Abstract
Kluyveromyces marxianus CCT 7735 shows potential for producing ethanol from lactose; however, its low ethanol tolerance is a drawback for its industrial application. The first aim of this study was to obtain four ethanol-tolerant K. marxianus CCT 7735 strains (ETS1, ETS2, ETS3, and ETS4) by adaptive laboratory evolution. The second aim was to select among them the strain that stood out and to evaluate metabolic changes associated with the improved ethanol tolerance in this strain. The ETS4 was selected for displaying a specific growth rate higher than the parental strain under ethanol stress (122%) and specific ethanol production rate (0.26 g/g/h) higher than those presented by the ETS1 (0.22 g/g/h), ETS2 (0.17 g/g/h), and ETS3 (0.17 g/g/h) under non-stress condition. Further analyses were performed with the ETS4 in comparison with its parental strain in order to characterize metabolic changes. Accumulation of valine and metabolites of the citric acid cycle (isocitric acid, citric acid, and cis-aconitic acid) was observed only in the ETS4 subjected to ethanol stress. Their accumulation in this strain may have been important to increase ethanol tolerance. Furthermore, the contents of fatty acid methyl esters and ergosterol were higher in the ETS4 than in the parental strain. These differences likely contributed to enhance ethanol tolerance in the ETS4.
Key points
• K. marxianus ethanol-tolerant strains were selected by adaptive laboratory evolution.
• Valine and metabolites of the TCA cycle were accumulated in the ETS4.
• High contents of fatty acids and ergosterol contributed to enhance ethanol tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Energy crisis and environmental issues have raised the demand for alternative renewable energies such as ethanol, the most commercialized biofuel in the world (Taylor 2008; Mussatto et al. 2010; Valdivia et al. 2016). Currently, ethanol is commonly produced from sugarcane and corn, thus creating a competition between the production of biofuels and food. Ethanol production from other feedstocks such as whey and its derivatives have been considered an alternative to increase its production and reducing the environmental impact of their disposal (González-Siso 1996; Prazeres et al. 2012; Parashar et al. 2016). Kluyveromyces marxianus strains isolated from dairy environments can convert lactose to ethanol. Kluyveromyces marxianus CCT 7735 has stood out as a good ethanol producer from whey permeate (Diniz et al. 2013; Silveira et al. 2005) and a mixture of sugarcane bagasse hydrolysate and ricotta whey (Ferreira et al. 2015). In this strain, the increase of the expression of the lactose permease genes under hypoxia leads to a higher specific lactose consumption rate and specific ethanol production rate (de Paiva et al. 2019). Nevertheless, in contrast to Saccharomyces cerevisiae, a yeast widely used for ethanol production, K. marxianus is less tolerant to the ethanol, which is a drawback for its use. The K. marxianus CCT 7735 growth is severely impaired in ethanol concentrations above 4% (v/v) (Costa et al. 2014; Silveira et al. 2005).
Ethanol alters cell membrane structures by affecting the lipid-water interface, increasing lipid interdigitations, and reducing membrane thickness (Dong et al. 2015; Henderson and Block 2014; Ma and Liu 2010). Protein structure/function are also targets of ethanol (Ma and Liu 2010; Stanley et al. 2010). Most studies that assess cellular responses to ethanol in yeasts have focused on S. cerevisiae (Ma and Liu 2010; Stanley et al. 2010; Snoek et al. 2016).
Changes in ergosterol content, the major sterol in yeast membranes, as well as in fatty acids content and composition are important to counteract the damages provoked by ethanol in S. cerevisiae (Stanley et al. 2010; Vanegas et al. 2012; Caspeta and Nielsen 2015). Diniz et al. (2017) observed that both ergosterol and unsaturated fatty acids contents did not change upon ethanol exposure in K. marxianus. Since in S. cerevisiae, the ergosterol and unsaturated fatty acids levels increase in response to ethanol, the authors claimed that these differences might be related to the low ethanol tolerance of K. marxianus.
Moreover, the accumulation of metabolites as proline, leucine, alanine, glutamate, valine, and the tricarboxylic acid (TCA) cycle-related metabolites confers cellular protection against ethanol stress (Zhao and Bai 2009; Ma and Liu 2010; Stanley et al. 2010; Lourenço et al. 2013; Ohta et al. 2015; Kim et al. 2016).
Over the last years, adaptive laboratory evolution (ALE) has been widely used to obtain yeast strains tolerant to stress conditions (Barrick et al. 2009; Dragosits and Mattanovich 2013; Caspeta et al. 2014; LaCroix et al. 2017). Herein, we selected ethanol-tolerant strains of K. marxianus (ETS) by ALE. The genome analysis, as well as metabolic and fatty acids profiles of unstressed and ethanol-stressed cells provided insights into the mechanisms involved in the acquisition of ethanol tolerance in K. marxianus.
Materials and methods
Yeast strain, maintenance, and culture media
Kluyveromyces marxianus CCT 7735 used in this work, previously designated as UFV-3, belongs to the culture collection of Microbial Physiology of the Department of Microbiology at the Federal University of Viçosa (UFV). It is stored in the Tropical Cultures Collection André Tosello Foundation, São Paulo, Brazil.
For the adaptive laboratory evolution (ALE) experiments, K. marxianus CCT 7735 strains were cultivated in a synthetic defined medium (SD), SD agar, and SDE. SD consisted of g/L: yeast nitrogen base—YNB—without amino acids (Sigma Chemical Co., MO, USA) 6.7, and lactose 20. To prepare the SD agar medium, agar (15 g/L) was added to the SD medium. The SDE medium was prepared by adding ethanol (4% v/v) to the SD medium. For chemostat cultivation, we used the defined medium described by Verduyn et al. (1992) containing 20 g/L of lactose (CBS) and CBSE, in which ethanol (4% v/v) was added to the CBS medium.
