Abstract
Escherichia coli is the most widely used heterologous protein expression system. However, this system remains a challenge due to the low solubility of proteins, insufficient yield, and inclusion body formation. Numerous approaches have sought to address these issues. The use of a fusion tag is one of the most powerful strategies for obtaining large amounts of heterologous protein in E. coli expression system. Here, recent advances in fusion tags that increase the expression of proteins are reviewed. In addition, proposed concepts for designing peptide tags to increase protein expression are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proteins are important biomolecules that perform numerous cellular functions and are essential for industrial and therapeutic applications, such as biotechnology (Sharma et al. 2019; Singh et al. 2019) and biotherapeutic medicines (Lagassé et al. 2017), and to further the understanding of biology. However, obtaining proteins from natural sources is limited, concerning protein quantity, ease of separation, and costs for the feasible applications (Geisse and Fux 2009). In contrast, genetic engineering methods can allow cells to synthesize sufficient quantities of the proteins of interest, even heterologous proteins, which can be purified for use in key research or for industrial and therapeutic applications (Ahmad et al. 2019; Khan et al. 2016; Priyadarshini and Singh 2019). Many heterogeneous systems, including bacteria (Ma et al. 2018; Mathiesen et al. 2008; Vavrová et al. 2010), filamentous fungi (Nevalainen et al. 2005), yeast (Cereghino and Cregg 2000; Juturu and Wu 2018; Madzak 2015), insect cells (Le et al. 2018; van Oers et al. 2014), plants (Fahad et al. 2015), and mammalian cell lines (Arena et al. 2019; Lee et al. 2015a), have been engineered for the expression of heterologous proteins. More recently, microalgae have been considered a promising host for recombinant protein production (Bañuelos-Hernández 2017; Gangl et al. 2015; Specht et al. 2010). These various expression systems have advantages and disadvantages, and can be selected according to the characteristics of the protein to be expressed.
Escherichia coli is one of the most preferred heterologous expression systems. It is easy to handle, fast growing with doubling times as short as 20 min in rich media (Fossum et al. 2007), amenable to high-density cultivation up to 100 g dry weight cells per liter in the controlled fed-batch mode (Khalilzadeh et al. 2004; Knorre et al. 1991), and capable of a high yield of recombinant protein production that is promoted by established methods of genetics and compatible molecular tools (Rosano and Ceccarelli 2014; Sørensen and Mortensen 2005). However, there are disadvantages in the E. coli expression system, especially for producing biopharmaceuticals. These disadvantages include lack of eukaryotic post-translational modifications, low solubility, improper protein folding, inclusion body formation, endotoxin issues, and poor secretion (Baeshen et al. 2014). These drawbacks are being addressed by advances in biotechnology, thus allowing E. coli to remain an attractive protein expression system (Redwan et al. 2015). Affinity tags have greatly assisted the efficient purification of proteins of interest (Saraswat et al. 2013), whereas solubility tags are still a trial-and-error experience and the passenger protein can be differentially affected by several fusion tags (Costa et al. 2014; Paraskevopoulou and Falcone 2018). For this reason, fusion tags to enhance heterologous protein expression and solubility have continued to be investigated and developed (Costa et al. 2013; Nguyen et al. 2019b). The acidity of fusion tags has substantially improved the solubility of fusion proteins (Costa et al. 2014; Paraskevopoulou and Falcone 2018). Thus, acidic fusion tags (i.e., negatively charged tags at physiological pH) have been theoretically designed and selected.
On the other hand, inclusion body formation has been recently considered a potentially exploitable phenomenon (Rinas et al. 2017) and a tag that is likely to cause aggregation has been developed (Yadav et al. 2016).
An N-terminal fusion tag can take advantage of efficient translation initiation sites on the tag (Malhotra 2009). Specifically, the folding free energy in the region between − 25 and +35 of messenger RNA (mRNA) has the greatest impact on the efficiency of prokaryotic translation (Seo et al. 2013). Thus, it is worthwhile to tune the nucleotide sequence around the translation initiation region (TIR) for translation efficiency.
Here, we consider fusion tags that enhance protein expression in E. coli. In addition, the impacts of recently discovered or designed peptide tags on protein expression are reviewed. We hope this information will be helpful in designing new fusion tags. Scheme 1 summarizes the strategies for the effective production of heterologous proteins in E. coli using expression-enhancing tags.
Schematic flowchart summarizing the strategies for heterologous protein production using expression enhancing tags through E. coli expression system. In general, when a heterologous gene is expressed in E. coli, the gene of interest is expressed using an expression vector with a promoter, an antibiotic resistance gene for selection, and a His-tag for purification. If the protein is not produced as a soluble form, then several conditions should be taken into consideration to facilitate the soluble production of target protein. The soluble expression can be increased by either lowering the incubation temperature or decreasing the amount of inducer such as IPTG or by both. If the gene consists of rare codons in E. coli or the expressed protein forms disulfide bonds, a specific E. coli strain can be used accordingly. Alternatively, solubility-enhancing tags can be used to induce soluble expression of the gene of interest. If not expressed, either the codon optimization of the target gene for E. coli or fusion of an expression-enhancing tag with the target gene is taken account. In some cases, insolubility-enhancing tags are intentionally used to get an insoluble protein. The insoluble protein can be refolded in denaturing or mild solubilizing conditions to regain the soluble form. Generally, heterologous proteins are expressed in the cytoplasm of E. coli, but certain tags transport a passenger protein to the compartments other than the cytoplasm, such as membranes, periplasm, or extracellular
Expression, solubility, and purification of proteins
Fusion tags increase the expression level and/or the solubility of proteins. These tags, which comprise proteins or peptides linked to target proteins, help to achieve natural folding leading to a proper functional activity or to achieve a higher level for a protein that is poorly expressed. Affinity tags are used along with the corresponding affinity binding ligand to allow rapid and efficient purification of proteins, in turn leading to increased production yield of the expressed protein (Malhotra 2009). When expressed in combination with soluble fusion tags and affinity tags, such as 6×His-tag for purification, the presence of fusion tags is undesirable because they can potentially impair biological function and interfere with proper structural analysis or immunogenicity (Costa et al. 2014; Malhotra 2009; Young et al. 2012). Therefore, removal of the fusion tag is a necessary step in the downstream process. Removal can be done by adding a protease recognition sequence between the fusion tag and the target protein, so that the tag can be removed by a protease if necessary (Costa et al. 2014; Yadav et al. 2016).
There is a strategy to further improve the soluble expression in combination with a soluble-enhancing fusion tag in E. coli. Simply lowering the incubation temperature and/or the concentration of the inducer, isopropyl β-d-1-thiogalactopyranoside (IPTG), reduces protein misfolding by decreasing the translation rate, which consequently avoids aggregation of proteins into inclusion bodies (Nguyen et al. 2017; Sorensen and Mortensen 2005).
Various engineered E. coli expression strains can be used for proper folding of protein. Engineered E. coli stains available for proper aim are summarized on the website of the Wolfson Center for Applied Structural Biology (http://wolfson.huji.ac.il/expression/bac-strains-prot-exp.html). Special engineered E. coli strains include Arctic Express strains to address the common bacterial gene expression hurdle of protein insolubility (Agilent Technologies); Shuffle T7 (NEB), BL21trxB (Novagen), and Origami (Novagen) to form disulfide bonds in the cytoplasm for proper folding of proteins; Rosetta (Novagen) and BL21CodonPlus (Stratagene) to enhance the expression of eukaryotic proteins that contain codons rarely used in E. coli; C41(DE3) and C43(DE3) (Lucigen), to express toxic and membrane proteins from all classes of organisms; and Lemo21(DE3) to allow for tunable expression of difficult clones. ClearColi®BL21(DE3) Electrocompetent Cells (Lucigen) are the first commercially available competent cells with a modified lipopolysaccharide (LPS, Lipid IVA) that does not trigger the endotoxic response in mammalian cells (Mamat et al. 2015). The combination of the aforementioned factors and trial-and-error tuning has led to the successful production of proteins of interest.
Protein fusion tags that enhance expression
Various solubility-enhancing tags are available: the well-known maltose-binding protein (MBP), glutathione S-transferase (GST) (Harper and Speicher 2011), N-utilization substance A (NusA) (Costa et al. 2013), thioredoxin (Trx) (Savitsky et al. 2010), small ubiquitin-related modifier (SUMO) (Butt et al. 2005), Fh8 (Costa et al. 2013), and some other tags. Table 1 summarizes the solubility-enhancing tags and compares their molecular weights, pI values, and GRAVY values. The GRAVY value of a protein is a measure of its hydrophobicity or hydrophilicity (Kyte and Doolittle 1982). The values range from − 2 to +2 for most proteins, and in simple terms, positive GRAVY values indicate hydrophobicity; negative values mean hydrophilicity (Isalan et al. 2013). Commonly used solubility-enhancing tags have a negative GRAVY value.
MBP is one of the most frequently used fusion tags to enhance the solubility of the expressed protein. MBP acts as a molecular chaperone to aid the accurate folding of a fusion protein. It is believed to interact with hydrophobic amino acid residues present in unfolded proteins to prevent aggregation or proteolysis (Needle and Waugh 2014; Sachdev and Chirgwin 2000). GST is a naturally occurring 26-kDa protein. The use of GST fusion proteins has been successful in both nuclear magnetic resonance (NMR) and crystallography structure determinations (Harper and Speicher 2011). GST is a relatively poor solubility enhancer compared with the other commonly used fusion tags (Bernier et al. 2018). NusA is a 55-kDa protein that slows down translation at transcriptional pauses, offering more time for protein folding, and also stabilizes the passenger protein during translation (Costa et al. 2014). NusA is reported to attract chaperones, such as GroEL, in E. coli (Douette et al. 2005). Trx is a 12-kDa intracellular thermostable protein of E. coli. Trx is commonly employed as a fusion tag to improve solubility of the protein of interest by virtue of its intrinsic solubility and by taking advantage of its natural oxido-reductase activity responsible for the reduction of disulfide bonds through thio-disulfide exchange (Costa et al. 2014). SUMO is a small protein (approximately 11 kDa) found in yeast and vertebrates. It promotes the proper folding and solubility of its target proteins, possibly by exerting chaperoning effects (Costa et al. 2014). Fh8 is a small protein (8 kD) with extensive homology with 8-kDa calcium-binding proteins. It is comparable with MBP, NusA, and Trx concerning soluble protein production, in which GST tag does not improve solubility of the target protein (Costa et al. 2012). Considering the pI value, commonly used fusion tags exhibit a pI between 4 and 5, whereas GST has a rather higher value of 6.51 (Table 1).
