however, it is possible that this difference is due to a position effect resulting from the tef1 promoter being introduced into different loci on the chromosome of A. oryzae. It is also possible that expression rates are actually equal, but the mRNAs of these fatty acid synthesis-related genes are less stable than the tef1 mRNA. The length of the A. oryzae tef1 promoter used in this study was sufficient to attained enhanced expression of other A. oryzae genes in two previous studies (Kitamoto et al. 1998; Marui et al. 2010). However, it is possible that these genes contain enhancer elements that are absent in the fatty acid synthesis-related genes. Further optimization of expression of these genes may result in greater increases in productivities of fatty acids and triglycerides through enhanced expression of enzyme functions in A. oryzae. Although we have found that the tef1 promoter showed the strongest expression, other promoters such as those of amyB (α-amylase gene) (Matsushita-Morita et al. 2011) and pgk (phosphoglycerate kinase gene) (Sakai et al. 2012) could be used in the presence of maltose and glucose, respectively. Further to enhance the fatty acid productivity, integration into other loci using other marker genes could also be examined as a trial.
To date, attempts have been made to increase fatty acid and triglyceride production in Mucor and Escherichia coli (Courchesne et al. 2009; Meng et al. 2009; Zhang et al. 2007). In the fungus Mucor, a 2.5-fold increase in accumulation of intracellular total lipid was attained by overexpression of a malic enzyme gene (Zhang et al. 2007). To test this idea in A. oryzae, we generated a strain with enhanced expression of two malic enzyme genes (AO090011000876 and AO090038000621) by replacing their promoters with tef1 promoters of A. oryzae. However, in contrast to the result in Mucor, no increase in productivity of fatty acids was observed (data not shown). Overexpressing the acetyl-CoA carboxylase gene in a Hansenula spp. of yeast resulted in a 1.4-fold increase in fatty acid productivity (Ruenwai et al. 2009). The increased productivity in this Hansenula strain was about 50 mg/g of dry cell which is approximately four times higher than the maximal productivity we attained in this study (12 mg/g of lyophilized hyphae in the enhanced fatty acid synthase gene expression strain). In bacteria, productivity of 1.0 to 2.5 g of fatty acid/l was attained in E. coli by combining overexpression of a thioesterase gene with deletion of fatty acid degradation-related genes (Handke et al. 2011; Lu et al. 2008; Steen et al. 2010). This is 15- to 38-fold higher fatty acid productivity than the enhanced fatty acid synthase expression strain of A. oryzae that accomplished the largest productivity in this study. Additional research reported that co-overexpression of acetyl-CoA carboxylase and malic enzyme in E. coli resulted in a 5.6-fold increase in fatty acid productivity relative to the parental strain (Meng et al. 2011). This corresponded to 285 mg fatty acids/g of dry cell which is 4.3-fold higher than the maximum obtained in this study.
The fungus Mortierella isabellina has been reported to produce approximately 6 g of triglyceride/l of flask culture at 30 °C for 120 h (Chatzifragkou et al. 2010). In comparison, our strain with enhanced expression of the fatty acid synthase gene produced 1.23 g triglyceride/l of flask culture at 30 °C for 120 h. Although there are differences in culture media and growth rate, triglyceride productivity of M. isabellina appears to be 4.9-fold higher than we obtained with the enhanced fatty acid synthase expression strain of A. oryzae.
Our results indicated that enhancing expression of the fatty acid synthase genes in A. oryzae yielded a significant increase in production of both fatty acids and triglycerides. The influence of several other genes involved in fatty acid and triglyceride biosynthesis was also investigated. The maximal productivities we obtained in this study are less than those obtained in other microorganisms. However, the culture conditions used for this study have not been optimized, and other growth conditions may be more suitable for production of fatty acids. Changes in the composition of the media (such as changing concentrations of carbon and nitrogen sources and their ratio) and cultivation period could significantly improve fatty acid and triglyceride production by A. oryzae. As was observed in E. coli, further increases in productivity may be achieved in A. oryzae through introduction of multiple genetic changes. It will be particularly interesting to see the results of further research directed at determining the effects of combining overexpression of fatty acid biosynthetic genes with disruptions of genes related to fatty acid degradation.