Determination of specific growth rate and dry weight
The cell growth was monitored by measurement of optical density at 600 nm (OD600), using a UV-visible spectrophotometer (BECKMAN DU series 600). Biomass and specific growth rate were determined as previously described by da Silveira et al. (2018).
Adaptive laboratory evolution (ALE): yeast strains, growth media, and cultivation conditions
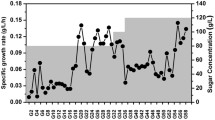
For the ALE experiments, K. marxianus CCT 7735 was cultivated in the SD agar medium and incubated at 37 °C for 48 h. After incubation, four colonies of K. marxianus CCT 7735 were randomly selected and grown for 1 day in 125-mL Erlenmeyer flasks containing 25 mL of SD medium at 37 °C with a stirring rate of 200 rpm (rpm) in an orbital incubator. Subsequently, the cells were harvested at 12,000 g at 4 °C for 5 min, diluted into fresh medium to give an OD600 of about 0.2. The four selected colonies were cultivated in 125-mL Erlenmeyer flasks containing 25 mL of SD plus 4% (v/v) ethanol (SDE), at 37 °C and 200 rpm up to reach 4–7 generations (exponential phase). One aliquot of the culture was diluted into a fresh SDE medium and transferred to a new Erlenmeyer flask containing SDE at OD600 of 0.2 and grown under the conditions aforementioned. The pH of medium was set to 5.5 with 3 M KOH before the outset of cultivation. Passage cultures were performed under ethanol stress during 85 days, the period in which there was a significant increase (above 50%) in the specific growth rate (Fig. 1). This period corresponded in ETS1, ETS2, ETS3, and ETS4 to 340, 310, 328, and 335 number of generations, respectively. Samples were taken every 10–20 generations to determine changes in specific growth rates under ethanol stress. Subsequently, the ethanol tolerant strains obtained by ALE were stored in YNB culture medium with 50% glycerol (v/v) at −80 °C.
Adaptive laboratory evolution of K. marxianus CCT 7735 under ethanol stress (4% v/v). The specific growth rates of the ethanol-tolerant strains (ETS1, ETS2, ETS3, and ETS4) under ethanol stress were higher than of the parental strains (P1, P2, P3, e P4). “*” indicates statistically results according to Student’s t test (P value < 0.05)
Physiological characterization
The physiological characterization of ETSs and parental strains were carried out by the determination of both kinetic and fermentative parameters. These strains were cultured in 50 mL of SD medium in 250-mL Erlenmeyer flasks and incubated at 37 °C, 200 rpm for 24 h, and initial pH of 5.5. Aliquots of 2 mL were collected every 1 h for the first 12 h. These samples were centrifuged at 4 °C, 16,000 g for 5 min, and then the supernatants were filtered (pore size 0.22 μm) and stored at −20 °C.
Lactose and ethanol quantification
The lactose consumption and ethanol production of ETSs and parental strain were determined by high-performance liquid chromatography (HPLC)—chromatograph Shimadzu TA-20 (Kyoto, Japan) coupled to refractive index detector and column ion exchange Rezex ROA-Organic acid H+ 8% (300 × 7.8 mm, Phenomenex, California, USA), temperature of 45 °C, using 0.005 M sulfuric acid as mobile phase, flow rate of 0.6 mL/min, and injection volume of 10 μL. Lactose and ethanol quantification was obtained by calibration curves using external standards.
Determination of fermentation parameters
The ethanol yield (YP/S ethanol, g/g) was determined by the angular coefficient from a linear regression of the plot ethanol concentration (g/L) versus lactose consumption (g/L). Cell biomass yield (YX/S, g/g) was determined by the angular coefficient from a linear regression of the plot dry cell weight (DCW) versus lactose consumption (g/L). Specific ethanol production rate (qp, g/g/h) was determined by multiplying the respective yield by the specific growth rate (μ, per hour). Specific lactose consumption rate (qs, g/g/h) was obtained dividing μ by YX/S. Linear regressions were adjusted at the exponential growth phase. Ethanol volumetric productivity (Qp ethanol, g/L/h) was calculated by maximum ethanol production divided by time.
Chemostat cultivation
Both ETS and parental strain were cultivated in a chemostat in order to identify the metabolic changes in response to ethanol. Exponentially growing cells in CBS medium were inoculated with an initial OD600 of 0.2 into 2 L of CBS medium in a 5 L Labfors 5 bioreactor system from INFORS HT (Bottmingen, Switzerland). Air flow was set to 1 vessel volume per minute (vvm), and the dissolved oxygen (DO) levels were kept > 15% throughout the experiments. Both temperature and pH were kept constant at 37 °C and 5.5 by adding either NaOH (10% w/v) or H2SO4 (10% v/v), consequently. The cultivation started as a batch culture with a stirring rate of 200 rpm. When the lactose concentration reached 5 g/L, the chemostat mode was started. The chemostat was operated at a dilution rate of 0.05 (per hour); the feed medium was the same described for the batch phase, except for lactose concentration, 5 g/L instead 20 g/L. Sampling for intracellular (n = 5) and extracellular (n = 3) metabolites as well as fatty acids methyl esters (FAMEs) (n = 3) analysis were performed in steady-state through at least two residence times (two culture volumes of fresh medium). After sampling, an ethanol pulse in steady state was carried out to reach the final ethanol concentration of 4% (v/v). Subsequently, the feed medium was changed from CBS to CBSE. The steady state was reached when the cell mass was kept constant after two residences times and then samples were withdrawn in steady state for intracellular and extracellular metabolites as well as FAMEs analysis.
Sampling and extraction procedures for metabolite analysis
For intracellular metabolite analysis, we adopted the protocol described by Smart et al. (2010) and Villas-Bôas and Bruheim (2007). Samples containing intracellular metabolites were freeze-dried and concentrated using a VirTis freeze-dryer from SP Scientific (Newtown Square, PA, USA). The freeze-dried samples were stored at −80 °C for posterior analysis.