Human protein disulfide isomerase I (PDI) catalyzes the formation of native disulfide bonds of secretory proteins in the endoplasmic reticulum, and has a vital role in protein folding. Loss of PDI activity has been associated with the pathogenesis of numerous disease states (Galligan and Petersen 2012). PDI is composed of four thioredoxin-like domains (abb′a′). Two contain redox-active catalytic sites (a and a′) and two do not (b and b′) (Byrne et al. 2009; Galligan and Petersen 2012; Song et al. 2013). The b′a′ domain of PDI (PDIb′a′) is the smaller functional unit of PDI. Both PDI and PDIb′a′ have been used to enhance the soluble cytoplasmic expression of recombinant proteins in E. coli (Song et al. 2013; Do et al. 2014; Nguyen et al. 2014; Nguyen et al. 2016; Nguyen et al. 2017; Nguyen et al. 2019a).
Another study (Guo et al. 2018) reported that at low temperatures MBP was a very efficient fusion tag for soluble mouse leukemia inhibitory factor (mLIF) expressed in E. coli cytoplasm. The MBP-mLIF fusion exhibited bioactivity without removal of MBP (Guo et al. 2018). Human vascular endothelial growth factor (hVEGF), human fibroblast growth factor 21 (hFGF21), and human oncostatin M (OSM) were expressed at 18 °C with above 90% solubility, respectively, in E. coli by MBP-tagging at the N-terminus of the target protein, where 6×His-tag was positioned at the N-terminus of MBP (Nguyen et al. 2016; Nguyen et al. 2017; Nguyen et al. 2019a). The fused proteins were successfully purified and all were stable after tag cleavage, displaying similar bioactivity to counterparts produced in mammalian cells or commercially available. Multiple comparisons of various protein fusion tags, such as Trx, SUMO, GST, MBP, NusA, PDI, and PDIb′a′, with the target protein revealed MBP as one of the most effective enhancers of the expression and solubility of target protein (Nguyen et al. 2016; Nguyen et al. 2017; Nguyen et al. 2019a). At 37 °C, only the MBP and PDI tags could enhance the solubility of corresponding fused hFGF21 to greater than 60%, whereas the other fusion proteins were primarily expressed as inclusion bodies (Nguyen et al. 2017). The expression level tended to be higher as the size of fusion tag decreased, due to an alleviated metabolic burden on E. coli compared with larger fusion tags (Paraskevopoulou and Falcone 2018). In contrast, the solubility was increased as the size of fusion tag increased at 37 °C due to a chaperone-like function of larger fusion tags (Costa et al. 2014; Nguyen et al. 2019a). Lowering the temperature to 18 °C enhanced the solubility of all the expressed constructs to more than 80% in the case of PDIb′a′ (31 kDa), MBP (40 kDa), NusA (55 kDa), and PDI (55 kDa) (Nguyen et al. 2017). These results indicate that larger fusion tags caused more soluble expression of hFGF21 independent of temperature, while smaller fusion tags, such as Trx, SUMO, and GST including the 6×His tag, induce soluble expression at a lower temperature (18 °C).
The combination of the MBP tag with the 10×His tag and their specific positions had a strong impact on the expression and solubility levels of hydrophobic bovine R9AP protein (bR9AP) as well as on the purification yield of the fusion protein (Bernier et al. 2018). N-Terminal positioning of the 10×His tag impaired MBP-induced expression by 50%, but purity only increased by 10% compared with N-terminal position of MBP. In contrast, N-terminal positioned 10×His tag enhanced the purity of GST-fused bR9AP by 15% without hampering the expression level. MBP has been used in combination with a poly-His tag because of the low binding of MBP to the amylose resin and the resulting poor purity of the fusion protein (Nguyen et al. 2016; Sun et al. 2011). Therefore, the combination of MBP and His tags needs to be optimized for each position and passenger protein. If the tags are positioned on either side, more than one protease is required to remove both tags and the resulting procedure will be time-consuming. Hence, a careful design of the fusion proteins is a necessary step where many parameters must be considered to optimize and simplify the procedures (Bernier et al. 2018).
The HEHEHE-tag is a modified 6×His tag in which every second histidine residue is replaced by a more hydrophilic glutamate, which can work as an affinity tag in place of the 6×His tag (Tolmachev et al. 2010). The HE-MBP (Pyr) tag that is 7×(HE) tagging at N-terminus of truncated maltotriose-binding protein from Pyrococcus furiosus was compared with the 6×His-MBP tag (Han et al. 2018; Sun et al. 2011). HE-MBP (Pyr) enhanced solubility by 1.6- to 2.7-fold for passenger proteins that exhibited either same or higher expression levels compared with those by the 6×His-MBP. HE-MBP (Pyr) even expressed the protein (CdiGMP026) with 40% solubility, whereas 6×His-MBP did not express the protein. There were little differences in expression levels between 6×His-MBP (Pyr) and 6×His-MBP, whereas the former exhibited higher solubility than the latter. HE-MBP (Pyr) also showed superior solubility and expression level compared with those for 6×His-MBP (Pyr).
Another notable carbohydrate-binding protein used as a fusion tag is the cellulose-binding domain. The CBDs can contain 30–180 amino acids, and exist as a single, double, or triple domain in one protein (Levy and Shoseyov 2002; Shoseyov et al. 2006). Cellulose is an economical support matrix for large-scale protein purification. Thus, CBD has been widely used as an affinity tag in expression and purification. In terms of protein expression using CBDs, many studies have demonstrated that CBD has a beneficial effect on the expression of passenger proteins when fused to either C- or N-terminal of them (Levy and Shoseyov 2002). CBD has been reported as a fusion partner that can enhance the soluble expression in E. coli (Murashima et al. 2003; Klocke et al. 2005; Xu and Foong 2008). A recent study showed N-terminal CBD fusion enhanced soluble expression of fructosyl peptide oxidase in E. coli and its simultaneous purification and immobilization (Chen et al. 2019). CBD is a β-hairpins structure with short amino acid residues (110 a.a) which are considered to fold with two-state kinetics (Murashima et al. 2003). If so, this feature could help the passenger protein be soluble through readily folding of CBD.
Superfolder green fluorescent protein (sfGFP) is a mutant of wild-type GFP (wtGFP) with pronounced solubility and fast folding ability. sfGFP is being developed as a fusion tag to facilitate protein solubility (Pedelacq et al. 2006). A recent study successfully obtained soluble anti-influenza PB2 scFv using sfGFP as the N-terminus tag. PB2 scFv with 60% solubility was obtained at 18 °C in E. coli (Liu et al. 2019). Both proteins exhibited biological activity in the presence and absence of sfGFP. Using sfGFP as the N-terminal tag, heterologous molecules ranging from peptides to complex proteins were successfully expressed and secreted extracellularly with functional conformations (Zhang et al. 2017). The beta-barrel structure and net negative charges of sfGFP play important roles in its auto-extracellular secretion property (Zhang et al. 2017). This is evident in autotransporters which have a sec signal peptide at the N-terminus, the target protein in the middle, and a beta-barrel structure at the C-terminus (Rojas-Lopez et al. 2018). The C-terminal beta-barrel structure facilitates the extracellular secretion of target proteins (Knowles et al. 2009).
The mature form of β-fructofuranosidase (β-FFase) isolated from Arthrobacter arilaitensis NJEM01 was expressed as a soluble protein in the cytoplasm of E. coli (Chu et al. 2014). β-FFase (Ffu) consists of a mature β-FFase comprising 495 residues and a signal peptide of 53 residues. When expressed in E. coli as a total sequence including the signal sequence, Ffu are highly secretory proteins. The feature of Ffu has spurred interest in developing novel fusion tags (Cheng et al. 2017). By using the Ffu fusion system, CARDS TX, VEGFR-2, RVs, and Omp85 were successfully expressed in the soluble form and secreted in the periplasmic space in E. coli, whereas MBP fusions led to a mass of inclusion bodies, representing a slight advantage when compared with the direct expression of the studied proteins (Cheng et al. 2017). One of the disadvantages of the industrial application of the E. coli expression system is the lack of extracellular protein production in the absence of a secretory system. Successful secretion of proteins of interest into the periplasm or their excretion into the culture medium is more beneficial compared with their intracellular expression. Compared with the cytoplasm, the periplasm, which has lower levels of proteases and is a more oxidative environment, facilitates the precise formation of disulfide bonds and protects target proteins against proteolytic degradation (de Marco 2009). Extracellular protein secretion can also simplify the protein purification process.
