Abstract
One of the main reasons for the bacterial resistance to antibiotics is caused by biofilm formation of microbial pathogens during bacterial infections. Salmonella enterica and Vibrio harveyi are known to form biofilms and represent a major health concern worldwide, causing human infections responsible for morbidity and mortality. The current study aims to investigate the effect of purified sulfated polysaccharides (SPs) from Chlamydomonas reinhardtii (Cr) on planktonic and biofilm growth of these bacteria. The effect of Cr-SPs on bacterial planktonic growth was assessed by using the agar well diffusion method, which showed clear zones ranging from 13 to 26 mm in diameter from 0.5 to 8 mg/mL of Cr-SPs against both the bacteria. Time-kill activity and reduction in clonogenic propagation further help to understand the anti-microbial potential of Cr-SPs. The minimum inhibitory concentration of Cr-SPs against S. enterica and V. harveyi was as low as 440 μg/mL and 490 μg/mL respectively. Cr-SPs inhibited bacterial cell attachment up to 34.65–100% at 0.5–8 mg/mL in S. enterica and V. harveyi respectively. Cr-SPs also showed 2-fold decrease in the cell surface hydrophobicity, indicating their potential to prevent bacterial adherence. Interestingly, Cr-SPs efficiently eradicated the preformed biofilms. Increased reduction in total extracellular polysaccharide (EPS) and extracellular DNA (eDNA) content in a dose-dependent manner demonstrates Cr-SPs ability to interact and destroy the bacterial EPS layer. SEM analysis showed that Cr-SPs effectively distorted preformed biofilms and also induced morphological changes. Furthermore, Cr-SPs also showed anti-quorum-sensing potential by reducing bacterial urease and protease activities. These results indicate the potential of Cr-SPs as an anti-biofilm agent and will help to develop them as alternative therapeutics against biofilm-forming bacterial infections.
Key points
• Cr-SPs not only inhibited biofilm formation but also eradicated preformed biofilms.
• Cr-SPs altered bacterial cell surface hydrophobicity preventing biofilm formation.
• Cr-SPs efficiently degraded eDNA of the EPS layer disrupting mature biofilms.
• Cr-SPs reduced activity of quorum-sensing-mediated enzymes like protease and urease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Food can get infected by pathogenic microorganisms such as bacteria, parasites, and viruses or through toxicants that can occur at any phase from production to processing into final form of product. Foodborne diseases can be severe in many cases, leading to death of the affected individual (Boyd 2000; Adley and Ryan 2016). According to the 2019 analysis of World Health Organization (WHO), 0.42 million people die each year due to consumption of contaminated food. Also, diarrheal diseases are one of the most common forms of food-related illness, causing over 550 million people to fall sick every year and 0.23 million death every year (World Health Organization 2019; Bhaskar 2017; IFT 2004). About 60% of food diseases are caused by bacteria. Food provides nutrients that support the growth of pathogenic microorganisms and serve as a good vehicle for transport of these foodborne pathogens. Foodborne diseases caused by bacteria can be categorized as infection, toxicoinfections, and intoxication. Few examples of bacteria causing infections are Staphylococcus aureus, Clostridium botulinum, Vibrio spp. Salmonella spp., and Listeria (Giannella 1996).
The current study focuses on foodborne infections caused especially by Vibrio harveyi and Salmonella enterica. S. enterica is one of the most common causes of salmonellosis foodborne infections. It is a flagellated, Gram-negative facultative anaerobe that can enter the host through vehicles like animal harbor (i.e., meat, poultry, eggs, milk), contaminated fruits or vegetables and contamination of food during processing to prepare ready to eat products, and improper cooking. Salmonellosis can lead to gastroenteritis, septicemia, and enteric fevers (Giannella 1996). Globally, each year, over 80 million cases of foodborne salmonellosis are reported (Shannon et al. 2010). The symptoms of salmonellosis are nausea, vomiting, diarrhoea, abdominal pain, myalgia, and headache that begin after 6 to 48 h after the bacteria enters the host system (Giannella 1996). Infections caused by salmonella are life-threatening in severe cases; therefore, antibiotics with lesser side effects are the immediate need (Giannella 1996).
Sea foods that are commonly consumed are squids, fishes, crabs, shellfish, and shrimp. These sea foods are prone to contamination by different species of Vibrio. One among these species is Vibrio harveyi. Wound infections, gastroenteritis, and septicaemia are caused by pathogenic Vibrio. It is a Gram-negative, rod-shaped, motile, luminous facultative anaerobic organism (Thongkao and Sudjaroen 2017). The organism enters human host systems through contaminated water and food especially inadequately cooked seafood and similar products (Thongkao and Sudjaroen 2017; Osunla and Okah 2017). The symptoms of this infection are nausea, abdominal cramping, vomiting, fever, chills, and diarrhoea, which are visible within the first 24 h of encounter with the pathogen. People with low immune systems are likely to be affected severely by them. V. harveyi causes skin infections in cases of open wound when exposed to seawater (Chochlakis et al. 2019).
As ingestion of pathogenic bacteria is one of the major causes of foodborne infections, it is generally treated using antibiotics. Use of antibiotics has increased in recent times due to increasing occurrences of foodborne infections. MDR strains of Salmonella have shown resistance to antibiotics like ampicillin and chloramphenicol. WHO recommends to use a third-generation antibiotics, such as ciprofloxacin of the fluoroquinolone group to treat salmonella-related infection. However, a reduced susceptibility towards this drug was reported when tested over 300 isolates obtained from bloodstream infections collected from 2007 to 2012 in a study carried out in Ghana (Eibach et al. 2016; Al-Emran et al. 2016). Similarly, a combination of third-generation antibiotics like cephalosporin (ceftazidime, cefotaxime, ceftriaxone) and tetracycline or one of its analogs (doxycycline) is used to treat Vibrio infections in non-cholera adults (Stevens et al. 2014). Antibiotic resistance has vastly raised due to selective pressure among bacteria (Singh 2017). One of the major reasons for this is biofilm-forming ability of the microorganisms. Biofilm confers resistance against antibiotics by its multi-layered defense mechanism which involves various mechanisms like limiting antibiotic penetration, persistent cell population, poor growth, and stress responses (Stewart 2002). Another major reason is modifications among the genes that reside in biofilm, which results in decreased sensitivity against the antibiotics, thereby developing resistance among them (Dibyajit et al. 2019).
Bacteria form biofilms through a process of cell-cell communication known as quorum sensing. As a response to population density among the microbial cells, the quorum-sensing (QS) mechanism is activated, enabling microbial communication. This quorum-sensing mechanism plays a role in regulation of virulence production and biofilm formation in many of the bacterial species (Paluch et al. 2020). In Salmonella spp., this mechanism is mediated by autoinducers Al-I, AI-2, and AI-3 (Walters and Sperandio 2006). The AI-1 system in Salmonella spp. is different from others due to the presence of sidA protein, a homologous of LuxR protein generally present in other bacteria. sidA detects the presence of acyl homoserine lactone (AHL) produced by other microbes unlike LuxR in other bacteria (Almeida et al. 2016). The AI-2 system in Salmonella spp. is operated by the LuxS-Lsr system. The LuxS protein synthesizes signaling molecule which is internalized by Lsr operon products (Xavier and Bassler 2005). The signal molecule AI-3 is detected by products of qseBC operon and qseE gene and allows communication between bacteria and mammalian hosts (Hughes and Sperandio 2008). Similarly, the quorum-sensing mechanism in Vibrio harveyi is regulated by three autoinducers (AIs). Along with its role in virulence and biofilm production, QS in this organism is also responsible for bioluminescence phenomenon, type III secretion (TTS), and protease production. The three AIs are CAI-1, HAI-1, and AI-2. While two of these autoinducers are known, i.e., AI-2 is a furanosyl-borate-diester and HAI-1 is an acyl homoserine lactone, the structure of CAI-1 is still not known. The two-component phospho-relay cascade system that controls the production of LuxR protein in V. harveyi is involved in generating response to the AIs (Waters and Bassler 2006).