The obtainment of extracellular metabolites was performed according to Smart et al. (2010). The samples were freeze-dried on a VirTis freeze-dryer (SP Scientific, Newtown Square, PA, USA) and stored at −80 °C. Twenty microliters of 2, 3, 3, 3-d4-alanine (internal standard) were added to both intracellular and extracellular samples.
Derivatization and GC-MS analysis
The metabolites were derivatized as described by Smart et al. (2010). The derivatized samples were analyzed by using a GC-MS using a GC7890 gas chromatograph coupled to a MSD5975 mass spectrometer (Agilent Technologies, Santa Clara, CA, USA).
Metabolite identification and data analysis
The deconvolution of GC-MS chromatograms and metabolite identification was carried out by the AMDIS software using an in-house MCF mass spectral library. The GC peak values of selected reference ions of identified metabolites were used to determine the relative abundance by ChemStation (Agilent Technologies, Santa Clara, CA, USA). The data analysis and normalization of both intracellular and extracellular metabolites were carried out as described Smart et al. (2010). The entire data-mining, normalization, and statistical analysis were automated in the R software as described by Aggio et al. (2011) and Smart et al. (2010).
Fatty acids methyl esters analysis
For FAME analysis, the samples from the chemostat in steady state were freeze-dried on a VirTis freeze-dryer (SP Scientific, Newtown Square, PA, USA). Direct-transesterification method was performed according to Lepage and Roy (1986) with modifications. The freeze-dried samples were weighed into glass tubes, 1 and 5 mg of parental strain, and ETS, respectively. The internal standard concentration (tridecanoic acid—C13:0) dissolved in 2 mL of methanol-toluene 4:1 (v/v) ranged from 50 to 300 μg. Finally, the samples were injected into GC-MS using a GC7890 gas chromatograph (Agilent Technologies, Santa Clara, CA) coupled to a MSD5975 mass spectrometer (Agilent Technologies, Santa Clara, CA, USA).
Ergosterol analysis
Ergosterol of both ethanol-tolerant and parental strains was extracted according to Diniz et al. (2017) with some modifications. The frozen samples were freeze-dried on a LIOTOP L101 freeze-dryer (Liobras, São Carlos, SP, Brazil). Freeze-dried cells were weighed (40–50 mg) and subjected to extraction. The ergosterol content determination was performed by high-performance liquid chromatography (HPLC)—chromatograph Shimadzu TA-20 (Kyoto, Japan) coupled to SPD-10A UV–visible detector at 282 nm and column Kinetex RP-C18 column (250 × 4.6 mm, 5 μm, Phenomenex, California, USA), at 30 °C, using 98% (v/v) methanol solution as the mobile phase, flow rate of 1 mL/min, and an injection volume of 10 μL. Ergosterol was quantified by a calibration curve obtained from external standards (0.01 to 1.0 mg/mL). The data were normalized with biomass.
Pathways analysis
We used MetaboAnalyst (www.metaboanalyst.ca), a web-based data analysis platform, for pathway analysis to predict and compare the activity of different pathways in ETS and parental strain. This tool uses metabolomics data, as well as lists of genes or KEGG orthologs to support integrative analysis, using the relative abundances of intracellular and extracellular metabolite profiles to predict which pathways may be active in the yeast. Graphical representations of these results were performed by the R software 3.2.5 (www.r-project.org).
DNA-seq analysis
The whole genomes of both parental (P4) and ethanol-tolerant (ETS4) strains were sequenced. Total DNA was extracted and purified according to Promega Wizard® Genomic DNA Purification Kit (Wisconsin, USA). DNA samples were sequenced by GenOne Biotechnologies (Rio de Janeiro, Brazil) using the Illumina HiSeq2500 platform using the 2 × 150 paired-end method, with an insert size of 350 bp. Paired-end reads were quality-analyzed by FastQC v.0.71 (Andrews 2010). The P4 strain genome was assembled by MIRA v4.0.2 (Chevreux et al. 1999; Chevreux et al. 2004), by mapping to the previously available K. marxianus CCT 7735 genome (Silveira et al. 2014). Genome assembly quality metrics were computed by QUAST v.4.6.3 (Gurevich et al. 2013) and REAPR v.1.0.18 (Hunt et al. 2013). Protein-coding genes were predicted ab initio by BRAKER2 (Altschul et al. 1990; Lomsadze et al. 2005; Stanke et al. 2006; Stanke et al. 2008; Ter-Hovhannisyan et al. 2008; Camacho et al. 2009; Hoff et al. 2016) and by the homology-based predictor GeMoMa v.1.5.3 (Keilwagen et al. 2016; Keilwagen et al. 2018), using the previous genome annotation as reference. EVidenceModeler v.1.1.1 (Haas et al. 2008) was used to generate an annotation consensus. Genome assembly and annotation completeness was evaluated by BUSCO v.3.0.2 (Simão et al. 2015; Waterhouse et al. 2018) against the Saccharomycetales dataset. To detect mutations in the ETS4, the reads were aligned to the P4 strain assembly by BWA-MEM v.0.7.17 (Li 2013). Alignments were filtered by SAMtools v.1.9.52 (Li et al. 2009) to a minimum mapping quality of 60. Variant calling was performed by Freebayes v.1.2.0–4-gd15209e (Garrison and Marth 2012), and the resulting VCF file was filtered by VCFtools v.0.1.17 (Danecek et al. 2011) and VCFlib (Garrison 2018) according to the dDocent pipeline (Puritz et al. 2014a, b; Puritz et al. 2014a, b). The detected variants were annotated by SnpEff v.4.3 (Cingolani et al. 2012) using a custom database with a 900 bp flank in the gene sequences included. The gene variants were then identified by DIAMOND v.0.9.21 (Buchfink et al. 2015) searches against K. marxianus UniProt datasets (The UniProt Consortium 2008). GO terms assigned by Blast2GO v.5.2 (Conesa et al. 2005; Conesa and Götz 2008; Götz et al. 2008; Götz et al. 2011) and plotted by REVIGO (Supek et al. 2011). The whole-genome sequencing data has been deposited at NCBI under the BioProject PRJNA553996. Genome quality assessment results and software parameters can be found on Supplementary Information (Tables S3 and S4).