Mistic is an unusual Bacillus subtilis integral membrane protein (approximately 13 kDa) that folds autonomously into the membrane, bypassing the cellular translocon machinery (Roosild et al. 2005). The N-terminal fusion of Mistic peptide enhances the expression of membrane proteins in a functionally active conformation by facilitating autonomous folding into the bacterial membrane, even though it is a highly hydrophilic peptide lacking any candidate signal sequence (Kefala et al. 2007). N-Terminal fusion of Mistic peptide reportedly enhanced the production of G protein-coupled receptor (GPCR) proteins in E. coli cell-free systems, resulting in sufficient protein production for structure-function studies (Lyukmanova et al. 2012). Eukaryotic type I rhodopsins were expressed by linking between N- and C-terminal Mistic domains in E. coli. Lee et al. (2015b) initially fused Mistic tag to the N-terminus of algal rhodopsin, but failed to express the protein to the E. coli membrane. Next, Mistic was fused to either side of rhodopsin (Mistic-rhodopsin-Mistic) and was successfully expressed in the E. coli membrane. However, expression levels were still low compared with that of the bacterial-origin counterpart (Lee et al. 2015b). Likewise, the authors tested various eukaryotic rhodopsin and eukaryotic membrane proteins, including ARI, CSRB, ARII, and human melanopsin (Lee et al. 2015b). Mistic comprises four helical fragments, all of which can replace full-length Mistic as N-terminal fusions to achieve overexpression of a human GPCR in E. coli, although with different effects on quantity and quality of the protein produced (Marino et al. 2015). To elucidate the function of Mistic, the authors found Mistic homologs that have a highly conserved motif with characteristics of a Shine-Dalgarno (SD) sequence upstream of yugO, which coded for a K+ channel. The removal of SD sequences reduced the expression of YugO (Marino et al. 2015). The authors suggested that the presence of N-terminal sequences at the mRNA level, rather than the ability of the translated product to interact with the membrane, is important for high expression levels of Mistic-tagged protein constructs.
Antimicrobial peptides (AMPs) can disrupt microbial membranes and are liable for proteolytic degradation. Thus, they are often produced as a fusion partner in heterologous hosts to neutralize their toxicity and increase their expression levels (Li 2011; Li et al. 2011). Since the effects of the fusion tags on different target AMPs differ between targets, it is difficult to say that one fusion tag is superior to others. However, in terms of the production of recombinant LL-37 in E. coli, Trx is better than GST (Li 2011). The former displayed the highest absolute yield of the fused peptide among 13 different carrier proteins tested (Bogomolovas et al. 2009), and the latter yielded inefficient or failed peptide production due to high susceptibility to proteolytic degradation (Li 2011). SUMO is an excellent carrier protein and favors the soluble production of small hydrophobic peptides including AMPs. The SUMO tag takes advantage of the subsequent release of the passenger due to a highly specific SUMO protease, although SUMO requires additional affinity tags for purification, such as the 6×His tag (Li 2011; Li et al. 2011; Mo et al. 2018; Wei et al. 2018; Kim et al. 2019).
ThiS is a smaller sulfur-carrier protein involved in thiamin synthesis, which is conserved among most prokaryotic species. Its structure is similar to ubiquitin (Ub) and SUMO, which are used as soluble enhancing fusion tags in prokaryotic expression. ThiS-fused insulin A and B chains and EGFP (N-terminal fusion for insulin and C-terminal fusion for EGFP) were overexpressed in inclusion bodies (Uversky et al. 2013). The prokaryotic ubiquitin-like protein MoaD falls in the category of insoluble expression (Yuan et al. 2014). Compared with SUMO, both tags are more acidic and more likely hydrophobic (Table 1). Insoluble expression is believed to be more efficient than soluble fusion in masking the toxic effects of peptides and protecting them from degradation. In case of AMPs, if there is no cysteine residue, refolding is not necessary to recover activity. Thus, the use of inclusion body inducing fusion tags can be an alternative to the use of a solubility tag to produce a high quantity of AMPs (Lee et al. 2000; Zorko and Jerala 2010; Park et al. 2012).
Intrinsically disordered proteins (IDPs) are natively unfolded proteins that feature biased amino acid compositions and low sequence complexity, with a low proportion of hydrophobic amino acids and a high proportion of hydrophilic amino acids (Wright and Dyson 2015). They account for a large part of the cellular proteome and are key molecules in controlling intracellular signaling by a flexible network of protein interactions (Wright and Dyson 2015). Because of their structural features, IDPs display extreme sensitivity toward proteolytic cleavage, and denaturing conditions can be used without having to refold the protein (Graether 2019). Thus, the aggregation-prone tags can be applied to produce IDPs (Goda et al. 2015; Hwang et al. 2012). Solubility tags have been used for expression of IDPs, but caution is needed because IDPs are vulnerable to proteolytic degradation due to the unfolded structures (Graether 2019). The fusion tags available for insoluble expression, which include TrpΔLE, ketosteroid isomerase (KSI), PruE, and PagP, have been well documented in a review (Hwang et al. 2014). MBP has been used to successfully express GPCRs in E. coli (Furukawa and Haga 2000; Locatelli-Hoops et al. 2013). However, the low-level expression is still an obstacle for biophysical studies of GPCRs. Thus, the ability of E. coli to produce large amounts of eukaryotic proteins as inclusion bodies has been exploited for the high-level production of GPCRs (McCusker et al. 2008). KSI and TrpΔLE are commonly used for bacterial expression of membrane protein along with MBP and GST tag (Caroccia et al. 2011; Potetinova et al. 2012). In addition, Npro is the N-terminal autoprotease of the classical swine fever viral nucleocapsid protein and it aggregates a passenger protein to inclusion body in E. coli. Npro and its variant EDDIE allows the passenger protein to be released from the C-terminal end of the autoprotease by self-cleavage, leaving the target protein with an authentic N-terminus during refolding process (Achmuller et al. 2007). Thus, these expression systems can be useful for high-level production of recombinant toxic peptides and proteins in E. coli avoiding the need for chemical or enzymatic removal of the fusion tag (Achmuller et al. 2007).
Peptide fusion tags that enhance expression
The advantage of short peptide tags for expression is that the amino acid sequence is generally 15 residues or less, which does not severely interfere with the protein structure or impair its activity when fused to the protein of interest (Carson et al. 2007; Kato et al. 2007; Nguyen et al. 2019b). Therefore, it may be not necessary to remove the fusion tags for the protein application other than therapeutic proteins, in contrast to the case of a protein fusion tag. Short peptide tags can also reduce the metabolic burden of protein expression on the host compared with the large-sized tags (Zhao et al. 2013). Expression-enhancing peptide tags are shown in Table 2 along with comparison values of molecular weights, pI values, GRAVY values, and tag positions compared among peptide tags.
The S Tag is commercially available and is usually known as an affinity tag for purification. RNase S is a subtilisin cleavage product of native RNase A with modified RNase activity, in which the first N-terminal 15 amino acids (residues 1–15; KETAAAKFEREHMDS, S•Tag™) of a high affinity linked S-peptide comprising 20 amino acids are involved in the formation of functional complexes with the remaining part of S-protein (residues 21–124) (Kim and Raines 1993; Yadav et al. 2016). Even if it is suitable for purification of the S Tag tagged proteins by binding to the immobilized S-proteins as a ligand via S Tag, it also improved the solubility of a passenger protein. The NtrX protein was overexpressed as an S Tag fusion protein induced by l-arabinose in the E. coli strain BL21AI (Assumpcao et al. 2007). Human erythropoietin (EPO) fused with MBP and 6×His-MBP could not be recovered due to its aggregation after removal of tags, but when fused to 6×His-S-tag, it refolded as a functional protein even after the removal of the 6×His-S-tag (Grunina et al. 2017).
The many studies that have addressed poly-ionic tags as enhancers of protein solubility in heterologous protein expression were recently reviewed (Paraskevopoulou and Falcone 2018). In contrast, polycationic tags may be prone to aggregation when fused to the net negatively charged passenger protein at physiological pH, and vice versa, due to intramolecular attractive electrostatic interaction between the proteins and the peptide tags (Kim et al. 2012). Proteins are least soluble at the ambient pH equal to their pI, where they display a zero net charge. Thus, changing the net charge into negative or positive improves the solubility of proteins (Lawrence et al. 2007). Based on the pI value of the protein of interest, using a fusion tag capable of inducing a repulsive electrostatic interaction between the proteins may provide sufficient time for the correct folding of the protein, and consequently prevent protein aggregation (Kato et al. 2007; Paraskevopoulou and Falcone 2018; Zhang et al. 2004). Aggregation-prone Ig variable-type domains derived from three human membrane glycoproteins can reportedly be solubilized during overexpression in the E. coli cytoplasm by extension of the domain N- or C-terminus with highly acidic peptides (net negative charge > − 6) made from T7B variants with the additional acidic residues derived from bacteriophage T7 (Zhang et al. 2004). Other studies described that polylysine or polyarginine tags fused to C-terminus of the protein of interest more likely improve solubility than when fused to the N-terminus (Park et al. 2003; Hage et al. 2015; Islam et al. 2015; Nautiyal and Kuroda 2018).
Hirose et al. (2011) constructed seven proteins combined with 12 kinds of data-driven designed tags (DDTs) based on highly frequent sequence property patterns in an experimentally assessed protein solubility dataset in a wheat germ cell-free system. The 12 DDTs (six for solubility and six for insolubility) were fused to the N-terminus region of the passenger proteins. Among six solubility tags, Glu and Ser-rich solubility tags only enhanced solubility toward one protein that displayed the least solubility among seven proteins. In contrast, among the insolubility tags, Arg, Lys, and Gly were frequently present in DDTs and these insolubility tags induced four of the five proteins, which are soluble in the form of fusion-free, to be insoluble proteins. Based on the patterns of the insolubility tags, they are rich in positive charge, especially Arg and amino acids (Ala, Gly, and Ser) with a tiny side chain located at each side of Arg and the [AGS]-R-[AGS] motif is considered to be an important characteristic for increasing protein insolubility (Hirose et al. 2011).