One of the strategies to combat antibiotic resistance in bacteria is to inhibit quorum sensing. This can be achieved in several ways. One approach is inhibition of synthesis of signaling molecules like blocking of proteins of Lux operon (Lade et al. 2014). Second method involves competitive inhibition of signal as well as receptor molecules as described by Ni et al. (2009). Enzymatic degradation of synthetic molecule or inactivation is also one of the methods of inhibiting quorum sensing (Delago et al. 2016). Another approach given by Rampioni et al. is blocking of the signaling cascades. For example, this is achieved by blocking of AI-receptor complexes (Rampioni et al. 2014). Nanoparticles are believed to act as quorum quenchers by inhibiting the synthesis of autoinducer or degrading AI-receptor proteins, which results in decreased production of biofilm components and other virulence factors such as elastase and pigment (Radzig et al. 2013; Garcia-Lara et al. 2015). Other inhibitors include bacterial products like norspermidine, and secondary metabolites from plants like catechins and enzymes (animal products) like acylases, lactonases, and oxidoreductase (Paluch et al. 2020). However, most of these research approaches are still in in vitro lab trials and there is an urgent need of natural antibiotics which can eradicate preformed biofilms.
A lot of focus has shifted recently to research in the field of bioactive compounds from natural resources for treating these bacterial biofilms (Dibyajit et al. 2019). An important bioactive compound that is effective against bacterial biofilms is algal sulfated polysaccharides (SPs) (Patel 2012). These are naturally present in marine algae. In fresh water algae, they are produced when subjected to stress (Seedevi et al. 2016). Recent research has uncovered the potential of Chlamydomonas reinhardtii (Cr) to produce SPs under stress conditions (Vishwakarma et al. 2019). These Cr-SPs are known to have potent properties as good anti-cancer, antioxidant, and neuroprotective agents (Kamble et al. 2018; Vishwakarma et al. 2019; Choudhary et al. 2018; Panigrahi et al. 2019). However not much work has been reported on combating biofilms of these bacteria. The aim of the current study is to evaluate the anti-biofilm and anti-quorum-sensing potential of Cr-SPs against these foodborne infection causing bacteria as a step towards developing a novel anti-biofilm drug aimed at both prevention and cure of these illnesses.
Materials and methods
Algal source and culturing conditions
A wild strain of Cr CC-124 was procured from the Chlamydomonas Genetic Centre, Duke University, USA. The algal culture was maintained at neutral pH 7 in tris acetate phosphate (TAP) medium at 25 °C with continuous illumination of 300-μmol photons m−2s in shaker incubator as mentioned by Sirisha et al. (2014).
Extraction of sulfated polysaccharides and its purification
CC-124 cells were maintained in TAP medium at 25 °C under 300-μmol photon white light illumination for 72 h in an incubator shaker. As the cells reach their stationary phase, they were collected by centrifuging at 1100×g for 5 min at 25 °C and subjected to a modified hot water extraction method of SPs (Kamble et al. 2018). The solvent was then evaporated by using rota evaporation at 60 °C, rpm 114, and 250 mbar. The dried crude extract was dissolved in autoclaved distilled water and purified using Q-Sepharose (GE Healthcare) anion exchange column chromatography using a NaCl gradient of 0–3 M. The elutes were then tested for their biochemical composition using standard protocols (Dodgson and Price 1962; Dubois et al. 1956), and the elutes which have highest carbohydrate and sulfate content were pooled and were used to analyze their anti-bacterial and anti-biofilm properties.
Bacterial strains and culture conditions
Two strains Salmonella enterica (MTCC No.9844) and Vibrio harveyi (MTCC No.7771) that are causative agents for many bacterial related food diseases were used in this study. These strains were procured from Microbial Type Culture Collection and Gene Bank (MTCC), Chandigarh, India. Growth Medium 3 (GM3) was used for maintenance and growth of these microorganisms at a temperature of 37 °C for 24 h.
Agar well diffusion method
Overnight-grown bacterial culture was spread on a GM3 agar plate. Wells were punched and different concentrations of Cr-SP (0–32 mg/mL) were loaded in these wells. The plates were incubated at 37 °C for 24 h. Post-incubation, the diameter of the clear zones of inhibited bacterial growth around the well was measured. This diameter is recorded and reported in millimeters (Teanpaisan 2016; Yan et al. 2017). This test helps to detect the potential of Cr-SPs to inhibit the growth of the bacteria.
Time-kill and clonogeny assay
Time-kill assay was performed to assess the in vitro potential of Cr-SPs to inhibit bacterial growth over a period of time. A total of 1010CFU/mL bacterial cells were treated with Cr-SPs in varying concentrations (0–8 mg/mL) and incubated at 37 °C for 24 h, 120 rpm. Bacterial growth was measured at 595 nm spectrophotometrically at 0 h till 48 h. The time-kill analysis was performed by plotting concentration on X-axis and absorbance on Y-axis (Vishwakarma and Vavilala 2019; Pinto et al. 2017). For clonogeny assay, bacterial cultures were treated with different concentrations of Cr-SPs for 24 h; post-incubation, 100 μL was spread onto GM3 agar plates. These plates were incubated for 24 h at 37 °C, and the colony count was recorded (Zhou et al. 2012).
Minimum inhibitory concentration and minimum bactericidal concentration assay
Minimum inhibitory concentration (MIC) is defined as the minimum concentration of the compound required to reduce visible growth of the bacteria. The modified broth micro-dilution method was used to assay the minimum concentration of Cr-SPs that inhibits as well as kills the bacteria. In a microtiter plate, 106 CFU/mL bacterial cells were allowed to grow with different concentrations of Cr-SPs (0–8 mg/mL) for 24 h. Post-incubation, MTT assay was performed to assess the viability of the bacterial cells. Viability was determined by plotting Cr-SPs concentrations vs percentage viability. While minimum bactericidal concentration (MBC) is the minimum concentration of the compound required to kill the bacteria. In order to check the bactericidal effect, the cells were supplied with fresh medium for 24 h post-Cr-SPs treatment and viability was checked (Bazargani and Jens 2015; Getahun et al. 2004).
Anti-biofilm potential of Cr-SPs
The potential of Cr-SPs to inhibit biofilm formation and to eradicate the preformed biofilms was tested by performing the following experiments.
Biofilm inhibition
The biofilm inhibition potential of Cr-SPs was assessed using a modified microtiter plate crystal violet quantification assay as described below. The principle of the assay involves that the positively charged dye penetrates and binds to the negatively charged bacterial cell surface molecules and polysaccharides present in the extracellular polysaccharide (EPS) layer of mature biofilm, thereby giving an information on the density of the attached bacterial cells. Bacterial cultures were grown till an O.D. of 0.5–0.6 and treated with different concentrations of Cr-SPs (0 to 8 mg/mL) for 24 h at 37 °C at 120 rpm. Post-incubation, the wells were washed with autoclaved distilled water twice and dried for 15 min at 37 °C. Furthermore, the cells were stained with 1% crystal violet which was prepared in 10% ethanol for 30 min at room temperature. After staining, the cells were washed with distilled water twice and the plate was dried at 37 °C for 15 min. Furthermore, 200 μL of absolute ethanol was added and incubated at room temperature for 10 min and was spectrophotometrically quantified at 595 nm (Bazargani and Jens 2015; Graziano et al. 2015Di Martino et al. 2003; Sasirekha et al. 2015).
Cell surface hydrophobicity assay
The bacterial adherence to hydrocarbons (BATH) assay was used to evaluate the cell surface hydrophobicity (CSH). Overnight-grown culture of bacteria was taken and inoculated in fresh medium and incubated at 37 °C till the O.D. of bacterial cell suspension reaches 0.6 to 0.8 (measured at 600 nm). At this O.D, the cells were harvested and treated with Cr-SPs at MIC and D-MIC (double-minimum inhibitory concentration) concentration for 24 h at 37 °C in an incubator shaker at 120 rpm. Post-incubation, the cell density was recorded spectrophotometrically at 600 nm. Furthermore, 1 mL of toluene was added to 1 mL of cell suspension and vortexed vigorously for 2 min. The mixture was allowed to stand for 10 min to aid separation of the phases. The aqueous phase density was recorded spectrophotometrically at 600 nm (Chari et al. 2014; Sorongon et al. 1991).
Biofilm eradication
To evaluate the ability of Cr-SPs’ ability to eradicate preformed biofilms, this assay was performed. Overnight grown bacterial cultures were reinoculated and incubated at 37 °C till it reaches 0.5 O.D. Further the culture were treated with 15-mM hydrogen peroxide at 37 °C, 120 rpm for 24 h allowing them to form biofilms. The preformed bacterial biofilms were then treated with different concentrations of Cr-SPs (0.5–8 mg/mL), incubated at 37 °C, 120 rpm for 24 h. Post-incubations, crystal violet staining was performed. The results were analyzed by calculating percentage eradication and plotted on Y-axis against concentration of Cr-SPs (Geier et al. 2008; Trentin Dda et al. 2011).