Results
Physiological characterization of ethanol tolerant strains
We used four different populations from individual clones of K. marxianus CCT 7735 wild-type in order to select strains more tolerant to ethanol stress by ALE. The specific growth rates of ETS1, ETS2, ETS3, and ETS4 increased above 125, 58.3, 90, and 122.2%, respectively, compared to their respective parental strains.
Beyond mutations that lead to the acquisition of the desirable phenotype in microbial cells evolved by ALE, trade-off phenotypes have been reported. To address this issue, we evaluated whether the ETSs had an alteration in their fermentative capacity. In general, we observed that the physiological parameters of ETS and parental strain were similar (Table 1); thereby, their fermentative capacities were not altered. However, ETS1 and ETS4 displayed the highest maximum ethanol production and volumetric ethanol productivities (Table 1). In addition, we verified that the specific ethanol production rate presented by ETS4 was higher than those obtained by other ETSs (Table 1); therefore, we chose this strain for further analysis.
Genomic alterations in the ethanol-tolerant K. marxianus CCT 7735 strain
We aligned the evolved strain (ETS4) reads to the parental (P4) genome to search mutations that could be related to the phenotype of ethanol tolerance. In total, we found 19 variant types such as single nucleotide variants (SNVs), insertions and deletions (InDels), and mixed mutations. These variants were predicted to have 103 different effects, occurring in 99 different genes. We then classified the variants according to SNPeff annotation, and observed that many mutations occurred in regulatory regions, such as regions directly upstream or downstream of a coding sequence (CDS) (Table S6). We also identified GO terms that are over- or under-represented in the data (Fig. S4, Fig. S5 and Fig. S6). In “Molecular Function” category, the GO terms “catalytic activity”, “nucleic acid binding”, and “transcription regulatory region RNA binding” were enriched. In “Biological Function” category, we found the enriched terms “nitrogen compound binding”, “alcohol metabolism”, “endosomal transport”, and “protein glutathionylation”. In “Cellular Component” category, the enriched terms were “membrane” and “integral component of membrane”. A spreadsheet containing all predicted genes and their corresponding mutations is available as Online Resource 1.
Metabolic changes caused by ethanol exposure
We identified 155 intracellular metabolites and 55 unknown metabolites from chemostat cultures of ETS4 and P4 strain under both ethanol and non-ethanol stress (Table S1). For the exometabolome, 145 metabolites and 47 unknown metabolites were identified under the aforementioned conditions (Table S2). The intracellular and extracellular profiles of metabolites were compared by principal component analysis (PCA). The four groupings formed on both intracellular and extracellular metabolites PCA plots (Fig. S2A and Fig. S2B) are composed of two strains under non-ethanol stress (P4 and ETS4) and ethanol stress (P4 stress and ETS4 stress). All groups were separated by principal components 1 (PC1) and 2 (PC2), which accounted for the total variance of 61.30% and 14.09% for intracellular metabolites, 90.45% and 4.24% for extracellular metabolites, respectively. Notably, we observed that the P4 stress strain was in a distinct cluster for both intracellular and extracellular metabolites (Fig. S2) and that the ETS4 stress was close to ETS4/P4 cluster (Fig. S2A). Indeed, we observed some ellipses overlap on ETS4 and ETS4 stress clusters. Importantly, PC1 contains the higher total variance value, indicating that the components significantly influenced the separation of the strains. β-alanine, cystathionine, threonine, proline, and alanine (intracellular metabolites); and malonic acid, leucine, 2-hydroxybutiric acid, lactic acid, and nicotinic acid (extracellular metabolites) were the most influential metabolites that contributed positively to PC1. We observed higher levels of those metabolites in P4, ETS4, and ETS4 stress, suggesting that their decrease in the parental strain subjected to the ethanol stress seems to be related to its lower ethanol tolerance compared to ETS4.
We performed a hierarchical cluster analysis (HCA) to analyze the different groups of intracellular and extracellular metabolites (Fig. 2). Regardless of type strain and stress condition, the intracellular metabolites identified were similar; however, their relative abundance was group dependent. Most of the intracellular amino acids and derivatives, amine/amide compounds, and organic acids had their relative abundance strongly reduced in the P4 strain under ethanol stress (Fig. 2a and b). Meanwhile, the extracellular abundance of those metabolites remarkably increased upon ethanol exposure (Fig. 2c and d), suggesting that ethanol-induced a leakage of metabolites in the parental strain by altering the permeability of its membrane. Interestingly, the relative abundance of most intracellular amino acids and derivative also decreased under stress in ETS4 but to a lesser degree than in the P4 strain under stress (Fig. 2a). Otherwise, most of amine/amide compounds and organic acids in ETS4 had their relative abundance enhanced under ethanol stress (Fig. 2b), indicating that they are likely related to its improved tolerance to ethanol. Consistent with this, we observed that the ethanol stress led to a decrease in the biomass of both parental and ETS4; however, this drop was higher in the parental strain (Table S5). Taken together, these results indicate that the membrane permeability of the ethanol-tolerant strain was less affected by ethanol compared to the parental strain.