Pandey et al. (2014) reported successful overexpression of transmembrane segments 1–3 of the apelin receptor (AR_TM1–3) in the C41(DE3) strain of E. coli using an AT-rich 5′ gene tag previously reported to enhance cell-free expression yields. The BL21(DE3) strain was not suitable for expression of AR_TM1–3 whether untagged or with any of the four tags, as pronounced cell death was observed after induction, whereas all four AT-rich tagged AR_TM1–3 constructs revealed AR_TM1–3 expression in the C41(DE3) strain, though target protein expression was not observed without AT-rich tags. The mRNA binding cleft of the ribosome can only accommodate single-stranded mRNA (Mustoe et al. 2018). Thus, translation initiation requires unfolding of the TIR, so AT-rich codon in TIR increases translation efficiency by decreasing the energy penalty imposed on mRNA to be unfolded (Haberstock et al. 2012; Mustoe et al. 2018).
Zhao et al. (2019) examined the effects of the hydrophobicity and net charge of self-assembling amphipathic peptides [SAP: (AEAEAKAK)2] on the protein expression. A variant of SAP that Lys is replaced with His revealed a multifunctional tag with the ability to benefit the expression, purification, thermostability, and activity of recombinant proteins (Zhao et al. 2018). The changes in the hydrophobicity of SAP have a tendency to aggregate the protein into inclusion bodies. The SAP variants with Ile and Leu in place of Ala induced the fused GFP to form inclusion bodies, whereas the SAP variants with Phe, Pro, and Gly in place of Ala reduced the soluble expression levels of the fused GFP compared with that of SAP. Moreover, the positive net charge of SAP was more efficient for protein expression than with a negative charge, although the type of hydrophilic residues has little effect on the efficiency of the SAPs (Zhao et al. 2019). In addition, other variants of SAP, including 18A (EWLKAFYEKVLEKLKELF), ELK16 (LELELKLKLELELKLK), L6KD (LLLLLLKD), and GFIL8 (GFILGFIL), induce the formation of active protein aggregates that retain comparable specific activities with the native counterparts in E. coli when these tags are terminally fused to target proteins (García-Fruitós et al. 2005; Wu et al. 2011; Xing et al. 2011). They were developed as a cleavable self-aggregating tag by intein-mediated cleavage (Xing et al. 2011). Compared with the classical process for isolation and refolding of the inclusion body, SAP tags induced aggregated proteins to take advantage of the self-cleavage in the aggregate state and eliminate the refolding process, thus shortening the downstream process of protein production (Lin et al. 2015). Because these SAP tags are aggregation-prone tags, they can lower the soluble expression level of a passenger protein compared with that without the tag, so these systems are beneficial for the expression and recovery of proteins or peptides that are toxic to E. coli and are sensitive to proteolytic degradation.
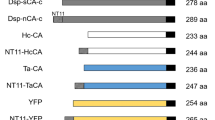
The NT11 tag recently discovered has been exploited as an expression-enhancing tag (Nguyen et al. 2019b). NT11 is the first 11 amino acid residues of a duplicated carbonic anhydrase (dCA) from Dunaliella species. dCA is comprised of two CA domains that have structural similarity with alpha-type CA. When separately expressed in E. coli as N-terminal and C-terminal domains, the former domain (dCA-N) showed a very high soluble expression level without CA activity whereas the latter domain (dCA-C) displayed a very low expression level with CA activity (Ki et al. 2012). In some native multidomain proteins, the N-terminal domain acts to enhance solubility for their downstream domains (Kim et al. 2007) and the N-terminal residues play key roles in stability, solubility, or function of the protein (Gaudry et al. 2012). In addition, the folding free energy in the region between − 25 and +35 of the mRNA was reported to have the greatest impact on prokaryotic translation efficiency (Seo et al. 2013). Thus, the first 12 amino acid residues may be important for protein expression. Subsequently, the first 11 amino acid sequences (NT11;VSEPHDYNYEK), except for the start codon methionine of dCA-N, were selected and the effect of NT11 on the expression of dCA-NT11ΔC, whose first 10 amino acid residues had been replaced with NT11, was examined. As a result, the solubility of dCA-NT11ΔC was increased more than 2-fold that of dCA-C (Ki et al. 2016). The ability of NT11 to improve protein expression was also confirmed through fusion with other proteins (Nguyen et al. 2019b). NT11 was fused to the N-terminus of a protein with a low soluble expression level, including Ta-CA and yellow fluorescence protein (YFP), and a protein with insoluble expression, including Hc-CA and dCA-ΔC. The NT11 increased total protein expression in E. coli by 1.1-, 6.9-, and 7.6-fold for Hc-CA, Ta-CA, and YFP, respectively. Moreover, NT11 enhanced the soluble expression of Hc-CA, Ta-CA, and YFP by 1.7-, 5.0-, and 3.2-fold, respectively. The dCA-ΔC was expressed as an insoluble form, but NT11 enhanced the solubility by more than 10% of the total expression. Furthermore, NT11 increased the protein yield without interfering with protein function and structure, indicating that tag cleavage is not required. The mechanism by which NT11 enhances protein expression is unclear. However, as mentioned above, the first N-terminal amino acid residues are on the TIR, so it is worthwhile to calculate the folding free energy of the nucleotide sequences of − 25 to + 35 of proteins with or without NT11. Based on the calculated ΔGUTR value using the UTR Designer quantitative prediction method for proper codon optimization around the 5′- proximal coding sequence (Seo et al. 2013), increasingly higher negative values of ΔGUTR are associated with increasingly higher expression levels. However, this prediction does not guarantee soluble expression. In the view of translation efficiency, NT11 produced a beneficial effect for translation by shifting ΔGUTR values of − 5.69 for dCA-ΔC, − 7.44 for Hc-CA, − 4.39 for Ta-CA, and − 4.34 for YFP into − 8.89 for all. It is not surprising that NT11 displayed a minimal effect on the expression for Hc-CA because it already has a higher negative ΔGUTR value, indicating it has potential for high expression in E. coli, even though it is insoluble (Nguyen et al. 2019b). Commonly used fusion tags generally reveal high negative ΔGUTR values, between − 12 and − 7, whether they induce soluble or insoluble expression. However, GST and sfGFP represent − 3.64 and − 4.32, respectively, suggesting that other factors may act on protein expression. NT11 could be a promising method for improving protein expression in E. coli, although the effects of NT11 as a fusion tag on more proteins should be required.
Outlook
Generally, commercially available protein fusion tags have a pI value of 4 to 6. Proteins with these pI values display a negative charge at neutral pH. CBD, on the other hand, has a high pI value (9.16), manifesting a positive charge at neutral pH. Thus, these fusion tags are hydrophilic under physiological conditions. This feature may help solubilize a passenger protein (Wilkinson and Harrison 1991). More than anything, the chaperone-like activity of tag proteins aids the correct folding of a passenger protein, leading to soluble expression of it, which is prominent in larger protein tags such as NusA, MBP, and PDI (Costa et al. 2014; Nguyen et al. 2017). The SUMO tag has been the most widely used in recent years. Although small in size, this tag aids the correct folding of the target protein by chaperone-like functions as well (Chen et al. 2015). In addition, most peptide tags tend to promote insoluble expression, suggesting that peptides are not sufficient to play a chaperone-like function to aid proper folding for the soluble expression of proteins (Hirose et al. 2011).
The protein or peptide fusion tag can increase the expression of the protein as a soluble or insoluble form depending on a passenger protein. In this regard, it is believed that similar net charges between the carrier and passenger will induce repulsion between the two, lowering the protein folding rate, thereby leading to proper folding for soluble expression (Paraskevopoulou and Falcone 2018). From these findings, an appropriate tag can regulate the expression of a passenger protein. Especially, the advantage of the peptide tag is that it can be designed to control the expression of the passenger protein (Hirose et al. 2011; Pandey et al. 2014). The hydrophobicity, hydrophilicity, and net charge of a fusion tag can be tuned to express a target protein as either a soluble or insoluble form (Zhao et al. 2019). As mentioned above, though most peptide tags tend to promote insoluble expression, there is room for development as a solubility-enhancing peptide tag because the N-terminus is important for the transcription and solubility of proteins. The secondary structure of the TIR is very important for the efficiency of transcription initiation (Mustoe et al. 2018). Therefore, optimizing the TIR sequence around the start codon to which the ribosome binds is also a method for promoting expression efficiency (Seo et al. 2013; Nguyen et al. 2019b). Lowering the folding free energy of TIR on mRNA sequences to have an unstable structure (such as AT-rich codon optimization) may promote translation efficiency, resulting in an increase in expression level. Although promoting translation efficiency does not mean a soluble expression, an increased expression level can proportionally increase the rate of soluble expression (Nguyen et al. 2019b). In contrast, 3′-untranslated region (3′UTR) engineering can be used to improve soluble expression by decreasing the stability of mRNA (López-Garrido et al. 2014; Song et al. 2016), which in turn leads to low local concentrations of polypeptide and thereby the possibility of proper folding of protein may be increased (Song et al. 2016). However, in order to produce a heterologous protein that is difficult to express in E. coli, such as eukaryotic membrane-binding proteins and IDPs, promoting translation efficiency by controlling TIR region may be a feasible strategy for expressing such intractable proteins (Pandey et al. 2014).
Since the peptide fusion tag accounts for a small part over the larger passenger protein, the peptide fusion tag does not affect the structure or activity of the expressed passenger protein, and thus can be used without removing the tag in cases of protein structure analysis or industrial enzyme use. However, it is desirable that expression-promoting peptide tags are removed when applied to therapeutic proteins, given the fact that a variety of commercially available peptide affinity tags are epitopes (Randolph 2012).
Membrane-binding proteins can be expressed using the Mistic tag that translocates the passenger protein to the membrane during its expression (Kefala et al. 2007; Lee et al. 2015b). However, most of the membrane-binding proteins have been expressed as inclusion bodies due to their hydrophobicity, even if they are fused to the MBP or GST tag (McCusker et al. 2008). Usually, the aggregation-prone tags have been used for obtaining a high-level yield of protein (Caroccia et al. 2011; Potetinova et al. 2012). However, refolding of these proteins is a very intractable challenge. Therefore, it is favorable to induce soluble expression if possible.