Quantification of extracellular polysaccharide layer
Bacterial cells were allowed to form biofilms, and then the cells were collected by centrifugation at 8499×g for 5 min at 25 °C. Furthermore, the cell pellet was treated with MIC and D-MIC of Cr-SPs for 24 h at 37 °C, 120 rpm, along with untreated controls. Post-incubation, the cell suspension was treated with 10% TCA and equal volume of acetone and incubated at 4 °C overnight. The mixture was centrifuged at 33995×g for 10 min at 25 °C. The weight of treated pellet was compared with the control and used for percentage EPS calculation (Chari et al. 2014).
Extracellular DNA quantification in Cr-SP-treated bacterial biofilms
A modified method suggested by Wang et al. (2011) was used for quantification of the extracellular DNA (eDNA) present in bacterial biofilms. The bacterial cells were allowed to form biofilms and further treated with Cr-SPs for 24 h. A 96-well microtiter plate was used to carry out this experiment. Post-incubation, the plates were kept at 4 °C for an hour and 1 μL of 0.5M EDTA was added per well. The biofilm contents of each well were collected and centrifuged. The pellet obtained was dissolved in 50mM Tris HCl (pH 8), and eDNA was extracted by adding equal amount of phenol:chloroform:isoamylalcohol (PCI) (25:24:1). To the aqueous phase, three volumes of absolute ethanol and one-tenth volume of sodium acetate (pH 5.2) were added and incubated at − 20 °C overnight. eDNA was collected by centrifugation at 18000×g at 4 °C for 20 min, washed with 70% ethanol, dried, and dissolved in 10μL TE buffer. An absorbance ratio of A260/A280 was used to check the purity and concentration of the eDNA obtained.
Biofilm metabolic activity
A modified MTT assay was used to assay metabolic activity of bacterial biofilms. Briefly, 106CFU/mL cells were treated with hydrogen peroxide to initiate biofilm formation, and furthermore, these preformed biofilms were treated with Cr-SPs for 24 h. The wells were washed using sterile media afterwards and treated with 20μg/mL MTT and incubated at 37 °C for 3 h. The formazan crystals were dissolved using di methyl sulfoxide and read spectrophotometrically at 570 nm (Bazargani and Jens 2015).
Anti-quorum-sensing potential of Cr-SPs
In order to check the quorum-quenching potential of Cr-SPs the following experiments were performed.
Swimming–swarming assay
Bacteria were treated with MIC and D-MIC of Cr-SPs, and their motility was determined. For S. enterica swarming assay, 0.3% GM3 agar plate was used to seed 10 μL of the culture at the center of the agar plates, while 0.5% GM3 agar was used to check its swimming potential (Kearns 2010; Kim et al. 2003), while for V. harveyi, 0.5% Luria Bertanii media with 2% NaCl (LBS) media were used to evaluate its swimming ability and 1.5% of GM3 for swarming motility (Manuel et al. 2009). Bacterial cells without treating with Cr-SPs serve as controls in this experiment.
Protease assay
Protease quantification is done by using 1% casein solution as a substrate. In this reaction, 1 mL of 1% casein solution was added to 1 mL of cell supernatant and incubated at room temperature for 10 min. Then of 0.4N TCA was added to the reaction mixture and incubated at 40 °C for 10 min. The solution was centrifuged at 8499×g for 5 min. To 1 mL of the supernatant, 5 mL of 0.4M sodium carbonate and 1 mL and 1mL Folin’s reagent were added, mixed well, and incubated at 40 °C for 20 min. The protease activity was then measured spectrophotometrically at 680 nm (Dalal 2015).
Urease assay
To determine bacterial urease activity in control and Cr-SPs treated cells, bacteria were treated with MIC and D-MIC of Cr-SPs and incubated at 37 °C for 24 h. Post-incubation, the supernatant was collected by centrifugation at 8499×g for 5 min at 25 °C. Two percent of urea was used as a substrate to quantify the urease activity. Approximately 0.1 mL of bacterial cell supernatant was incubated with 0.5 mL of substrate at 37 °C for 3 h in a water bath. Post-incubation, 0.1 mL of Nessler’s reagent was added to these tubes, mixed well, and incubated for 5 min at room temperature. The amount of urease produced was then quantified spectrophotometrically at 530 nm (Kauffmann and Moller 1955; MacFaddin 1980).
Scanning electron microscopy analysis
Bacterial cultures were grown in GM3 liquid medium overnight at 37 °C. Biofilms were allowed to form on treated coverslips by treating the cultures with 15mM H2O2. After 24 h, the cultures were treated with MIC of Cr-SPs, and the control (untreated biofilm) was incubated at 37 °C for another 24 h. Following this, the coverslips were washed thrice with PBS and fixed with 2.5% of glutaraldehyde for 12 h and were subjected to dehydration using graded ethanol (50–100%) and dried in desiccator for 24 h. The dried biofilms (control and treated) were coated with platinum and observed under scanning electron microscope (FEI Quanta 200 (XT Microscope Control) at a magnification of × 20,000) (Yan et al. 2017).
Statistical analysis
All the experiments were performed in triplicates while maintaining experimental duplicates each time. All the data collected were processed for statistical analysis systems using Originpro 8.5, and comparisons to proper controls were done. Furthermore, the results obtained were then analyzed using one-way analysis of variance (ANOVA), and Turkey’s method was done for pairwise comparison among the groups.
Results
Extraction and purification of sulfated polysaccharides from C. reinhardtii
Sulfated polysaccharides from C. reinhardtii were extracted using the hot water method and purified using anion exchange column chromatography. The extract was assessed for its biochemical composition and was found to contain 75% total carbohydrate, 33% sulfate, 42% uronic acid, and a very low protein content of ~ 5%. FTIR analysis of this extract was found to have characteristic side chains of algal sulfated polysaccharides (Kamble et al. 2018). NMR analysis of Cr-SPs clearly showed the structural characterization of algal polysaccharides (Panigrahi et al. 2019). These results clearly indicate that the extract is enriched with polysaccharides which are sulfated. This extract is further used to evaluate its antibacterial and anti-biofilm properties.
Antibacterial effect of Cr-SPs on bacterial growth by using the agar well diffusion method
Antibacterial activity of Cr-SPs against S. enterica and V. harveyi was tested using the agar well diffusion assay. Cr-SPs showed good antibacterial activity against both the bacteria tested (Table.1). Increased clear zones in the diameter ranging from 1 to 22 mm were observed with increasing concentrations of Cr-SPs from 0 to 32 mg/mL. These results clearly showed that Cr-SPs have the potential to inhibit the growth of these bacteria (Table 1).
Effect of Cr-SPs on bacterial time-kill and clonogeny assay
In vitro evaluation of Cr-SPs on the growth of these bacteria was determined by using the time-kill assay. The growth of the bacteria was assessed over a period of 0–48 h in the presence of Cr-SPs in a concentration-dependent manner. Cr-SPs started to show a reduction in growth from 0.5 mg/mL. At 1 mg/mL of Cr-SPs, both organisms showed complete growth inhibition at 24 h, while from 2 to 8 mg/mL, a complete killing of these bacteria were observed in the third hour of their growth. A decline in their growth curve can be clearly seen (Fig. 1a and b). These results indicate that there is a reduction in the microbial population in presence of Cr-SPs over a time period. Clonogeny results indicate that the colony-forming ability of S. enterica and V. harveyi was severely hampered by Cr-SPs in a dose-dependent manner. At 0.5mg/mL concentration of Cr-SPs, there was 43% and 50% of colony-forming ability of these bacteria, while at 2–4 mg/mL concentration, it has drastically dropped to 0% and 0.63% in S. enterica and V. harveyi (Fig. 1c and d) respectively, indicating that Cr-SPs not only inhibit the growth of these bacteria but also prevent their colony-forming ability.
Effect of different concentrations of Cr-SPs on time-dependent killing of a S. enterica and b V. harveyi.  0 mg/mL.
0 mg/mL.  0.5 mg/mL.
0.5 mg/mL.  1 mg/mL.
1 mg/mL.  2 mg/mL.
2 mg/mL.  4 mg/mL.
4 mg/mL.  8 mg/mL. c Inhibition of colony-forming units of S. enterica by Cr-SPs. d Inhibition of colony-forming units of V. harveyi by Cr-SPs.
8 mg/mL. c Inhibition of colony-forming units of S. enterica by Cr-SPs. d Inhibition of colony-forming units of V. harveyi by Cr-SPs.  24 h.