Hierarchical cluster analysis of intracellular metabolites grouped based on chemical properties: (a) amino acids and derivatives; and (b) amine/amide and organic acids; and extracellular metabolites groups: (c) amino acids and derivatives; and (d) amine/amide and organic acids. P4: parental strain under non-ethanol stress; P4 stress: parental strain under ethanol stress; ETS4: ethanol-tolerant strain under non-ethanol stress; and ETS4 stress: ethanol-tolerant strain under ethanol stress. A dendrogram was added to the left side to reorder the group of metabolites according to the set of abundance values and show the correlation among them. Metabolites with similar abundance were placed close to each other and metabolites with different abundance were placed farther apart under ethanol and non-ethanol stress. The highest (green color) and lowest (red color) abundance values are shown on row-Z score
Remarkably, the amino acid valine and some organic acids of the citric acid cycle (TCA cycle) such as isocitric acid, citric acid, and cis-aconitic acid had their levels increased intracellular and extracellularly. Therefore, those metabolites seem to have been secreted due to metabolic overflow, indicating that the metabolic fluxes of the pathways involved with their synthesis was enhanced (Fig. 2). In agreement with the valine accumulation (Fig. 2), we detected changes in the gene encoding the acetolactase synthase small subunit, ILV6 (single substitution downstream of CDS), which is related to the branched-chain amino acid biosynthetic process (Fig. S5). We analyzed the relative abundance of intracellular and extracellular metabolite profiles under ethanol and non-ethanol stress to generate a comparative metabolic pathway analysis (Fig. S3). TCA cycle was affected in ETS4 under ethanol stress (Fig. S4, Fig. S5, Online Resource 1), which is consistent with the accumulation of isocitric acid, citric acid, and cis-aconitic acid (TCA intermediates).
Alterations in both membrane fatty acids and ergosterol content
The membrane fatty acid abundance in the ETS4 was remarkably higher than in the P4 strain. Notably, monounsaturated fatty acids were more abundant under both non- and ethanol stress in the P4 strain (Fig. 3a and b). Their abundance was also higher in the ETS4 that had not been exposed to ethanol; however, decreased upon ethanol stress. According to those results, pathways associated with fatty acid metabolism and biosynthesis, unsaturated fatty acids biosynthesis, and glycerolipid metabolism in the P4 strain were less affected upon ethanol stress than in the ETS4 (Fig. S3). In addition, we detected that the long-chain FAMEs were more abundant in the two strains regardless of the stress condition (Fig. 3b). These results are consistent with the fact that most common fatty acids from yeast phospholipids bilayers are long-chain fatty acids such as palmitic acid (C16:0), palmitoleic acid (C16:1), stearic acid (C18:0), and oleic acid (C18:1). Additionally, their abundance was highest in the ETS4. However, their abundance decreased once the strains were subjected to ethanol stress.
Fatty acids methyl esters profiles of P4 strain and ETS4 under non- and ethanol stress. (a) total of FAMEs (saturated, monounsaturated, and polyunsaturated); and (b) length of FAMEs: medium-chain fatty acids (MCFA) ranging from 6 to 12 carbons; long-chain fatty acids (LCFA) ranging from 13 to 21 carbons and very long chain fatty acids (VLCFA) > 21 carbons; and (c) palmitic acid (C16:0), stearic acid (C18:0), palmitoleic acid (C16:1), and oleic acid (C18:1) profiles. Relative abundance values from each sample were divided by value of biomass detected in P4 and ETS4 samples (specific relative abundance by biomass). “*” indicates statistically results according to Student’s t test (P value < 0.05)
Ergosterol plays a crucial role in response to ethanol stress in S. cerevisiae (Stanley et al. 2010; Vanegas et al. 2012). Ergosterol was more abundant in ETS4 than in P4 under both conditions (Fig. 4). In agreement with this result, we detected mutations in the gene RRI1, that encodes the COP9 signalosome complex subunit 5, which is associated with positive regulation of ergosterol biosynthesis. The predicted mutation is a deletion in the upstream region of the coding sequence.
Variants detected in the genome of the ethanol-tolerant strain were consistent with the FAMEs results. We identified in the ETS4 mutations in the genes KLMA_10136 (insertion upstream of CDS), and PXA2 (insertion downstream of CDS), which are respectively associated with the lipid metabolic process GO term (Fig. S5).
Discussion
Kluyveromyces marxianus CCT 7735 (UFV-3) is capable of converting lactose from whey permeate to ethanol efficiently under hypoxia, that is, with yields close to the theoretical value (Diniz et al. 2013; Silveira et al. 2005). Nevertheless, its low ethanol tolerance is the main drawback for its application at an industrial level. Herein, ALE was suitable to select K. marxianus CCT 7735 ethanol-tolerant strains (ETS), highlighting its efficiency in improving the robustness of yeasts (Mo et al. 2019).
Under stress conditions, the overproduction of metabolites can be associated with an adaptive response. Notably, the metabolic profiles were significantly different between the ETS4 and P4 strain. In addition, the ETS4 under ethanol stress, contrary to the P4 strain, displayed higher levels of the following metabolites both intracellular and extracellularly: valine and TCA cycle intermediates (isocitric acid, citric acid, and cis-aconitic acid). It has been reported in S. cerevisiae that valine accumulation enhances ethanol tolerance (Ohta et al. 2015). Based on this evidence, we propose that its accumulation also improved ethanol tolerance in the ETS4. Importantly, the TCA cycle was increased significantly in the ETS4 under ethanol stress (Fig. S3). Therefore, the accumulation of those metabolites seems to have contributed to the acquisition of ethanol tolerance by the ETS4. Consistent with our results, previous studies have related the accumulation of metabolites of the TCA cycle with ethanol tolerance in S. cerevisiae (Lourenço et al. 2013; Ohta et al. 2015; Ming et al. 2019).
The yeast cellular membrane is the main target of ethanol. Its integrity is compromised upon ethanol exposure leading to metabolite leakage (Dong et al. 2015; Henderson and Block 2014; Salgueiro et al. 1988). In the parental strain, the decrease of most intracellular metabolites and their increase extracellularly indicate a metabolite leakage. Notably, we did not observe this effect in the ETS4, suggesting that the ethanol has a less pronounced effect on its membrane compared to the parental strain (Fig. 2). Likely, this is associated with the fact that the ETS4 had a higher relative abundance of membrane fatty acids than the P4 strain (Fig. 3a), irrespective of the ethanol presence in culture medium.