In contrast, the insoluble expression is often preferred for some proteins that are vulnerable to proteolysis and do not require tertiary structure or have a loose structure. Inclusion body expression can increase protein yield and purification efficiency so that the protein obtained as the inclusion body can be easily recovered by centrifugation from cell lysates followed by solubilization under mild reconstitution conditions (Wei et al. 2018; Graether 2019). In addition, active protein aggregation tags have been recently developed, allowing simple recovery of a large amount of protein as a manageable aggregate (Zhou et al. 2012).
In E. coli, most proteins are expressed in the cytoplasm due to the lack of an extracellular secretion system. However, Ffu and sfGFP fusion tags will allow extracellular secretion of the passenger protein (Cheng et al. 2017; Zhang et al. 2017). A commercialized periplasmic-translocating tag that includes pelB and ompA can be used for periplasmic expression by N-terminal tagging (Sockolosky and Szoka 2013).
Low-temperature culture is almost always required, even when using the appropriate fusion tags to obtain soluble proteins. It is also important to select the appropriate E. coli strain. A variety of E. coli strains to meet various requirements have been developed. These include E. coli capable of forming disulfide bonds in the cytoplasm, good growth at low temperatures, expression of genes with rare codons of advanced organisms, and E. coli strains with poor protease activity and no lipopolysaccharide. In addition, E. coli is inexpensive, capable of rapid growth, and easy to handle and control. These attributes make E. coli the preferred host for heterologous protein production (Rosano and Ceccarelli 2014).
Conclusions
E. coli is the most powerful host for heterologous protein expression, and various methods and techniques have been developed for the expression and production of proteins of interest. In particular, the use of proteins and peptides as fusion tags is the surest method of increasing the expression of the desired protein. Knowing the physicochemical properties, including the pI, net charge, and GRAVY values of the protein of interest, is helpful in selecting or designing the appropriate fusion tag. By utilizing various bioinformatics tools, fusion tags can be designed at the mRNA level to enhance the translation efficiency toward downstream protein by optimizing the TIR. However, improving expression efficiency and increasing soluble expression are different issues. For the latter, it is important to induce proper protein folding, by lowering the rate of translation and folding in the cytoplasm, or using tags to help disulfide bonds. The aggregation-prone tags can be considered for expression of the protein of interest, including AMPs, IDPs, and GPCRs. Given the recent tendency to use multi-tags to increase purification efficiency, proper selection and arrangement of affinity tags in conjunction with expression enhancing tags are required. Since the tag may need to be removed, a protease recognition site is additionally inserted between the tag and the target protein so that the tag can be removed if necessary. Peptide tags may not be sufficient to perform chaperone-like functions that aid proper folding of a protein compared to larger protein tags, but there is room for development as solubility tags, because the N-terminus is important for transcription and solubility of proteins.
References
Achmuller C, Kaar W, Ahrer K, Wechner P, Hahn R, Werther F, Schmidinger H, Cserjan-Puschmann M, Clementschitsch F, Striedner G, Bayer K, Jungbauer A, Auer B (2007) N(pro) fusion technology to produce proteins with authentic N termini in E. coli. Nat Methods 4:1037–1043. https://doi.org/10.1038/nmeth1116
Ahmad T, Singh RS, Gupta G, Sharma A, Kaur B (2019) Chapter 15 Metagenomics in the search for industrial enzymes. Adv Enzyme Technol:419–451. https://doi.org/10.1016/b978-0-444-64114-4.00015-7
Arena TA, Chou B, Harms PD, Wong AW (2019) An anti-apoptotic HEK293 cell line provides a robust and high titer platform for transient protein expression in bioreactors. mAbs 11:977–986. https://doi.org/10.1080/19420862.2019.1598230
Assumpcao MC, de Souza EM, Yates MG, de Oliveira PF, Benelli EM (2007) Purification and characterisation of Azospirillum brasilense N-truncated NtrX protein. Protein Expr Purif 53:302–308. https://doi.org/10.1016/j.pep.2007.01.003
Baeshen NA, Baeshen MN, Sheikh A, Bora RS, Ahmed MMM, Ramadan HAI, Saini KS, Redwan EM (2014) Cell factories for insulin production. Microb Cell Factories 13:141. https://doi.org/10.1186/s12934-014-0141-0
Bañuelos-Hernández B (2017) Algevir: an expression system for microalgae based on viral vectors. Front Microbiol 8:1100. https://doi.org/10.3389/fmicb.2017.01100
Bernier SC, Cantin L, Salesse C (2018) Systematic analysis of the expression, solubility and purification of a passenger protein in fusion with different tags. Protein Expr Purif 152:92–106. https://doi.org/10.1016/j.pep.2018.07.007
Bogomolovas J, Simon B, Sattler M, Stier G (2009) Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr Purif 64:16–23. https://doi.org/10.1016/j.pep.2008.10.003
Butt TR, Edavettal SC, Hall JP, Mattern MR (2005) SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif 43:1–9. https://doi.org/10.1016/j.pep.2005.03.016
Byrne LJ, Sidhu A, Wallis AK, Ruddock LW, Freedman RB, Howard MJ, Williamson RA (2009) Mapping of the ligand-binding site on the b' domain of human PDI: interaction with peptide ligands and the x-linker region. Biochem J 423:209–217. https://doi.org/10.1042/BJ20090565
Caroccia KE, Estephan R, Cohen LS, Arshava B, Hauser M, Zerbe O, Becker JM, Naider F (2011) Expression and biophysical analysis of a triple-transmembrane domain-containing fragment from a yeast G protein-coupled receptor. Biopolymers 96:757–771. https://doi.org/10.1002/bip.21614
Carson M, Johnson DH, McDonald H, Brouillette C, Delucas LJ (2007) His-tag impact on structure. Acta Crystall Sec D, Biol Crystallogr 63:295–301. https://doi.org/10.1107/S0907444906052024
Cereghino JL, Cregg JM (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev 24:45–66. https://doi.org/10.1111/j.1574-6976.2000.tb00532.x
Chen X, Shi J, Chen R, Wen Y, Shi Y, Zhu Z, Guo S, Li L (2015) Molecular chaperones (TrxA, SUMO, Intein, and GST) mediating expression, purification, and antimicrobial activity assays of plectasin in Escherichia coli. Biotechnol Appl Biochem 62:606–614. https://doi.org/10.1002/bab.1303
Chen K-J, Wu Y-T, Lee C-K (2019) Cellulose binding domain fusion enhanced soluble expression of fructosyl peptide oxidase and its simultaneous purification and immobilization. Int J Biol Macromol 133:980–986. https://doi.org/10.1016/j.ijbiomac
Cheng C, Wu S, Cui L, Wu Y, Jiang T, He B (2017) A novel Ffu fusion system for secretory expression of heterologous proteins in Escherichia coli. Microb Cell Factories 16:231. https://doi.org/10.1186/s12934-017-0845-z
Chu J, Wu X, Wu B, Wang R, He B (2014) Characteristics of an organic solvent-tolerant β-fructofuranosidase from Arthrobacter arilaitensis NJEM01 and efficient synthesis of prebiotic kestose. J Agric Food Chem 62:5408–5411. https://doi.org/10.1021/jf5020523
Costa SJ, Almeida A, Castro A, Domingues L, Besir H (2012) The novel Fh8 and H fusion partners for soluble protein expression in Escherichia coli: a comparison with the traditional gene fusion technology. Appl Microbiol Biotechnol 97:6779–6791. https://doi.org/10.1007/s00253-012-4559-1
Costa SJ, Almeida A, Castro A, Domingues L, Besir H (2013) The novel Fh8 and H fusion partners for soluble protein expression in Escherichia coli: a comparison with the traditional gene fusion technology. Appl Microbiol Biotechnol 97:6779–6791. https://doi.org/10.1007/s00253-012-4559-1
Costa S, Almeida A, Castro A, Domingues L (2014) Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front Microbiol 5:63. https://doi.org/10.3389/fmicb.2014.00063
de Marco A (2009) Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Factories 8:26. https://doi.org/10.1186/1475-2859-8-26
Do BH, Ryu HB, Hoang P, Koo BK, Choe H (2014) Soluble prokaryotic overexpression and purification of bioactive human granulocyte colony-stimulating factor by maltose binding protein and protein disulfide isomerase. PLoS One 9:e89906. https://doi.org/10.1371/journal.pone.0089906
Douette P, Navet R, Gerkens P, Galleni M, Lévy D, Sluse FE (2005) Escherichia coli fusion carrier proteins act as solubilizing agents for recombinant uncoupling protein 1 through interactions with GroEL. Biochem Biophys Res Commun 333:686–693. https://doi.org/10.1016/j.bbrc.2005.05.164