24 h.  48 h. Experiment results are represented as mean of three independent experiments (p < 0.05)
48 h. Experiment results are represented as mean of three independent experiments (p < 0.05)
MIC and MBC assay
MTT assay was used to identify the minimum inhibitory as well as bactericidal concentration of Cr-SPs against S. enterica and V. harveyi. The cell viability was measured, and the MIC50 was found to be as low as 440 μg/mL and 490 μg/mL for S. enterica and V. harveyi respectively (Fig. 2a and c). The Cr-SPs showed significant bactericidal activity with 50% cell death at 1 mg/mL for both the organisms. Cr-SPs is a promising antibacterial agent, as it not only inhibits the growth of these foodborne illness causing bacteria but also has the potential to kill them (Fig. 2b and d).
Biofilm inhibition and cell surface hydrophobicity assay
Both the bacteria were allowed to grow with different concentrations of Cr-SPs, and the bacteria were subsequently checked for biofilm formation using the crystal violet assay. It was observed that Cr-SPs at a concentration of 1 mg/mL inhibited biofilms up to 50% in S. enterica and up to 44% in V. harveyi. Above 2 mg/mL, more than 90% biofilm inhibition was observed in both the organisms and a complete 100% inhibition at 4 mg/mL and 8 mg/mL respectively (Fig. 3a and b). Cell surface hydrophobicity is known to play an important role in biofilm formation, as cells that are rich in hydrophobic nature have maximum capability of forming biofilms. It was observed that for S. enterica, when treated with MIC and D-MIC of Cr-SPs, its CSH ability was decreased to 57.13% and 35.18% respectively as compared with 77.37% in control (Fig. 3c). Similarly, in the case of V. harveyi, the CSH of control is 80.5%, and it reduces to 62.84% at MIC and 45.84% at D-MIC of Cr-SPs (Fig. 3d). These results indicate that Cr-SPs reduce the hydrophobicity of the bacterial cells, thereby inhibiting biofilm formation.
Biofilm eradication and quantification of extracellular polysaccharide layer
To further check if Cr-SPs can distort preformed biofilms or not, the bacteria were allowed to form biofilms using oxidative stress. These preformed biofilms were then treated with different concentrations of Cr-SPs. It was observed that Cr-SPs effectively eradicated approximately 50% of biofilms at around 1.5 mg/mL. Also, more than 95% of biofilm was eradicated at concentration of 4 mg/mL and above (Fig. 4a and b). To further check if Cr-SPs affect the EPS layer, quantification of EPS was carried out. The results clearly showed that Cr-SPs effectively distort and reduce the EPS layer formed in these organisms. In the case of S. enterica, approximately 9-fold reduction of EPS was seen when treated with MIC and D-MIC of Cr-SPs (Fig. 4c). In the case of V. harveyi, 5-fold decrease in EPS quantity was observed in comparison with control (Fig. 4d). These results show that Cr-SPs efficiently eradicate preformed biofilms by distorting the EPS layer.
Effect of Cr-SPs on eradication of preformed biofilms in a  S. enterica and b
S. enterica and b  V. harveyi. The exo-polymeric substance quantification in c
V. harveyi. The exo-polymeric substance quantification in c  S. enterica and d
S. enterica and d  V. harveyi after Cr-SP treatment at MIC and D-MIC concentrations along with untreated control. The results are cumulative mean of three independent experiments (p < 0.05)
V. harveyi after Cr-SP treatment at MIC and D-MIC concentrations along with untreated control. The results are cumulative mean of three independent experiments (p < 0.05)
eDNA quantification from bacterial biofilms
eDNA is one of important constituents of EPS layer. It has a significant role in bacterial maturation and maintenance of biofilms. This assay is designed to quantify the eDNA present in bacterial biofilms. Here, the bacterial biofilms are formed by giving mild hydrogen peroxide stress to the cells for 24 h and then treating them with Cr-SPs for a time period of 24 h. The positive controls are untreated biofilms. Upon quantification, it was seen that there is a significant reduction in the total quantity of eDNA with increased Cr-SPs concentration; when compared with the control, in V. harveyi, there is a 2-fold decrease in eDNA quantity at 0.5 mg/mL, while in the case of S. enterica, a 3-fold reduction is observed at 2 mg/mL Cr-SPs (Fig. 5a and b). These results strongly support the action of Cr-SPs against these two bacterial biofilms. It is likely related to direct interaction of Cr-SPs with EPS layer and distorting it.
Effect of Cr-SPs on bacterial biofilm metabolic activity
Cr-SPs induced decreased metabolic activity in bacterial cells in biofilms. Metabolic activity was inhibited ranging from 78–14% at concentrations 0.5 to 4 mg/mL in the case of S. enterica and 75–52% at concentrations ranging from 0.5 to 2 mg/mL in the case of V. harveyi. At 8-mg/mL Cr-SPs, a complete inhibition of metabolic activity (0%) was observed in both the organisms (Table.2). The reason behind this is Cr-SPs at high concentrations not only completely eradicated the biofilm and but also killed the bacteria present within the biofilm (Table.2). The positive control cells (bacteria with preformed biofilms) showed 100% reduced metabolic activity, as the cells in a biofilm do not show metabolic activity, thereby escaping the effect of antibiotics which target actively metabolizing cells. The percentage inhibition of metabolic activity of Cr-SP-treated bacterial cells was calculated relative to positive control. These results indicate that the Cr-SPs are effective not only in eradicating biofilms but also in efficiently killing the bacteria present inside the biofilms.
Anti-quorum-sensing activity of Cr-SPs
Swimming–swarming assay
Flagellar motility plays an important role in initial attachment during biofilm development and also during detachment phase of biofilm, where they have to move to another target surface. Therefore, swimming–swarming motility is very important in mediating adherence of bacterial cells to biotic and abiotic surfaces. In the case of both S. enterica and V. harveyi, when compared with control, Cr-SP-treated bacteria showed significant reduction in swimming and swarming motility (Fig. 6), indicating its potential to both prevention and spreading of bacterial infections.
Protease assay
The quorum-sensing mechanism triggers protease production. Proteases help in infection process by hydrolyzing peptide bonds and degrading proteins that are important for carrying out vital functions of the human body. They play an active role against the host’s immune system by conferring protease-mediated cleavage of immunoglobulins. Therefore, the ability of Cr-SPs to inhibit protease enzyme production was spectrophotometrically quantified and it was found that D-MIC value of Cr-SPs showed 50% decreased protease activity in both the bacteria as compared with untreated controls (Fig. 7a).
Inhibition of protease and urease enzyme activities using Cr-SPs. a Decrease protease activity in S. enterica and V. harveyi after treatment with Cr-SPs.  Control.
Control.  MIC of Cr-SPs.
MIC of Cr-SPs.  D-MIC of Cr-SPs. b Decrease urease activity in S. enterica and V. harveyi after treatment with Cr-SPs.
D-MIC of Cr-SPs. b Decrease urease activity in S. enterica and V. harveyi after treatment with Cr-SPs.  Control.
Control.  MIC of Cr-SPs.
MIC of Cr-SPs.  D-MIC of Cr-SPs. Each bar represents mean value of three experimental sets (p < 0.05)
D-MIC of Cr-SPs. Each bar represents mean value of three experimental sets (p < 0.05)
Urease assay
The urease produced by these bacteria was quantified, and it was found that Cr-SPs effectively reduce urease production, making it difficult for the organism to survive in the intact biofilm. In S. enterica, the activity reduces ~ 71% at MIC value and ~ 38% at D-MIC value, while in the case of V. harveyi, the activity significantly reduces to 19% at MIC and ~ 4% at D-MIC (Fig. 7a). These protease and urease results clearly indicate that Cr-SPs are an effective quorum-sensing quenching molecule leading to reduced pathogenicity.
SEM visualization
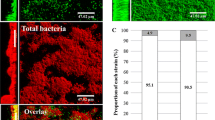
The surface structure and morphology of bacterial biofilms with and without Cr-SPs treatment were determined using SEM analysis. Control bacteria showed multiple layers of bacterial biofilms (Fig. 8a and c), while MIC Cr-SPs-treated bacterial biofilms showed remarkable decrease in number of adherent bacteria; decreased biofilm formation was observed in both the strains (Fig. 8b and d). Also, Cr-SPs-treated bacterial biofilms showed a loss of the bacterial original shape as observed by irregular and distorted bacterial cell walls. These results clearly indicate that Cr-SPs have potent anti-biofilm action against S. enterica and V. harveyi.