References
Akao T, Sano M, Yamada O, Akeno T, Fujii K, Goto K, Ohashi-Kunihiro S, Takase K, Yasukawa-Watanabe M, Yamaguchi K, Kurihara Y, Maruyama J, Juvvadi PR, Tanaka A, Hata Y, Koyama Y, Yamaguchi S, Kitamoto N, Gomi K, Abe K, Takeuchi M, Kobayashi T, Horiuchi H, Kitamoto K, Kashiwagi Y, Machida M, Akita O (2007) Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res 14(2):47–57
Andersen MR, Salazar MP, Schaap PJ, van de Vondervoort PJ, Culley D, Thykaer J, Frisvad JC, Nielsen KF, Albang R, Albermann K, Berka RM, Braus GH, Braus-Stromeyer SA, Corrochano LM, Dai Z, van Dijck PW, Hofmann G, Lasure LL, Magnuson JK, Menke H, Meijer M, Meijer SL, Nielsen JB, Nielsen ML, van Ooyen AJ, Pel HJ, Poulsen L, Samson RA, Stam H, Tsang A, van den Brink JM, Atkins A, Aerts A, Shapiro H, Pangilinan J, Salamov A, Lou Y, Lindquist E, Lucas S, Grimwood J, Grigoriev IV, Kubicek CP, Martinez D, van Peij NN, Roubos JA, Nielsen J, Baker SE (2011) Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res 21(6):885–897
Baker SE, Perrone G, Richardson NM, Gallo A, Kubicek CP (2012) Phylogenomic analysis of polyketide synthase-encoding genes in Trichoderma. Microbiology 158(1):147–154
Brown DW, Adams TH, Keller NP (1996) Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc Natl Acad Sci U S A 93(25):14873–14877
Chatzifragkou A, Fakas S, Galiotou-Panayotou M, Komaitis M, Aggelis G, Papanikolaou S (2010) Commercial sugars as substrates for lipid accumulation in Cunninghamella echinulata and Mortierella isabellina fungi. Eur J Lipid Sci Technol 112:1048–1057
Courchesne NM, Parisien A, Wang B, Lan CQ (2009) Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J Biotechnol 141(1–2):31–41
Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422(6934):859–868
Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438(7071):1105–1115
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274(5287):563–567
Handke P, Lynch SA, Gill RT (2011) Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab Eng 13(1):28–37
Hynes MJ, Murray SL (2010) ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans. Eukaryot Cell 9(7):1039–1048
Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S (2004) The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A 101(19):7329–7334
Kitamoto N, Matsui J, Kawai Y, Kato A, Yoshino S, Ohmiya K, Tsukagoshi N (1998) Utilization of the TEF1-alpha gene (TEF1) promoter for expression of polygalacturonase genes, pgaA and pgaB, in Aspergillus oryzae. Appl Microbiol Biotechnol 50(1):85–92
Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A 100(26):15670–15675
Lu X, Vora H, Khosla C (2008) Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab Eng 10(6):333–339
Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438(7071):1157–1161
Marui J, Ohashi-Kunihiro S, Ando T, Nishimura M, Koike H, Machida M (2010) Penicillin biosynthesis in Aspergillus oryzae and its overproduction by genetic engineering. J Biosci Bioeng 110(1):8–11
Matsushita-Morita M, Tada S, Suzuki S, Hattori R, Marui J, Furukawa I, Yamagata Y, Amano H, Ishida H, Takeuchi M, Kashiwagi Y, Kusumoto K (2011) Overexpression and characterization of an extracellular leucine aminopeptidase from Aspergillus oryzae. Curr Microbiol 62(2):557–564
Meng X, Yang J, Xu X, Zhang L, Nie Q, Xian M (2009) Biodiesel production from oleaginous microorganisms. Renew Energy 34:1–5
Meng X, Yang J, Cao Y, Li L, Jiang X, Xu X, Liu W, Xian M, Zhang Y (2011) Increasing fatty acid production in E. coli by simulating the lipid accumulation of oleaginous microorganisms. J Ind Microbiol Biotechnol 38(8):919–925
Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K (2008) A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol 45:878–889