Fahad S, Khan FA, Pandupuspitasari NS, Ahmed MM, Liao YC, Waheed MT, Sameeullah M, Darkhshan HS, Saud S, Hassan S, Jan A, Jan MT, Wu C, Chun MX, Huang J (2015) Recent developments in therapeutic protein expression technologies in plants. Biotechnol Lett 37:265–279. https://doi.org/10.1007/s10529-014-1699-7
Fossum S, Crooke E, Skarstad K (2007) Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli. EMBO J 26:4514–4522. https://doi.org/10.1038/sj.emboj.7601871
Furukawa H, Haga T (2000) Expression of functional M2 muscarinic acetylcholine receptor in Escherichia coli. J Biochem 127:151–161. https://doi.org/10.1093/oxfordjournals.jbchem.a022577
Galligan JJ, Petersen DR (2012) The human protein disulfide isomerase gene family. Hum Genom 6:6. https://doi.org/10.1186/1479-7364-6-6
Gangl D, Zedler JAZ, Rajakumar PD, Martinez EMR, Riseley A, Włodarczyk A, Purton S, Sakuragi Y, Howe CJ, Jensen PE, Robinson C (2015) Biotechnological exploitation of microalgae. J Exp Bot 66:6975–6990. https://doi.org/10.1093/jxb/erv426
García-Fruitós E, González-Montalbán N, Morell M, Vera A, Ferraz R, Arís A, Ventura S, Villaverde A (2005) Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb Cell Factories 4:27. https://doi.org/10.1186/1475-2859-4-27
Gaudry A, Lorber B, Neuenfeldt A, Sauter C, Florentz C, Sissler M (2012) Re-designed N-terminus enhances expression, solubility and crystallizability of mitochondrial protein. Prot Eng Des Sel 25:473–481. https://doi.org/10.1093/protein/gzs046
Geisse S, Fux C (2009) Chapter 15 Recombinant protein production by transient gene transfer into mammalian cells. Methods Enzymol 463:223–238. https://doi.org/10.1016/s0076-6879(09)63015-9
Goda N, Matsuo N, Tenno T, Ishino S, Ishino Y, Fukuchi S, Ota M, Hiroaki H (2015) An optimized Npro-based method for the expression and purification of intrinsically disordered proteins for an NMR study. Intrinsic Disord Proteins 3:e1011004. https://doi.org/10.1080/21690707.2015.1011004
Graether SP (2019) Troubleshooting guide to expressing intrinsically disordered proteins for use in NMR experiments. Front Mol Biosci 5:118. https://doi.org/10.3389/fmolb.2018.00118
Grunina TM, Demidenko AV, Lyaschuk AM, Poponova MS, Galushkina ZM, Soboleva LA, Cherepushkin SA, Polyakov NB, Grumov DA, Solovyev AI, Zhukhovitsky VG, Boksha IS, Subbotina ME, Gromov AV, Lunin VG, Karyagina AS (2017) Recombinant human erythropoietin with additional processable protein domains: purification of protein synthesized in Escherichia coli heterologous expression system. Biochemistry (Mosc) 82:1285–1294. https://doi.org/10.1134/S0006297917110062
Guo Y, Yu M, Jing N, Zhang S (2018) Production of soluble bioactive mouse leukemia inhibitory factor from Escherichia coli using MBP tag. Protein Expr Purif 150:86–91. https://doi.org/10.1016/j.pep.2018.05.006
Haberstock S, Roos C, Hoevels Y, Dötsch V, Schnapp G, Pautsch A, Bernhard F (2012) A systematic approach to increase the efficiency of membrane protein production in cell-free expression systems. Protein Expr Purif 82:308–316. https://doi.org/10.1016/j.pep.2012.01.018
Hage N, Renshaw JG, Winkler GS, Gellert P, Stolnik S, Falcone FH (2015) Improved expression and purification of the Helicobacter pylori adhesin BabA through the incorporation of a hexa-lysine tag. Protein Expr Purif 106:25–30. https://doi.org/10.1016/j.pep.2014.10.009
Han Y, Guo W, Su B, Guo Y, Wang J, Chu B, Yang G (2018) High-level expression of soluble recombinant proteins in Escherichia coli using an HE-maltotriose-binding protein fusion tag. Protein Expr Purif 142:25–31. https://doi.org/10.1016/j.pep.2017.09.013
Harper S, Speicher DW (2011) Purification of proteins fused to glutathione S-transferase. Methods Mol Biol 681:259–280. https://doi.org/10.1007/978-1-60761-913-0_14
Hirose S, Kawamura Y, Mori M, Yokota K, Noguchi T, Goshima N (2011) Development and evaluation of data-driven designed tags (DDTs) for controlling protein solubility. New Biotechnol 28:225–231. https://doi.org/10.1016/j.nbt.2010.08.012
Hwang PM, Pan JS, Sykes BD (2012) A PagP fusion protein system for the expression of intrinsically disordered proteins in Escherichia coli. Protein Expr Purif 85:148–151. https://doi.org/10.1016/j.pep.2012.07.007
Hwang PM, Pan JS, Sykes BD (2014) Targeted expression, purification, and cleavage of fusion proteins from inclusion bodies in Escherichia coli. FEBS Lett 588:247–252. https://doi.org/10.1016/j.febslet.2013.09.028
Isalan M, Chang KY, Yang J-R (2013) Analysis and prediction of highly effective antiviral peptides based on random forests. PLoS One 8:e70166. https://doi.org/10.1371/journal.pone.0070166
Islam MM, Nakamura S, Noguchi K, Yohda M, Kidokoro S-i, Kuroda Y (2015) Analysis and control of protein crystallization using short peptide tags that change solubility without affecting structure, thermal stability, and function. Cryst Growth Des 15:2703–2711. https://doi.org/10.1021/acs.cgd.5b00010
Juturu V, Wu JC (2018) Heterologous protein expression in Pichia pastoris: latest research progress and applications. ChemBioChem 19:7–21. https://doi.org/10.1002/cbic.201700460
Kato A, Maki K, Ebina T, Kuwajima K, Soda K, Kuroda Y (2007) Mutational analysis of protein solubility enhancement using short peptide tags. Biopolymers 85:12–18. https://doi.org/10.1002/bip.20596
Kefala G, Kwiatkowski W, Esquivies L, Maslennikov I, Choe S (2007) Application of Mistic to improving the expression and membrane integration of histidine kinase receptors from Escherichia coli. J Struct Funct Genom 8:167–172. https://doi.org/10.1007/s10969-007-9033-4
Khalilzadeh R, Shojaosadati SA, Maghsoudi N, Mohammadian-Mosaabadi J, Mohammadi MR, Bahrami A, Maleksabet N, Nassiri-Khalilli MA, Ebrahimi M, Naderimanesh H (2004) Process development for production of recombinant human interferon expressed in Escherichia coli. J Ind Microbiol Biotechnol 31:63–69. https://doi.org/10.1007/s10295-004-0117-x
Khan S, Ullah MW, Siddique R, Nabi G, Manan S, Yousaf M, Hou H (2016) Role of recombinant DNA technology to improve life. Int J Genom 2016:1–14. https://doi.org/10.1155/2016/2405954
Ki M-R, Kanth BK, Min KH, Lee J, Pack SP (2012) Increased expression level and catalytic activity of internally-duplicated carbonic anhydrase from Dunaliella species by reconstitution of two separate domains. Process Biochem 47:1423–1427. https://doi.org/10.1016/j.procbio.2012.05.005
Ki M-R, Nguyen TKM, Kim SH, Kwon I, Pack SP (2016) Chimeric protein of internally duplicated α-type carbonic anhydrase from Dunaliella species for improved expression and CO2 sequestration. Process Biochem 51:1222–1229. https://doi.org/10.1016/j.procbio.2016.05.013
Kim JS, Raines RT (1993) Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci 2:348–356. https://doi.org/10.1002/pro.5560020307
Kim CW, Han KS, Ryu KS, Kim BH, Kim KH, Choi SI, Seong BL (2007) N-terminal domains of native multidomain proteins have the potential to assist de novo folding of their downstream domains in vivo by acting as solubility enhancers. Protein Sci 16:635–643. https://doi.org/10.1110/ps.062330907
Kim S-G, Min W-K, Rho Y-T, Seo J-H (2012) Electrostatic interaction-induced inclusion body formation of glucagon-like peptide-1 fused with ubiquitin and cationic tag. Protein Expr Purif 84:38–46. https://doi.org/10.1016/j.pep.2012.04.010
Kim DS, Kim SW, Song JM, Kim SY, Kwon KC (2019) A new prokaryotic expression vector for the expression of antimicrobial peptide abaecin using SUMO fusion tag. BMC Biotechnol 19:13. https://doi.org/10.1186/s12896-019-0506-x
Klocke M, Mundt K, Idler F, Jung S, Backhausen JE (2005) Heterologous expression of enterocin A, a bacteriocin from Enterococcus faecium, fused to a cellulose-binding domain in Escherichia coli results in a functional protein with inhibitory activity against Listeria. Appl Microbiol Biotechnol 67:532–538. https://doi.org/10.1007/s00253-004-1838-5
Knorre WA, Deckwer WD, Korz D, Pohl HD, Riesenberg D, Ross A, Sanders E, Schulz V (1991) High cell density fermentation of recombinant Escherichia coli with computer-controlled optimal growth rate. Ann N Y Acad Sci 646(1 recombinant D):300–306. https://doi.org/10.1111/j.1749-6632.1991.tb18592.x
Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR (2009) Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7:206–214. https://doi.org/10.1038/nrmicro2069
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Lagassé HAD, Alexaki A, Simhadri VL, Katagiri NH, Jankowski W, Sauna ZE, Kimchi-Sarfaty C (2017) Recent advances in (therapeutic protein) drug development. F1000Res 6:113. https://doi.org/10.12688/f1000research.9970.1
Lawrence MS, Phillips KJ, Liu DR (2007) Supercharging proteins can impart unusual resilience. J Am Chem Soc 129:10110–10112. https://doi.org/10.1021/ja071641y