Discussion
Biofilms in the food industry represent a significant health and economic challenge. Their existence on surfaces of manufacturing machines results in corrosion which has a financial cost. At the same time, some of the bacterial species such as Bacillus spp. and Pseudomonas spp. secrete a range of lipolytic and proteolytic enzymes, which can potentially result in changes to taste (bitter) and odor (rancid). Such instances lead to the forcible junking of entire lots of manufacturing batches with financial implications. The long-term formation of biofilms on food manufacturing equipment also represents a public health issue. Biofilms are highly likely to contain either bacterial or fungal species that are known pathogens. These can potentially attack the immunocompromised—recipients among others. These pathogens are also capable of giving rise to diseases such as gastroenteritis (Escherichia coli, S. enterica) or lead to food intoxication (S. aureus, Bacillus cereus). An additional challenge with biofilms is their enhanced resistance to anti-microbial therapy, host immune response, and chemical disinfection. These challenges collectively have made biofilms a huge concern in industrial, clinical, and environmental settings (Serena et al. 2018; Camargo et al. 2017). To date, no antibiotic/anti-microbial agents have been developed for eliminating/treating biofilms, despite extensive research. In light of this, the only option left for researchers globally is to identify new strategies for inhibiting formation of biofilm, or developing a newer, modern class of natural antibiotics. In this context, phytochemicals are known to have anti-biofilm and anti-microbial characteristics against the broad spectrum of pathogenic organisms. They potentially play a key role in reducing the rise of drug resistance through their multi-targeted mechanism (Barbieri et al. 2017). Previous research has shown that phytochemicals can also increase the susceptibility of organisms to a variety of antibiotics (Lillehoj et al. 2018). Traditional medicinal systems in different parts of the world use many seaweed-derived medicines. These have been recorded as agents used to treat infections. Many of them have been investigated for their efficacy against multi-drug-resistant pathogens (Manikandan et al. 2011; Lu et al. 2019; Wagih et al. 2017). In the present study, the potential of sulfated polysaccharides that have been extracted from a green alga was tested for their efficacy against biofilms that were formed by Salmonella enterica and Vibrio harveyi.
In this study, it was shown that Cr-SPs exhibited efficient anti-microbial activity against both S. enterica and V. harveyi as observed by using the agar cup diffusion assay. With increased concentration of Cr-SPs, there was gradual increase in the clear zones, indicating that Cr-SPs are efficiently inhibiting the growth of both S. enterica and V. harveyi (Table.1). Moreover, the time-kill assay also showed that starting from 3 h, with increased concentrations of Cr-SPs, there is gradual decrease in the bacterial growth and complete growth inhibition was observed from 12 h till 48 h respectively (Fig. 1). Our results showed that Cr-SPs exhibited the strongest antibacterial activity at 440 μg/mL and 490 μg/mL MIC against S. enterica and V. harveyi respectively. Similarly, earlier reports from marine algae showed that fucoidan from brown algae possessed broad antibacterial spectrum against E. coli, Klebsiella pneumoniae, Vibrio cholera, Pseudomonas aeuroginosa, etc. Carrageenans showed bactericidal activity at 2500 μg/mL against S. enterica, Salmonella typhimurium, and S. aureus. However, ϒ-carrageenans showed no effect even at high dose of 5000 μg/mL. Cr-SPs showed efficient antibacterial activity at 440 and 490 μg/mL, which is much better than carrageenan (Jun et al. 2018). Also, prior studies have also shown that sulfated polysaccharide fractions from Sargassum kjellmanianum, Lachemilla angustata var. longissima, Laminaria japonica, Ecklonia cava Sargassum fulvellum L. angustata, and Eisenia bicyclis have shown efficient anti-microbial properties against a diverse range of bacteria (Ale et al. 2011; Caccamese et al. 1981; Horikawa et al. 1999). In particular, some of the studies have reported that Gram-positive bacteria are more sensitive to SPs than Gram-negative bacteria. This is potentially attributable to the differences in their cell wall structure and composition (Zapopozhets et al. 1995; Li et al. 2010; Pierre et al. 2011; Choi et al. 2015). Bioactive compounds attribute their antibacterial potential to their ability to permeabilize the bacterial cell membranes. This results in the widening of the pores and subsequent leakage of intracellular macromolecules such as proteins and nucleotides eventually leading to cell death (Joon et al. 2018).
S. enterica and V. harveyi are known to form firm biofilms, which are difficult to eradicate and are more resistant to anti-microbials. Therefore, there is an urgent need to develop novel bioactive compounds that can eradicate/distort preformed biofilms. In the current study, the anti-biofilm potential of Cr-SPs against S. enterica and V. harveyi biofilms was evaluated. It was found that there is a positive correlation between the concentration of Cr-SPs and biofilm inhibition with both the bacteria tested (Fig. 3). Interestingly, Cr-SPs effectively dissolved preformed biofilms of these bacteria (Fig. 4). Dose-dependent reduction in total EPS in the extrapolymeric substance revealed that Cr-SPs probably interacts with EPS and destroy the biofilms (Fig. 4c and d). Furthermore, scanning electron microscopy results clearly showed that MIC of Cr-SPs distorted the preformed biofilms and also significantly changed the morphology of the bacteria (Fig. 8). Earlier studies on Fucoidan from a brown alga from Sargassum wightii also showed efficient anti-biofilm activity against Staphylococcus epidermidis (Marudhupandi and Kumar 2013). Similarly, SPs from marine algal species effectively inhibited the growth of various Gram-positive and Gram-negative bacteria. They further concluded that increasing the concentration of SPs might inhibit biofilm formation (Jiao et al. 2011). Moreover, our current study showed that with increase concentration of Cr-SPs, there is a significant decrease in eDNA quantity (Fig. 5). It is known that eDNA is an important component in developing and stabilizing the biofilm in both Gram-positive and Gram-negative bacteria (Rattiyaphorn et al. 2019; Nguyen and Burrows 2014; Lappann et al. 2010; Whitchurch et al. 2002; Kim et al. 2017; Das et al. 2014; Liao et al. 2014). It was also shown earlier that in Pseudomonas aeruginosa eDNA helps in stabilizing the biofilm, and if the cells are treated with DNAse, biofilms were found to be disrupted easily in early stages but not in matured biofilms. Similarly, in S. aureus biofilms, eDNA was found to play a crucial role in cross-linking with beta toxin and helps in matrix formation (Montanaro et al. 2011). Our results clearly indicate that Cr-SPs are a promising anti-biofilm agent probably targeting the eDNA and disrupting the preformed biofilms.
A key mechanism through which bacteria communicate with one another is quorum sensing. This has been shown through multiple studies that quorum sensing is crucial for formation of biofilms. During quorum sensing, bacteria release a set of chemical signals. These signals go on to bind to receptors of another bacteria. This eventually helps in transcription of genes that are responsible for bioluminescence, virulence, biofilm formation, competence, antibiotic production, etc. This can occur inter- and intra-species (Packiavathy et al. 2011; Rutherford and Bassler 2012). Inhibiting the virulence factors like protease and urease could enable the host immune system to combat disease (Mahsa et al. 2015). It is known that urease hydrolyses urea to ammonia, which increase the pH of the surrounding environment leading to stone formation or apatites in urinary bladder (Jones et al. 1990; Marathe et al. 2018). Thus, inhibiting the urease activity is a crucial target in the development of drugs for treating urease-positive bacterial infections (Follmer 2010). Similarly, protease enzyme produced by these bacteria can enhance pathogenicity by degrading immunoglobulins, thus enabling the bacteria to evade host defense mechanism, thereby establishing infection (Aravindraja et al. 2017). The current study results showed that at MIC of Cr-SPs, there is a significant decrease in both urease and activities in both the organisms, probably leading to their reduced pathogenicity (Fig. 7). Earlier reports with plant extracts like naphthoquinone from Diospyrus lotus, allicin from garlic, curcumin, fluoroquinolones, and linoleic acid showed inhibition of urease activity in Proteus mirabilis (Abdullah et al. 2016; Marathe et al. 2018; Rauf et al. 2017; Mahsa et al. 2015; Prywer and Torzewska 2012). Similarly, phytol from Piper betel, Vanillic acid from Actinidia deliciosa, and Bisabolol from Padina gymnospora showed efficient protease inhibition in Serratia marcescens (Srinivasan et al. 2016; Sethupathy et al. 2016, 2017). The quorum-sensing mechanism is used by the bacteria to regulate biofilm formation and maturation. They help them access nutrient-rich environment compared with the non-biofilm-producing neighbors/normal flora. In various microorganisms, the mechanism and timing of biofilm production differ; however, the QS plays a role in dictating the timing of occurrence of the biofilm process (Ruby 1996, Nadell and Bassler 2011). The communication between the bacterial population is much stronger when they are associated with each other, i.e., in biofilms. Due to these reasons, QS is recognized as a key role player in virulence behavior and biofilm formation. Hence, targeting QS is a promising way to tackle this problem of pathogenicity. Targeting QS mechanisms involves altering a signal molecule. It is presumed that these therapies targeting QS will not be prone to resistance like the other targets of traditional antibiotics that focus on inhibition or killing of bacteria. Thus, these therapeutics could potentially have a longer shelf life compared with others. This is the reason we need to target the QS system of bacteria to interfere with their pathogenicity and biofilm-forming ability to solve the challenge of antibiotic resistance (Rutherford and Bassler 2012). Moreover, our current studies clearly showed that Cr-SPs can potentially serve as a useful tool for controlling pathogenicity and curbing the infection by targeting the quorum-sensing pathways of these bacteria. Additionally, better understanding of the molecular mechanism of quorum-quenching potential of Cr-SPs would help to develop them as antipathogenic compounds, which would prevent bacterial infections while not altering the normal microbiota of humans and would therefore prevent the development of resistant strains.