Mogensen J, Nielsen HB, Hofmann G, Nielsen J (2006) Transcription analysis using high-density micro-arrays of Aspergillus nidulans wild-type and creA mutant during growth on glucose or ethanol. Fungal Genet Biol 43(8):593–603
Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latgé JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O’Neil S, Paulsen I, Peñalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Córdoba S, Rodríguez-Peña JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sánchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438(7071):1151–1156
Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wösten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25(2):221–231
Ruenwai R, Cheevadhanarak S, Laoteng K (2009) Overexpression of acetyl-CoA carboxylase gene of Mucor rouxii enhanced fatty acid content in Hansenula polymorpha. Mol Biotechnol 42(3):327–332
Sakai K, Kinoshita H, Nihira T (2012) Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl Microbiol Biotechnol 93(5):2011–2022
Schüller HJ, Förtsch B, Rautenstrauss B, Wolf DH, Schweizer E (1992) Differential proteolytic sensitivity of yeast fatty acid synthetase subunits alpha and beta contributing to a balanced ratio of both fatty acid synthetase components. Eur J Biochem 203(3):607–614
Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463(7280):559–562
Tamano K, Satoh Y, Ishii T, Terabayashi Y, Ohtaki S, Sano M, Takahashi T, Koyama Y, Mizutani O, Abe K, Machida M (2007) The beta-1,3-exoglucanase gene exgA (exg1) of Aspergillus oryzae is required to catabolize extracellular glucan, and is induced in growth on a solid surface. Biosci Biotechnol Biochem 71(4):926–934
Tamano K, Sano M, Yamane N, Terabayashi Y, Toda T, Sunagawa M, Koike H, Hatamoto O, Umitsuki G, Takahashi T, Koyama Y, Asai R, Abe K, Machida M (2008) Transcriptional regulation of genes on the non-syntenic blocks of Aspergillus oryzae and its functional relationship to solid-state cultivation. Fungal Genet Biol 45(2):139–151
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Vongsangnak W, Olsen P, Hansen K, Krogsgaard S, Nielsen J (2008) Improved annotation through genome-scale metabolic modeling of Aspergillus oryzae. BMC Genomics 9:245
Yamada O, Lee BR, Gomi K (1997) Transformation system for Aspergillus oryzae with double auxotrophic mutations, niaD and sC. Biosci Biotechnol Biochem 61:1367–1369
Zhang Y, Adams IP, Ratledge C (2007) Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153(7):2013–2025
Acknowledgments
We appreciate the daily research-related paperwork by Ms. Debra Lee and Ms. Ana Lundeby at PNNL. We are also grateful to Dr. Osamu Yamada (National Research Institute of Brewing, Japan) for giving us information on the DNA sequences of mutations at niaD and sC loci, respectively, of A. oryzae RIB40 niaD− sC− strain that is called NS4 generally. And we thank Dr. Katsuya Gomi (Tohoku University) for providing A. oryzae RIB40 ΔligD::ptrA niaD− sC− strain, a derivative of NS4, to construct both NS4DLP and NS4DLDP. This work was performed by the grant-in-aid of the Ministry of Economy, Trade, and Industry, Japan. Also this work was supported by the grant-in-aid of the Department of Energy, USA.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 136 kb)
Rights and permissions
About this article
Cite this article
Tamano, K., Bruno, K.S., Karagiosis, S.A. et al. Increased production of fatty acids and triglycerides in Aspergillus oryzae by enhancing expressions of fatty acid synthesis-related genes. Appl Microbiol Biotechnol 97, 269–281 (2013). https://doi.org/10.1007/s00253-012-4193-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4193-y