Le LTM, Nyengaard JR, Golas MM, Sander B (2018) Vectors for expression of signal peptide-dependent proteins in Baculovirus/insect cell systems and their application to expression and purification of the high-affinity immunoglobulin gamma Fc receptor I in complex with its gamma chain. Mol Biotechnol 60:31–40. https://doi.org/10.1007/s12033-017-0041-8
Lee JH, Kim JH, Hwang SW, Lee WJ, Yoon HK, Lee HS, Hong SS (2000) High-level expression of antimicrobial peptide mediated by a fusion partner reinforcing formation of inclusion bodies. Biochem Biophys Res Commun 277:575–580. https://doi.org/10.1006/bbrc.2000.3712
Lee JS, Grav LM, Lewis NE, Faustrup Kildegaard H (2015a) CRISPR/Cas9-mediated genome engineering of CHO cell factories: application and perspectives. Biotechnol J 10:979–994. https://doi.org/10.1002/biot.201500082
Lee KA, Lee S-S, Kim SY, Choi AR, Lee J-H, Jung K-H (2015b) Mistic-fused expression of algal rhodopsins in Escherichia coli and its photochemical properties. Biochim Biophys Acta (BBA) 1850:1694–1703. https://doi.org/10.1016/j.bbagen.2015.04.002
Levy I, Shoseyov O (2002) Cellulose-binding domains. Biotechnol Adv 20:191–213. https://doi.org/10.1016/s0734-9750(02)00006-x
Li Y (2011) Recombinant production of antimicrobial peptides in Escherichia coli: a review. Protein Expr Purif 80:260–267. https://doi.org/10.1016/j.pep.2011.08.001
Li JF, Zhang J, Zhang Z, Kang CT, Zhang SQ (2011) SUMO mediating fusion expression of antimicrobial peptide CM4 from two joined genes in Escherichia coli. Curr Microbiol 62:296–300. https://doi.org/10.1007/s00284-010-9705-3
Lin Z, Zhao Q, Xing L, Zhou B, Wang X (2015) Aggregating tags for column-free protein purification. Biotechnol J 10:1877–1886. https://doi.org/10.1002/biot.201500299
Liu M, Wang B, Wang F, Yang Z, Gao D, Zhang C, Ma L, Yu X (2019) Soluble expression of single-chain variable fragment (scFv) in Escherichia coli using superfolder green fluorescent protein as fusion partner. Appl Microbiol Biotechnol 103:6071–6079. https://doi.org/10.1007/s00253-019-09925-6
Locatelli-Hoops SC, Gorshkova I, Gawrisch K, Yeliseev AA (2013) Expression, surface immobilization, and characterization of functional recombinant cannabinoid receptor CB2. Biochim Biophys Acta (BBA) 1834:2045–2056. https://doi.org/10.1016/j.bbapap.2013.06.003
López-Garrido J, Puerta-Fernández E, Casadesús J (2014) A eukaryotic-like 3′ untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res 42:5894–5906. https://doi.org/10.1093/nar/gku222
Lyukmanova EN, Shenkarev ZO, Khabibullina NF, Kulbatskiy DS, Shulepko MA, Petrovskaya LE, Arseniev AS, Dolgikh DA, Kirpichnikov MP (2012) N-terminal fusion tags for effective production of g-protein-coupled receptors in bacterial cell-free systems. Acta Nat 4:58–64
Ma RJ, Wang YH, Liu L, Bai LL, Ban R (2018) Production enhancement of the extracellular lipase LipA in Bacillus subtilis: effects of expression system and Sec pathway components. Protein Expr Purif 142:81–87. https://doi.org/10.1016/j.pep.2017.09.011
Madzak C (2015) Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl Microbiol Biotechnol 99:4559–4577. https://doi.org/10.1007/s00253-015-6624-z
Malhotra A (2009) Chapter 16 Tagging for protein expression. Methods Enzymol 463:239–258. https://doi.org/10.1016/s0076-6879(09)63016-0
Mamat U, Wilke K, Bramhill D, Schromm AB, Lindner B, Kohl TA, Corchero JL, Villaverde A, Schaffer L, Head SR, Souvignier C, Meredith TC, Woodard RW (2015) Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb Cell Factories 14:57. https://doi.org/10.1186/s12934-015-0241-5
Marino J, Bordag N, Keller S, Zerbe O (2015) Mistic’s membrane association and its assistance in overexpression of a human GPCR are independent processes. Protein Sci 24:38–48. https://doi.org/10.1002/pro.2582
Mathiesen G, Sveen A, Piard JC, Axelsson L, Eijsink VGH (2008) Heterologous protein secretion by Lactobacillus plantarum using homologous signal peptides. J Appl Microbiol 105:215–226. https://doi.org/10.1111/j.1365-2672.2008.03734.x
McCusker EC, Bane SE, O'Malley MA, Robinson AS (2008) Heterologous GPCR expression: a bottleneck to obtaining crystal structures. Biotechnol Prog 23:540–547. https://doi.org/10.1021/bp060349b
Mo Q, Fu A, Lin Z, Wang W, Gong L, Li W (2018) Expression and purification of antimicrobial peptide AP2 using SUMO fusion partner technology in Escherichia coli. Lett Appl Microbiol 67:606–613. https://doi.org/10.1111/lam.13079
Murashima K, Kosugi A, Doi RH (2003) Solubilization of cellulosomal cellulases by fusion with cellulose-binding domain of noncellulosomal cellulase engd from Clostridium cellulovorans. Proteins 50:620–628. https://doi.org/10.1002/prot.10298
Mustoe AM, Corley M, Laederach A, Weeks KM (2018) Messenger RNA structure regulates translation initiation: a mechanism exploited from bacteria to humans. Biochemistry 57:3537–3539. https://doi.org/10.1021/acs.biochem.8b00395
Nautiyal K, Kuroda Y (2018) A SEP tag enhances the expression, solubility and yield of recombinant TEV protease without altering its activity. New Biotechnol 42:77–84. https://doi.org/10.1016/j.nbt.2018.02.006
Needle D, Waugh DS (2014) Rescuing aggregation-prone proteins in Escherichia coli with a dual His(6)-MBP tag. Methods Mol Biol 1177:81–94. https://doi.org/10.1007/978-1-4939-1034-2_7
Nevalainen KM, Te'o VS, Bergquist PL (2005) Heterologous protein expression in filamentous fungi. Trends Biotechnol 23:468–474. https://doi.org/10.1016/j.tibtech.2005.06.002
Nguyen MT, Koo BK, Thi Vu TT, Song JA, Chong SH, Jeong B, Ryu HB, Moh SH, Choe H (2014) Prokaryotic soluble overexpression and purification of bioactive human growth hormone by fusion to thioredoxin, maltose binding protein, and protein disulfide isomerase. PLoS One 9:e89038. https://doi.org/10.1371/journal.pone.0089038
Nguyen MT, Krupa M, Koo BK, Song JA, Vu TT, Do BH, Nguyen AN, Seo T, Yoo J, Jeong B, Jin J, Lee KJ, Oh HB, Choe H (2016) Prokaryotic soluble overexpression and purification of human VEGF165 by fusion to a maltose binding protein tag. PLoS One 11:e0156296. https://doi.org/10.1371/journal.pone.0156296
Nguyen AN, Song J-A, Nguyen MT, Do BH, Kwon GG, Park SS, Yoo J, Jang J, Jin J, Osborn MJ, Jang YJ, Thi Vu TT, Oh H-B, Choe H (2017) Prokaryotic soluble expression and purification of bioactive human fibroblast growth factor 21 using maltose-binding protein. Sci Rep 7:16139. https://doi.org/10.1038/s41598-017-16167-x
Nguyen MT, Prima MJ, Song JA, Kim J, Do BH, Yoo J, Park S, Jang J, Lee S, Lee E, Novais MP, Seo HB, Lee SY, Cho ML, Kim CJ, Jang YJ, Choe H (2019a) Prokaryotic soluble overexpression and purification of oncostatin M using a fusion approach and genetically engineered E. coli strains. Sci Rep 9:13706. https://doi.org/10.1038/s41598-019-50110-6
Nguyen TKM, Ki MR, Son RG, Pack SP (2019b) The NT11, a novel fusion tag for enhancing protein expression in Escherichia coli. Appl Microbiol Biotechnol 103:2205–2216. https://doi.org/10.1007/s00253-018-09595-w
Pandey A, Sarker M, Liu X-Q, Rainey JK (2014) Small expression tags enhance bacterial expression of the first three transmembrane segments of the apelin receptor. Biochem Cell Biol 92:269–278. https://doi.org/10.1139/bcb-2014-0009
Paraskevopoulou V, Falcone F (2018) Polyionic tags as enhancers of protein solubility in recombinant protein expression. Microorganisms 6:47. https://doi.org/10.3390/microorganisms6020047
Park SH, Mrse AA, Nevzorov AA, Mesleh MF, Oblatt-Montal M, Montal M, Opella SJ (2003) Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J Mol Biol 333:409–424. https://doi.org/10.1016/j.jmb.2003.08.048
Park SY, Park SH, Choi SK (2012) Active inclusion body formation using Paenibacillus polymyxa PoxB as a fusion partner in Escherichia coli. Anal Biochem 426:63–65. https://doi.org/10.1016/j.ab.2012.04.002
Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. https://doi.org/10.1038/nbt1172
Potetinova Z, Tantry S, Cohen LS, Caroccia KE, Arshava B, Becker JM, Naider F (2012) Large multiple transmembrane domain fragments of a G protein-coupled receptor: biosynthesis, purification, and biophysical studies. Biopolymers 98:485–500. https://doi.org/10.1002/bip.22122
Priyadarshini P, Singh B (2019) Chapter 15 Computational resources and techniques in enzyme research. Adv Enz Technol:453–468. https://doi.org/10.1016/b978-0-444-64114-4.00016-9
Randolph TW (2012) The two faces of His-tag: immune response versus ease of protein purification. Biotechnol J 7:18–19. https://doi.org/10.1002/biot.201100459
Redwan EM, Baeshen NA, Saini KS, Ramadan HAI, Ahmed MMM, Bora RS, Al-Hejin AM, Baeshen MN (2015) Production of biopharmaceuticals in E. coli: current scenario and future perspectives. J Microbiol Biotechnol 25:953–962. https://doi.org/10.4014/jmb.1412.12079
Rinas U, Garcia-Fruitós E, Corchero JL, Vázquez E, Seras-Franzoso J, Villaverde A (2017) Bacterial inclusion bodies: discovering their better half. Trends Biochem Sci 42:726–737. https://doi.org/10.1016/j.tibs.2017.01.005