In summary, the current study demonstrated the potential of Cr-SPs in eradicating the bacterial biofilms through a combination of spectroscopy and microscopy analyses. It was observed that in the presence of Cr-SPs, the bacteria are unable to adhere to the surface. Interestingly, Cr-SPs were found to be effective in dissolving the preformed biofilms. The data also suggest that Cr-SPs are also potentially targeting the quorum-sensing pathways of these bacteria as indicated by the low urease and protease activity and preventing biofilm formation. SEM analysis further confirmed that in the presence of Cr-SPs, the firm bacterial biofilms got distorted with profound morphological changes. These results showed that Cr-SPs have the ability to both prevent and treat bacterial diseases, and with further validation, they can act as alternative natural therapeutics for eradication of bacterial biofilm infections.
References
Abdullah MAA, El-baky RMA, Hassan HA, Abdelhafez E, Abuo-Rahma G (2016) Fluoroquinolones as urease inhibitors: anti Proteus mirabilis activity and molecular docking studies. Am J Microbiol Res 4:81–84
The nature and extent of foodborne disease. In: J. Barros-Velázquez (ed) Antimicrobial food packaging; chapter 1. Academic Press, San Diego pp 1–10
Ale MT, Mikkelsen JD, Meyer AS (2011) Important determinants for Fucoidan bioactivity: a critical review of structure-function relations and extraction methods for fucose-containing sulfated polysaccharides from brown seaweeds. Mar Drugs 9:2106–2130
Al-Emran HM, Eibach D, Krumkamp R, Ali M, Baker S, Biggs HM, Bjerregaard-Andersen M, Breiman RF, Clemens JD, Crump JA, Espinoza LMC, Deerin J, Dekker DM, Sow AG, Hertz JT, Im J, Ibrango S, von Kalckreuth V, Kabore LP, Konings F, Løfberg SV, Meyer CG, Mintz ED, Montgomery JM, Olack B, Pak GD, Panzner U, Park SE, Razafindrabe JLT, Rabezanahary H, Rakotondrainiarivelo JP, Rakotozandrindrainy R, Raminosoa TM, Schütt-Gerowitt H, Sampo E, Soura AB, Tall A, Warren M, Wierzba TF, May J, Marks F (2016) A multicountry molecular analysis of Salmonella enterica Serovar Typhi with reduced susceptibility to ciprofloxacin in sub-Saharan Africa. Clin Infect Dis 62(suppl 1):S42–S46. https://doi.org/10.1093/cid/civ788
Almeida FA, Pimentel-Filho NJ, Pinto UM, Mantovani HC, Oliveira LL, Vanetti MCD (2016) Acyl homoserine lactone-based quorum sensing stimulates biofilm formation by Salmonella Enteritidis in anaerobic conditions. Arch Microbiol 199(3):475–486. https://doi.org/10.1007/s00203-016-1313-6
Aravindraja C, Valliammai A, Viszwapriya D, Pandian D, Karutha S (2017) Quorum sensing mediated virulence inhibition of an opportunistic human pathogen Serratia marcescens from unexplored marine sediment of Palk Bay through function driven metagenomic approach. Indian J Exp Biol 55:448–452
Barbieri R, Coppo E, Marchese A, Daglia M, Sobarzo-SE NSF, Nabavi SM (2017) Phytochemicals for human disease: an update on plant-derived compounds antibacterial activity. Microbiol Res 196:44–68
Bazargani MM, Jens R (2015) Anti-biofilm activity of essential oils and plant extracts against Staphylococcus aureus and E. coli biofilms. Food Control 61:156–164
Bhaskar SV (2017) Food borne diseases: disease burden. In: Food safety in the 21st century, 1st edn. Elsevier pp 3–12. https://doi.org/10.1016/B978-0-12-801773-900001-7
Boyd KM (2000) Disease, illness, sickness, health, healing and wholeness: exploring some elusive concepts. J Med Ethics: Medical Humanities 26:9–17. https://doi.org/10.1136/mh.26.1.9
Caccamese S, Azzolina R, Furnari G, Cormaci M, Grasso S (1981) Antimicrobial and antiviral activities of some marine algae from eastern Sicily. Bot Mar 24:365–368
Camargo AC, Woodward JJ, Call DR, Luís AN (2017) Listeria monocytogenes in food processing facilities, food contamination, and human listeriosis: the Brazilian scenario. Foodborne Pathog Dis 14:623–636
Chari Nithya, Felix L, Selvaraj K, Renganathan K, Dhamodharan B, Manivel A, Naiyf SA, Arunachalam C, Sulaiman AA, Nooruddin T(2014) Biofilm inhibitory potential of Chlamydomonas sp. extract against Pseudomonas aeruginosa. J Algal Biomass Util 5:74–81
Chochlakis D, Varveraki SP, Kostalas DM, Carouzou CK, Psaroulaki A (2019) Infection due to Vibrio harveyi and Photobacterium Damselae following injury into a marine environment. Biomed J Sci Tech Res 22(2):16462–16466
Choi SM, Jang EJ, Cha JD (2015) Synergistic effect between fucoidan and antibiotics against clinic methicillin-resistant Staphylococcus aureus. Adv Biosci Biotechnol 6:55731
Choudhary S, Save S, Vavilala LS (2018) Unravelling the inhibitory activity of Chlamydomonas reinhardtii sulphated polysaccharides against a-Synuclein fibrillation. Sci Report 8:5692
Dalal R (2015) Screening and isolation of protease producing bacteria from soil collected from different areas of Burhanpur region (MP) India. Int J Curr Microbiol App Sci 4:597–606
Das T, Sehar S, Koop L, Wong YK, Ahmed S, Siddiqui KS, Manefield M (2014) Influence of calcium in extracellular DNA mediated bacterial aggregation and biofilm formation. PLoS One 9:e91935
Delago A, Mandabi A, Meijler MM (2016) Natural quorum sensing inhibitors–small molecules, big messages. Isr J Chem 56(5):310–320. https://doi.org/10.1002/ijch.201500052
Dibyajit L, Sudipta D, Rachayeeta D, Rachayeeta D, Moupriya N (2019) Elucidating the effect of anti-biofilm activity of bioactive compounds extracted from plants. J Biosci 44:52
Di Martino P, Cafferini N, Joly B, Darfeuille-Michaud A (2003) Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res Microbiol 154:9–16
Dodgson KS, Price RG (1962) A note on the determination of the ester sulphate content of sulphated polysaccharides. Biochem J 84:106–110
Dubois M, Gilles KA, Hamilton JK (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Eibach D, Al-Emran HM, Dekker DM, Krumkamp R, Adu-Sarkodie Y, Espinoza LMC, Ehmen C, Boahen K, Heisig P, Im J, Jaeger A, von Kalckreuth V, Pak GD, Panzner U, Park SE, Reinhardt A, Sarpong N, Schütt-Gerowitt H, Wierzba TF, Marks F, May J (2016) The emergence of reduced ciprofloxacin susceptibility in Salmonella enterica causing bloodstream infections in rural Ghana. Clin Infect Dis 62(suppl 1):S32–S36. https://doi.org/10.1093/cid/civ757
Follmer C (2010) Ureases as a target for the treatment of gastric and urinary infections. J Clin Pathol 63:424–430
Geier H, Mostowy S, Cangelosi GA, Behr MA, Timothy EF (2008) Autoinducer-2 triggers the oxidative stress response in Mycobacterium avium, leading to biofilm formation. Appl Environ Microbiol 74:1798–1804
Garcia-Lara B, Saucedo-Mora MA, Roldan-Sanchez JA, Perez-Eretza B, Ramasamy M, Lee J, Coria-Jimenez R, Tapia M, Varela-Guerrero VV, Garcia-Contreras R (2015) Inhibition of quorum-sensing-dependent virulence factors and biofilm formation of clinical and environmental Pseudomonas aeruginosa strains by ZnO nanoparticles. Lett Appl Microbiol 61(3):299–305. https://doi.org/10.1111/lam.12456
Getahun A, Abraham A, Alemayehu S, Solomon G, Bekele F, Dawit W, Håkan M (2004) Evaluation of direct colorimetric assay for rapid detection of rifampicin resistant Mycobacterium tuberculosis. J Clin Microbiol 42:871–873