In the P4 strain, the ethanol stress did not change the content of both palmitoleic and oleic acids (Fig. 3c). Likewise, Diniz et al. (2017) observed that the level of unsaturated fatty acids in the batch culture of K. marxianus CCT 7735 was not altered upon ethanol exposure. Remarkably, the relative abundances of both palmitoleic acid and oleic acids were higher in ETS4 than in P4 (Fig. 3c). In S. cerevisiae ethanol-stressed cells, high levels of palmitoleic acid and oleic acid have been considered an important adaptive response against ethanol to avoid the formation of interdigitations, as well as keeping the optimal thickness of yeast membranes (Dong et al. 2015; Henderson and Block 2014; Kim et al. 2016; Lahtvee et al. 2016). The fatty acids metabolism and biosynthesis of unsaturated fatty acids in the ETS4 were induced under both non- and ethanol stress (Fig. S3), indicating that the metabolism of mono-unsaturated fatty acids is upregulated to adapt to the stress caused by ethanol. In agreement with our results, Mo et al. (2019) observed that genes involved in membrane lipid biosynthesis were upregulated in the ethanol-tolerant K. marxianus FIM1 strain even when it was grown in an ethanol-free medium.
In S. cerevisiae, ergosterol concentration increases under ethanol stress, which is considered an important adaptive response to ethanol, because it reduces both lipid interdigitations and transition phase of phospholipid bilayers thereby promoting membrane stabilization (Prakash et al. 2011; Vanegas et al. 2012; Caspeta et al. 2014; Caspeta and Nielsen 2015; Liu et al. 2019). Contrary to S. cerevisiae, ergosterol concentration was reduced in both ethanol tolerant and parental strains. Nevertheless, the concentration of ergosterol was remarkably higher in the tolerant strain than in the parental strain, which seems to be related to its highest ethanol tolerance. Consistent with these results, genes related to ergosterol biosynthesis were upregulated in the strain K. marxianus FIM1 tolerant to high ethanol concentrations, highlighting its importance for standing up the ethanol stress. Indeed, Alvim et al. (2019) pointed out that the overexpression of gene encoding enzymes of the ergosterol biosynthesis pathway in K. marxianus is a promising metabolic engineering strategy to enhance its capacity to tolerate ethanol.
In conclusion, the ethanol-tolerant K. marxianus CCT 7735 strain accumulates valine and metabolites of the TCA cycle in response to ethanol. In addition, the contents of FAMEs and ergosterol were higher in this strain than in the parental strain, indicating that these differences may be associated with increased ethanol tolerance displayed by the ethanol-tolerant strain. As such, our findings helped to identify metabolic changes that contributed to ethanol tolerance in the tolerant strain obtained by adaptive laboratory evolution.
References
Aggio R, Villas-Bôas SG, Ruggiero K (2011) Metab: an R package for high-throughput analysis of metabolomics data generated by GC-MS. Bioinformatics 27:2316–2318. https://doi.org/10.1093/bioinformatics/btr379
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Alvim MCT, Vital CE, Barros E, Vieira NM, da Silveira FA, Balbino TR, Diniz RHS, Brito AF, Bazzolli DMS, de Oliveira Ramos HJ, da Silveira WB (2019) Ethanol stress responses of Kluyveromyces marxianus CCT 7735 revealed by proteomic and metabolomic analyses. Antonie van Leeuwenhoek Int J Gen Mol Microbiol 112:827–845. https://doi.org/10.1007/s10482-018-01214-y
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data
Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF (2009) Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247. https://doi.org/10.1038/nature08480
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. https://doi.org/10.1038/nmeth.3176
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421. https://doi.org/10.1186/1471-2105-10-421
Caspeta L, Nielsen J (2015) Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. MBio 6:1–9. https://doi.org/10.1128/mBio.00431-15
Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, Petranovic D, Nielsen J (2014) Altered sterol composition renders yeast thermotolerant. Science 346:75–78. https://doi.org/10.1126/science.1258137
Chevreux B, Wetter T, Suhai S (1999) Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol Proc Ger Conf Bioinforma 99:45–46
Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WEG, Wetter T, Suhai S (2004) Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14:1147–1159. https://doi.org/10.1101/gr.1917404
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff:SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. https://doi.org/10.4161/fly.19695
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:1–12. https://doi.org/10.1155/2008/619832
Conesa A, Götz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Costa DA, de Souza CJA, Costa PA, Rodrigues MQRB, dos Santos AF, Lopes MR, Genier HLA, Silveira WB, Fietto LG (2014) Physiological characterization of thermotolerant yeast for cellulosic ethanol production. Appl Microbiol Biotechnol 98:3829–3840. https://doi.org/10.1007/s00253-014-5580-3
da Silveira FA, Diniz RHS, Sampaio GMS, Brandão RL, da Silveira WB, Castro IM (2018) Sugar transport systems in Kluyveromyces marxianus CCT 7735. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol 4:211–223. https://doi.org/10.1007/s10482-018-1143-4
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
de Paiva LC, Diniz RHS, Vidigal PMP, de Oliveira Mendes TA, Santana MF, Cerdán ME, González-Siso MI, da Silveira WB (2019) Genomic analysis and lactose transporter expression in Kluyveromyces marxianus CCT 7735. Fungal Biol 123:687–697. https://doi.org/10.1016/j.funbio.2019.06.004