Rojas-Lopez M, Zorgani MA, Kelley LA, Bailly X, Kajava AV, Henderson IR, Polticelli F, Pizza M, Rosini R, Desvaux M (2018) Identification of the autochaperone domain in the type Va secretion system (T5aSS): prevalent feature of autotransporters with a β-helical passenger. Front Microbiol 8:2607. https://doi.org/10.3389/fmicb.2017.02607
Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S (2005) NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science 307:1317–1321. https://doi.org/10.1126/science.1106392
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. https://doi.org/10.3389/fmicb.2014.00172
Sachdev D, Chirgwin JM (2000) Fusions to maltose-binding protein: control of folding and solubility in protein purification. Methods Enzymol 326:312–321. https://doi.org/10.1016/s0076-6879(00)26062-x
Saraswat M, Musante L, Ravidá A, Shortt B, Byrne B, Holthofer H (2013) Preparative purification of recombinant proteins: current status and future trends. Biomed Res Int 2013:1–18. https://doi.org/10.1155/2013/312709
Savitsky P, Bray J, Cooper CDO, Marsden BD, Mahajan P, Burgess-Brown NA, Gileadi O (2010) High-throughput production of human proteins for crystallization: the SGC experience. J Struct Biol 172:3–13. https://doi.org/10.1016/j.jsb.2010.06.008
Seo SW, Yang JS, Kim I, Yang J, Min BE, Kim S, Jung GY (2013) Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab Eng 15:67–74. https://doi.org/10.1016/j.ymben.2012.10.006
Sharma S, Vaid S, Bhat B, Singh S, Bajaj BK (2019) Chapter 17 Thermostable enzymes for industrial biotechnology. Adv Enz Technol. https://doi.org/10.1016/b978-0-444-64114-4.00017-0
Shoseyov O, Shani Z, Levy I (2006) Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev 70:283–295. https://doi.org/10.1128/mmbr.00028-05
Singh RS, Singh T, Pandey A (2019) Chapter 1 Microbial enzymes—an overview. Adv Enz Technol. https://doi.org/10.1016/b978-0-444-64114-4.00001-7
Sockolosky JT, Szoka FC (2013) Periplasmic production via the pET expression system of soluble, bioactive human growth hormone. Protein Expr Purif 87:129–135. https://doi.org/10.1016/j.pep.2012.11.002
Song JA, Koo BK, Chong SH, Kwak J, Ryu HB, Nguyen MT, Vu TT, Jeong B, Kim SW, Choe H (2013) Expression and purification of biologically active human FGF2 containing the b'a' domains of human PDI in Escherichia coli. Appl Biochem Biotechnol 170:67–80. https://doi.org/10.1007/s12010-013-0140-3
Song J-W, Woo J-M, Jung GY, Bornscheuer UT, Park J-B (2016) 3′-UTR engineering to improve soluble expression and fine-tuning of activity of cascade enzymes in Escherichia coli. Sci Rep 6:29406. https://doi.org/10.1038/srep29406
Sørensen HP, Mortensen KK (2005) Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol 115:113–128. https://doi.org/10.1016/j.jbiotec.2004.08.004
Sorensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Factories 4:1. https://doi.org/10.1186/1475-2859-4-1
Specht E, Miyake-Stoner S, Mayfield S (2010) Micro-algae come of age as a platform for recombinant protein production. Biotechnol Lett 32:1373–1383. https://doi.org/10.1007/s10529-010-0326-5
Sun P, Tropea JE, Waugh DS (2011) Enhancing the solubility of recombinant proteins in Escherichia coli by using hexahistidine-tagged maltose-binding protein as a fusion partner. Methods Mol Biol 705:259–274. https://doi.org/10.1007/978-1-61737-967-3_16
Tolmachev V, Hofström C, Malmberg J, Ahlgren S, Hosseinimehr SJ, Sandström M, Abrahmsén L, Orlova A, Gräslund T (2010) HEHEHE-tagged affibody molecule may be purified by IMAC, is conveniently labeled with [99mTc(CO)3]+, and shows improved biodistribution with reduced hepatic radioactivity accumulation. Bioconjug Chem 21:2013–2022. https://doi.org/10.1021/bc1002357
Uversky VN, Yuan S, Xu J, Ge Y, Yan Z, Du G, Wang N (2013) Prokaryotic ubiquitin-like ThiS fusion enhances the heterologous protein overexpression and aggregation in Escherichia coli. PLoS One 8:e62529. https://doi.org/10.1371/journal.pone.0062529
van Oers MM, Pijlman GP, Vlak JM (2014) Thirty years of baculovirus-insect cell protein expression: from dark horse to mainstream technology. J Gen Virol 96:6–23. https://doi.org/10.1099/vir.0.067108-0
Vavrová Ľ, Muchová K, Barák I (2010) Comparison of different Bacillus subtilis expression systems. Res Microbiol 161:791–797. https://doi.org/10.1016/j.resmic.2010.09.004
Vollmer B, Schooley A, Sachdev R, Eisenhardt N, Schneider AM, Sieverding C, Madlung J, Gerken U, Macek B, Antonin W (2012) Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J 31:4072–4084. https://doi.org/10.1038/emboj.2012.256
Wang X, Zhou B, Hu W, Zhao Q, Lin Z (2015) Formation of active inclusion bodies induced by hydrophobic self-assembling peptide GFIL8. Microb Cell Factories 14:88. https://doi.org/10.1186/s12934-015-0270-0
Wei X, Wu R, Zhang L, Ahmad B, Si D, Zhang R (2018) Expression, purification, and characterization of a novel hybrid peptide with potent antibacterial activity. Molecules 23:1491. https://doi.org/10.3390/molecules23061491
Wilkinson DL, Harrison RG (1991) Predicting the solubility of recombinant proteins in Escherichia coli. Nat Biotechnol 9:443–448. https://doi.org/10.1038/nbt0591-443
Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signaling and regulation. Nat Rev Mol Cell Biol 16:18–29. https://doi.org/10.1038/nrm3920
Wu W, Xing L, Zhou B, Lin Z (2011) Active protein aggregates induced by terminally attached self-assembling peptide ELK16 in Escherichia coli. Microb Cell Factories 10:9. https://doi.org/10.1186/1475-2859-10-9
Xing L, Wu W, Zhou B, Lin Z (2011) Streamlined protein expression and purification using cleavable self-aggregating tags. Microb Cell Factories 10:42. https://doi.org/10.1186/1475-2859-10-42
Xu Y, Foong FC (2008) Characterization of a cellulose binding domain from Clostridium cellulovorans endoglucanase-xylanase D and its use as a fusion partner for soluble protein expression in Escherichia coli. J Biotechnol 135:319–325. https://doi.org/10.1016/j.jbiotec.2008.05.004
Yadav DK, Yadav N, Yadav S, Haque S, Tuteja N (2016) An insight into fusion technology aiding efficient recombinant protein production for functional proteomics. Arch Biochem Biophys 612:57–77. https://doi.org/10.1016/j.abb.2016.10.012
Young CL, Britton ZT, Robinson AS (2012) Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications. Biotechnol J 7:620–634. https://doi.org/10.1002/biot.201100155
Yuan S, Wang X, Xu J, Yan Z, Wang N (2014) Ubiquitin-like prokaryotic MoaD as a fusion tag for expression of heterologous proteins in Escherichia coli. BMC Biotechnol 14:5. https://doi.org/10.1186/1472-6750-14-5
Zhang Y-B, Howitt J, McCorkle S, Lawrence P, Springer K, Freimuth P (2004) Protein aggregation during overexpression limited by peptide extensions with large net negative charge. Protein Expr Purif 36:207–216. https://doi.org/10.1016/j.pep.2004.04.020
Zhang Z, Tang R, Zhu D, Wang W, Yi L, Ma L (2017) Non-peptide guided auto-secretion of recombinant proteins by super-folder green fluorescent protein in Escherichia coli. Sci Rep 7:6990. https://doi.org/10.1038/s41598-017-07421-3
Zhao X, Li G, Liang S (2013) Several affinity tags commonly used in chromatographic purification. J Anal Methods Chem 2013:581093. https://doi.org/10.1155/2013/581093
Zhao W, Liu L, Du G, Liu S (2018) A multifunctional tag with the ability to benefit the expression, purification, thermostability and activity of recombinant proteins. J Biotechnol 283:1–10. https://doi.org/10.1016/j.jbiotec.2018.07.005
Zhao W, Liu S, Du G, Zhou J (2019) An efficient expression tag library based on self-assembling amphipathic peptides. Microb Cell Factories 18:91. https://doi.org/10.1186/s12934-019-1142-9
Zhou B, Xing L, Wu W, Zhang X-E, Lin Z (2012) Small surfactant-like peptides can drive soluble proteins into active aggregates. Microb Cell Factories 11:10. https://doi.org/10.1186/1475-2859-11-10
Zorko M, Jerala R (2010) Production of recombinant antimicrobial peptides in bacteria. Methods Mol Biol 618:61–76. https://doi.org/10.1007/978-1-60761-594-1_5
Funding
This work was supported by the Marine Biomaterials Research Center as a grant from the Marine Biotechnology Program, funded by the Ministry of Oceans and Fisheries, South Korea. This work is also supported by the Basic Core Technology Development Program for the Oceans and the Polar Regions of the NRF (NRF-2015M1A5A1037054) and the Research Fellow Funding Grant of NRF (NRF-2018R1A6A3A11040793), and supported by a Korea University Grant for MR Ki.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ki, MR., Pack, S.P. Fusion tags to enhance heterologous protein expression. Appl Microbiol Biotechnol 104, 2411–2425 (2020). https://doi.org/10.1007/s00253-020-10402-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10402-8