Giannella RA (1996) Salmonella. In: Baron, S (ed) Medical Microbiology, 4th edn. University of Texas Medical Branch at Galveston, Galveston, Galveston (TX) pp 1–7
Graziano TS, Cuzzullin MC, Franco GC, Schwartz-Filho HO, de Andrade ED, Groppo FC, Cogo-Müller K (2015) Statins and antimicrobial effects: simvastatin as a potential drug against Staphylococcus aureus biofilm. PLoS One 10:e0128098
Horikawa M, Noro T, Kamei Y (1999) In vitro antimethicillin-resistant Staphylococcus aureus activity found in extracts of marine algae indigenous to the coastline of Japan. J Antibiot 52:186–189
Hughes DT, Sperandio V (2008) Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol 6(2):111–120. https://doi.org/10.1038/nrmicro1836
IFT (2004) Institute of Food Technologist. Bacteria associated with foodborne disease. Food Tech Mag 58:20–21
Jiao G, Yu G, Zhang J, Ewart SH (2011) Chemical structures and bioactivities of sulphated polysaccharides from marine algae. Mar Drugs 9:196–223
Jones BD, Lockatell CV, Johnson DE, Warren JW, Mobley HL (1990) Construction of a urease negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun 58:1120–1123
Jun JY, Jung MJ, Jeong IH, Yamazaki K, Kawai Y, Kim BM (2018) Antimicrobial and anti-biofilm activities of sulfated polysaccharides from marine algae against dental plaque bacteria. Mar Drug 16:301
Kamble P, Sanith C, Lopus M, Vavilala S (2018) Chemical characteristics, antioxidant and anticancer potential of sulfated polysaccharides from Chlamydomonas reinhardtii. J Appl Phycol 30:1641–1653
Kauffmann F, Moller U (1955) On amino acid decarboxylases of Salmonella types and on the KCN test. Acta Pathol Microbial Scand 36:173–178
Kearns DB (2010) A field guide to bacterial swarming motility. Nat Rev Microbiol 2010(8):634–644
Kim SH, Park C, Lee EJ, Banga WS, Kim YJ, Kim JS (2017) Biofilm formation of Campylobacter strains isolated from raw chickens and its reduction with DNase I treatment. Food Control 71:94–100
Kim W, Killam T, Sood V, Surette MG (2003) Swarm-cell differentiation in Salmonella enterica serovar typhimurium results in elevated resistance to multiple antibiotics. J Bacteriol 185:3111–3117
Marathe K, Bundale S, Nashikkar N, Upadhyay A (2018) Influence of linoleic acid on quorum sensing in Proteus mirabilis and Serratia marcescens. Bios Biotech Res Asia 15:661–670
Lade H, Paul D, Kweon JH (2014) Quorum quenching mediated approaches for control of membrane biofouling. Int J Biol Sci 10:550–565. https://doi.org/10.7150/ijbs.9028
Lappann M, Claus H, Van AT, Harmsen M, Elias J, Molin S, Vogel U (2010) A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol Microbiol 75:1355–1371
Li LY, Li LQ, Guo CH (2010) Evaluation of in vitro antioxidant and antibacterial activities of Laminaria japonica polysaccharides. J Med Plant Res 4:2194–2198
Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT (2014) Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366
Lillehoj H, Liu Y, Calsamiglia S, Fernandez-Miyakawa ME, Chi F, Cravens RL, Oh S, Gay CG (2018) Phytochemicals as antibiotic alternatives to promote growth and enhance host health. Vet Res 49:76
Lu WJ, Lin HJ, Hsu PH, Lai M, Chiu JY, Lin HTV (2019) Brown and red seaweeds serve as potential efflux pump inhibitors for drug-resistant Escherichia coli. Evid Based Complement Alternat Med 2019:1836982
MacFaddin J (1980) Biochemical tests for identification of medical bacteria, 2nd edn. Baltimore, Williams and Wilkins
Mahsa RO, Mohsen A, Mojtaba A, Shokri SK, Mozafari AN, Doghaheh PH (2015) Allicin from garlic inhibits the biofilm formation and urease activity of Proteus mirabilis in vitro. FEMS Microbiol Lett 362(9): fnv049. https://doi.org/10.1093/femsle/fnv049
Manikandan S, Ganesapandian S, Singh M, Sangeetha N, Kumaraguru AK (2011) Antimicrobial activity of seaweeds against multi drug resistant strains. Int J Pharmacol 7:522–526
Manuel M, Christian J, Susan MB, Aaron N, Sarah T, Joachim R, Karl EK, Andrew C, Stefan S (2009) A novel regulatory protein involved in motility of Vibrio cholerae. J Bacteriol 191:7027–7038
Marudhupandi T, Kumar TTA (2013) Antibacterial effect of fucoidan from Sargassum wightii against the chosen human bacterial pathogens. Int Curr Pharm J 2:156–158
Montanaro L, Alessandro P, Livia V, Ravaioli S, Campoccia D, Speziale P, Carla RA (2011) Extracellular DNA in biofilms. Int J Artificial Organs 34:824–831
Nadell CD, Bassler BL (2011) A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci 108:14181–14185
Nguyen UT, Burrows LL (2014) DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int J Food Microbiol 187:26–32
Ni N, Li M, Wang J, Wang B (2009) Inhibitors and antagonists of bacterial quorum sensing. Med Res Rev 29(1):65–124. https://doi.org/10.1002/med.20145Return
Osunla CA, Okoh AI (2017) Vibrio Pathogens: A Public Health Concern in Rural Water Resources in Sub-Saharan Africa. Int J Envt Res Public Health 14:1188. https://doi.org/10.3390/ijerph14101188
Packiavathy IASV, Pandian SK, Priya S, Ravi AV (2011) Inhibition of biofilm development of uropathogens by curcumin–an anti-quorum sensing agent from Curcuma longa. Food Chem 148:453–460
Panigrahi GP, Rane AR, Vavilala SL, Choudhary S (2019) Deciphering the anti-Parkinson’s activity of sulfated polysaccharides from Chlamydomonas reinhardtii on the α-synuclein mutants A30P, A53T, E46K, E57K and E35K. J Biochem 166:463–474
Patel S (2012) Therapeutic importance of sulfated polysaccharides from seaweeds: updating the recent findings. 3 Biotech 2:171–815
Paluch E, Rewak-Soroczyńska J, Jędrusik I, Mazurkiewicz E, Jermakow K (2020) Prevention of biofilm formation by quorum quenching. Appl Microbiol Biotechnol 104. https://doi.org/10.1007/s00253-020-10349-w
Pierre G, Sopena V, Juin C, Mastouri A, Graber M, Maugard T (2011) Antibacterial activity of a sulfated galactan extracted from the marine alga Chaetomorpha aerea against Staphylococcus aureus. Biotechnol Bioprocess Eng 16:937–945
Pinto NCC, Silva JB, Menegati LM, Guedes MCMR, Marques LB, Silva TPD, Melo RCN, Souza-Fagundes EM, Salvador MJ, Scio E, Fabri RL (2017) Cytotoxicity and bacterial membrane destabilization induced by Annona squamosa L. extracts. Anais Acad Bras Cienc 89:2053–2073
Prywer J, Torzewska A (2012) Effect of curcumin against Proteus mirabilis during crystallization of struvite from artificial urine. Evid Based Complement Alternat Med 3:1–7
Radzig MA, Nadtochenko VA, Koksharova OA, Kiwi J, Lipasova VA, Khmel IA (2013) Antibacterial effects of silver nanoparticles gram-negative bacteria: influence on the growth and biofilms formation, mechanisms of action. Colloid Surf B: Biointerfaces 102:300–306. https://doi.org/10.1016/j.colsurfb.2012.07.039
Rampioni G, Leoni L, Williams P (2014) The art of antibacterial warfare: deception through interference with quorum sensing–mediated communication. Bioorg Chem 55:60–68. https://doi.org/10.1016/j.bioorg.2014.04.005
Rattiyaphorn P, Chitchanok A, Sakawrat K, Taweechaisupapong S, Chareonsudjai P, Chareonsudjai S (2019) Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS One 14:e0213288