Diniz RHS, Rodrigues MQRB, Fietto LG, Passos FML, Silveira WB (2013) Optimizing and validating the production of ethanol from cheese whey permeate by Kluyveromyces marxianus UFV-3. Biocatal Agric Biotechnol 3:111–117. https://doi.org/10.1016/j.bcab.2013.09.002
Diniz RHS, Villada JC, Alvim MCT, Vidigal PMP, Vieira NM, Lamas-Maceiras M, Cerdán ME, González-Siso MI, Lahtvee PJ, da Silveira WB (2017) Transcriptome analysis of the thermotolerant yeast Kluyveromyces marxianus CCT 7735 under ethanol stress. Appl Microbiol Biotechnol 101:6969–6980. https://doi.org/10.1007/s00253-017-8432-0
Dong SJ, Yi CF, Li H (2015) Changes of Saccharomyces cerevisiae cell membrane components and promotion to ethanol tolerance during the bioethanol fermentation. Int J Biochem Cell Biol 69:196–203. https://doi.org/10.1016/j.biocel.2015.10.025
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution – principles and applications for biotechnology. Microb Cell Fact 12 VN-r:64. https://doi.org/10.1186/1475-2859-12-64
Ferreira PG, da Silveira FA, dos Santos RCV, Genier HLA, Diniz RHS, Ribeiro JI, Fietto LG, Passos FML, da Silveira WB (2015) Optimizing ethanol production by thermotolerant Kluyveromyces marxianus CCT 7735 in a mixture of sugarcane bagasse and ricotta whey. Food Sci Biotechnol 24:1421–1427. https://doi.org/10.1007/s10068-015-0182-0
Garrison E (2018) Vcflib: a simple C++ library for parsing and manipulating VCF files, + many command-line utilities
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing
González-Siso MI (1996) The biotechnological utilization of cheese whey: a review. Bioresour Technol 57:1–11. https://doi.org/10.1016/0960-8524(96)00036-3
Götz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. https://doi.org/10.1093/nar/gkn176
Götz S, Arnold R, Sebastián-León P, Martín-Rodríguez S, Tischler P, Jehl M-A, Dopazo J, Rattei T, Conesa A (2011) B2G-FAR, a species-centered GO annotation repository. Bioinformatics 27:919–924. https://doi.org/10.1093/bioinformatics/btr059
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR (2008) Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol 9:R7. https://doi.org/10.1186/gb-2008-9-1-r7
Henderson CM, Block DE (2014) Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomyces cerevisiae. Appl Environ Microbiol 80:2966–2972. https://doi.org/10.1128/AEM.04151-13
Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M (2016) BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS: table 1. Bioinformatics 32:767–769. https://doi.org/10.1093/bioinformatics/btv661
Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD (2013) REAPR: a universal tool for genome assembly evaluation. Genome Biol 14:R47. https://doi.org/10.1186/gb-2013-14-5-r47
Keilwagen J, Wenk M, Erickson JL, Schattat MH, Grau J, Hartung F (2016) Using intron position conservation for homology-based gene prediction. Nucleic Acids Res 44:e89–e89. https://doi.org/10.1093/nar/gkw092
Keilwagen J, Hartung F, Paulini M, Twardziok SO, Grau J (2018) Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinformatics 19:189. https://doi.org/10.1186/s12859-018-2203-5
Kim S, Kim J, Song JH, Jung YH, Choi IS, Choi W, Park YC, Seo JH, Kim KH (2016) Elucidation of ethanol tolerance mechanisms in Saccharomyces cerevisiae by global metabolite profiling. Biotechnol J 11:1221–1229. https://doi.org/10.1002/biot.201500613
LaCroix RA, Palsson BO, Fiest AM (2017) A model for designing adaptive laboratory evolution experiments. Appl Enviromental Microbiol 83:1–14. https://doi.org/10.1128/AEM.03115-16
Lahtvee P-J, Kumar R, Hallstrom BM, Nielsen J (2016) Adaptation to different types of stress converge on mitochondrial metabolism. Mol Biol Cell 27:2505–2514. https://doi.org/10.1091/mbc.E16-03-0187
Lepage G, Roy CC (1986) Direct transesterification of all classes of lipids in a one-step reaction. J Lipid Res 27:114–120
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Liu JF, Xia JJ, Nie KL, Wang F, Deng L (2019) Outline of the biosynthesis and regulation of ergosterol in yeast. World J Microbiol Biotechnol 35:1–8. https://doi.org/10.1007/s11274-019-2673-2
Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M (2005) Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33:6494–6506. https://doi.org/10.1093/nar/gki937
Lourenço AB, Roque FC, Teixeira MC, Ascenso JR, Sá-Correia I (2013) Quantitative 1 H-NMR-metabolomics reveals extensive metabolic reprogramming and the effect of the aquaglyceroporin FPS1 in ethanol-stressed yeast cells. PLoS One 8:e55439. https://doi.org/10.1371/journal.pone.0055439
Ma M, Liu ZL (2010) Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 87:829–845. https://doi.org/10.1007/s00253-010-2594-3
Ming M, Wang X, Lou D, Lian L, Zhang H, Gao W, Zhu B (2019) Molecular responses of Saccharomyces cerevisiae to ethanol stress using gas chromatography-mass spectrometry:216–221. https://doi.org/10.1039/c9mo00055k
Mo W, Wang M, Zhan R, Yu Y, He Y, Lu H (2019) Kluyveromyces marxianus developing ethanol tolerance during adaptive evolution with significant improvements of multiple pathways. Biotechnol Biofuels 12:1–15. https://doi.org/10.1186/s13068-019-1393-z
Mussatto SI, Dragone G, Guimarães PMR, Silva JPA, Carneiro LM, Roberto IC, Vicente A, Domingues L, Teixeira JA (2010) Technological trends, global market, and challenges of bio-ethanol production. Biotechnol Adv 28:817–830. https://doi.org/10.1016/j.biotechadv.2010.07.001