Rauf A, Uddin G, Siddiqui BS, Khan A, Farooq U, Khan F, Majid SB, Khan SB (2017) Bioassay guided isolation of novel and selective urease inhibitors from Diospyros lotus. Chin J Nat Med 15:865–870
Ruby EG (1996) Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624
Rutherford ST, Bassler BL (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor Perspect Med 2:a012427
Sasirekha B, Megha DM, Sharath Chandra MS, Soujanya R (2015) Study on effect of different plant extracts on microbial biofilms. Asian J Biotechnol 7:1–12. https://doi.org/10.3923/ajbkr.2015
Seedevi P, Moovendhan M, Viramani S, Shanmugam A (2016) Bioactive potential and structural characterization of sulphated polysaccharide from seaweed (Gracilaria corticata). Carbohydr Polym 155:516–524
Serena G, Coral GG, Elisa MM, Claudio JV, Felipe L (2018) Biofilms in the food industry: health aspects and control methods. Front Microbiol 9:898
Sethupathy S, Ananthi S, Selvaraj A, Shanmuganathan B, Vigneshwari L, Balamurugan K, Mahalingam S, Pandian S (2017) Vanillic acid from Actinidia deliciosa impedes virulence in Serratia marcescens by affecting S-layer, flagellin and fatty acid biosynthesis proteins. Sci Rep 7:16328
Sethupathy S, Shanmuganathan B, Kasi PD, Pandian SK (2016) Alpha-bisabolol from brown macroalga Padina gymnospora mitigates biofilm formation and quorum sensing controlled virulence factor production in Serratia marcescens. J Appl Phycol 28:1987–1996
Shannon EM, Jennie M, Elaine S, Frederick JA, Martyn K, Sarah JB, Timothy FJ, Aamir F, Robert M (2010) The global burden of nontyphoidal Salmonella Gastroenteritis. Clin Inf Dis 50:882–889. https://doi.org/10.1086/650733
Singh OV (2017) Foodborne pathogens and antibiotic resistance. Publisher: John Wiley & Sons Inc, Boston
Sirisha VL, Mahuya S, D’Souza SJ (2014) Menadione-induced caspase dependent programmed cell death in the green chlorophyte Chlamydomonas reinhardtii. J Phycol 50:587–601
Sorongon ML, Bloodgood RA, Burchard RP (1991) Hydrophobicity, adhesion, and surface-exposed proteins of gliding bacteria. App Envt Microbiol 57:3193–3199
Srinivasan R, Devi KR, Kannappan A, Pandian SK, Ravi AV (2016) Piper betle and its bioactive metabolite phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J Ethnopharmacol 193:592–603
Stevens DL, Bisno AL, Chambers HF, Dellinger EP, Goldstein EJ, Gorbach SL et al (2014) Practice guidelines for diagnosis and management of skin and soft tissue infections update by the Infectious Diseases Society of America. Clin Infect Dis 59(2):e10–e52
Stewart PS (2002) Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 292:107–113
Teanpaisan R (2016) Screening for antibacterial and biofilm activity in Thai medicinal plant extracts against oral microorganisms. J Tradit Complement Med 7:172–177
Thongkao K, Sudjaroen Y (2017) Vibrio harveyi, V. parahaemolyticus, and V. vulnificus detection in Thai shellfishes by the triplex PCR method. Ann Trop Med Public Health 10:417–422
Trentin Dda S, Giordani RB, Zimmer KR, da Silva AG, da Silva MV, Correia MT, Baumvol IJ, Macedo AJ (2011) Potential of medicinal plants from the Brazilian semi-arid region (Caatinga) against Staphylococcus epidermidis planktonic and biofilm lifestyles. J Ethnopharmacol 137:327–335
Vishwakarma J, Parmar V, Vavilala SL (2019) Nitrate stress induced bioactive sulfated polysaccharides from Chlamydomonas reinhardtii. Biomed Res J 6:7–16
Vishwakarma J, Vavilala SL (2019) Evaluating the antibacterial and antibiofilm potential of sulfated polysaccharides extracted from green algae Chlamydomonas reinhardtii. J Appl Microbiol 127:1004–1017
Wagih S, Reda G, Gehan I, Elzanaty ME (2017) Antibacterial activity of some seaweed extracts against multidrug resistant urinary tract bacteria and analysis of their virulence genes. Int J Curr Microbiol App Sci 6:2569–2586
Walters M, Sperandio V (2006) Quorum sensing in Escherichia coli and Salmonella. Int J Med Microbiol 296:125–131. https://doi.org/10.1016/j.ijmm.2006.01.04.1
Wang D, Jin Q, Xiang H, Wang W, Guo N, Zhang K, Tang X, Meng R, Feng H, Liu L, Wang X, Liang J, Shen F, Xing M, Deng X, Yu L (2011) Transcriptional and functional analysis of the effects of magnolol: inhibition of autolysis and biofilms in Staphylococcus aureus. PLoS One 6:e26833
Whitchurch CB, Tolker NT, Ragas PC, Mattick JS (2002) Extracellular DNA required for bacterial biofilm formation. Sci 295:1487
World Health Organization (2019) https://www.who.int/en/news-room/fact-sheets/detail/food-safety
Xavier KB, Bassler BL (2005) Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol 187(1):238–248. https://doi.org/10.1128/JB.187.1.238-248.2005
Waters CM, Bassler BL (2006) The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20(19):2754–2767. https://doi.org/10.1101/gad.1466506
Yan X, Gu S, Shi Y, Cui X, Wen S, Ge J (2017) The effect of emodin on Staphylococcus aureus strains in planktonic form and biofilm formation in vitro. Arch Microbiol 199:1267–1275
Zapopozhets TS, Besednova NN, Loenko IN (1995) Antibacterial and immunomodulating activity of fucoidan. Antibiot Chemother 40:9–13
Zhou Y, Kong Y, Kundu S, Cirillo JD, Liang H (2012) Antibacterial activities of gold and silver nanoparticles against Escherichia coli and Bacillus Calmette-Guerin. J Nanobiotechnol 10:19
Acknowledgments
This work is supported by Department of Atomic Energy, India.
Author contribution statement
Dr. Vavilala—concept, data analysis, and editing the manuscript; Ms. Vishwakarma—performed the experiments and analyzed the data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vishwakarma, J., V.L, S. Unraveling the anti-biofilm potential of green algal sulfated polysaccharides against Salmonella enterica and Vibrio harveyi. Appl Microbiol Biotechnol 104, 6299–6314 (2020). https://doi.org/10.1007/s00253-020-10653-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10653-5



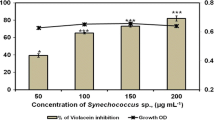


 S. enterica and c
S. enterica and c  V. harveyi and minimum bactericidal concentration of Cr-SPs against b
V. harveyi and minimum bactericidal concentration of Cr-SPs against b  S. enterica and d
S. enterica and d  V. harveyi. Three independent experiments of both MIC and MBC were considered, and mean + SE were plotted (p < 0.05)
V. harveyi. Three independent experiments of both MIC and MBC were considered, and mean + SE were plotted (p < 0.05)
 S. enterica and b
S. enterica and b  V. harveyi. The effects of MIC and D-MIC of Cr-SPs on cell surface hydrophobicity in c
V. harveyi. The effects of MIC and D-MIC of Cr-SPs on cell surface hydrophobicity in c  S. enterica and d
S. enterica and d  V. harveyi. Three independent experiments were considered, and results represent mean + SE (p < 0.05)
V. harveyi. Three independent experiments were considered, and results represent mean + SE (p < 0.05)

 S. enterica and b
S. enterica and b  V. harveyi. Each bar is a representative of mean + SE of three independent experiments (p < 0.05)
V. harveyi. Each bar is a representative of mean + SE of three independent experiments (p < 0.05)