Ohta E, Nakayama Y, Mukai Y, Bamba T, Fukusaki E (2015) Metabolomic approach for improving ethanol stress tolerance in Saccharomyces cerevisiae. J Biosci Bioeng 121:1–7. https://doi.org/10.1016/j.jbiosc.2015.08.006
Parashar A, Jin Y, Mason B, Chae M, Bressler DC (2016) Incorporation of whey permeate, a dairy effluent, in ethanol fermentation to provide a zero waste solution for the dairy industry. J Dairy Sci 99:1859–1867. https://doi.org/10.3168/jds.2015-10059
Prakash G, Varma AJ, Prabhune A, Shouche Y, Rao M (2011) Microbial production of xylitol from d-xylose and sugarcane bagasse hemicellulose using newly isolated thermotolerant yeast Debaryomyces hansenii. Bioresour Technol 102:3304–3308. https://doi.org/10.1016/j.biortech.2010.10.074
Prazeres AR, Carvalho F, Rivas J (2012) Cheese whey management: a review. J Environ Manag 110:48–68. https://doi.org/10.1016/j.jenvman.2012.05.018
Puritz JB, Hollenbeck CM, Gold JR (2014a) dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ 2:e431. https://doi.org/10.7717/peerj.431
Puritz JB, Matz MV, Toonen RJ, Weber JN, Bolnick DI, Bird CE (2014b) Demystifying the RAD fad. Mol Ecol 23:5937–5942. https://doi.org/10.1111/mec.12965
Salgueiro SP, Sá-Correia I, Novais JM (1988) Ethanol-induced leakage in Saccharomyces cerevisiae: kinetics and relationship to yeast ethanol tolerance and alcohol fermentation productivity. Appl Environ Microbiol 54:903–909
Silveira WB, Passos FJV, Mantovani HC, Passos FML (2005) Ethanol production from cheese whey permeate by Kluyveromyces marxianus UFV-3: a flux analysis of oxido-reductive metabolism as a function of lactose concentration and oxygen levels. Enzym Microb Technol 36:930–936. https://doi.org/10.1016/j.enzmictec.2005.01.018
Silveira WB, Diniz RHS, Cerdan ME, Gonzalez-Siso MI, de A Souza R, Vidigal PMP, Brustolini OJB, de Almeida Prata ERB, Medeiros AC, Paiva LC, Nascimento M, Ferreira EG, dos Santos VC, CRS B, TAR F, Colombo LT, Passos FML (2014) Genomic sequence of the yeast Kluyveromyces marxianus CCT 7735 (UFV-3), a highly lactose-fermenting yeast isolated from the Brazilian dairy industry. Genome Announc 2:e01136–e01114. https://doi.org/10.1128/genomeA.01136-14
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Smart KF, Aggio RBM, Van Houtte JR, Villas-Bôas SG (2010) Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat Protoc 5:1709–1729. https://doi.org/10.1038/nprot.2010.108
Snoek T, Verstrepen KJ, Voordeckers K (2016) How do yeast cells become tolerant to high ethanol concentrations? Curr Genet 62:475–480. https://doi.org/10.1007/s00294-015-0561-3
Stanke M, Schöffmann O, Morgenstern B, Waack S (2006) Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7:62. https://doi.org/10.1186/1471-2105-7-62
Stanke M, Diekhans M, Baertsch R, Haussler D (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644. https://doi.org/10.1093/bioinformatics/btn013
Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA (2010) The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol 109:13–24. https://doi.org/10.1111/j.1365-2672.2009.04657.x
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. https://doi.org/10.1371/journal.pone.0021800
Taylor G (2008) Biofuels and the biorefinery concept. Energy Policy 36:4406–4409. https://doi.org/10.1016/j.enpol.2008.09.069
Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M (2008) Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18:1979–1990. https://doi.org/10.1101/gr.081612.108
The UniProt Consortium (2008) The universal protein resource (UniProt). Nucleic Acids Res 36:D190–D195. https://doi.org/10.1093/nar/gkm895
Valdivia M, Galan JL, Laffarga J, Ramos JL (2016) Biofuels 2020: biorefineries based on lignocellulosic materials. Microb Biotechnol 9:585–594. https://doi.org/10.1111/1751-7915.12387
Vanegas JM, Contreras MF, Faller R, Longo ML (2012) Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biophys J 102:507–516. https://doi.org/10.1016/j.bpj.2011.12.038
Verduyn C, Postma E, Scheffers WA, van Dijken JP (1992) Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517. https://doi.org/10.1007/BF00270792
Villas-Bôas SG, Bruheim P (2007) Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal Biochem 370:87–97. https://doi.org/10.1016/j.ab.2007.06.028
Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM (2018) BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 35:543–548. https://doi.org/10.1093/molbev/msx319
Zhao XQ, Bai FW (2009) Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J Biotechnol 144:23–30. https://doi.org/10.1016/j.jbiotec.2009.05.001
Acknowledgments
We thank all the authors for contributing to this work.
Funding
This study was funded by the Brazilian Agencies: FAPEMIG (Foundation for Research Support of the State of Minas Gerais- grant number APQ-02825-16), CAPES (Coordination for the Improvement of Higher Education Personnel – Finance Code 001), and CNPq (National Science and Technology Development Council).
Author information
Authors and Affiliations
Contributions
WBS and FAS conceived and designed the research. FAS, DLOS, TRB, and LAL conducted the experiments. FAS and KWB contributed with the analytical tools. FAS, MAMF, RHSD, KWB, MMB, SGV, and WBS analyzed data and manuscript writing. FAS, KWB, MMB, and WBS revised and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 691 kb)
Rights and permissions
About this article
Cite this article
da Silveira, F.A., de Oliveira Soares, D.L., Bang, K.W. et al. Assessment of ethanol tolerance of Kluyveromyces marxianus CCT 7735 selected by adaptive laboratory evolution. Appl Microbiol Biotechnol 104, 7483–7494 (2020). https://doi.org/10.1007/s00253-020-10768-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10768-9