Abstract
A novel fed-batch strategy based on carbon/nitrogen (C/N) ratio in a microbial co-culture production medium broth was carried out in a biocalorimeter for improved production of poly (3-hydroxybutyrate) (PHB). Shake flask study suggested that the C/N ratio of 10 increased the yield of PHB by 2.8 times. Online parameters monitored during the C/N ratio of 10 in biocalorimeter (BioRC1e) indicated that the heat profile was maintained in the fed-batch mode resulting in a PHB yield of 30.3 ± 1.5 g/L. The oxy-calorific heat yield coefficient during the fed-batch strategy was found to be 394.24 ± 18.71 kJ/O2 due to the oxidative metabolism of glucose. The reported heat-based model adapted for PHB concentration prediction in the present fed-batch mode. The heat-based model has a Nash-Sutcliffe efficiency of 0.9758 for PHB prediction. PHB obtained by fed-batch-mode was characterized using gas chromatography-mass spectrometry (GC-MS) for the monomer-acid analysis, Thermogravimetric analysis (TGA) for thermal stability of PHB, and Fourier transform infrared spectroscopy (FT-IR) for confirmation of functional groups. Here, we establish a favorable C/N ratio for achieving optimal PHB yield and a predictive heat-based model to monitor its production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Poly (3-hydroxybutyrate) (PHB), an important member of polyhydroxyalkanoates (PHAs) family, is a biodegradable thermoplastic biopolymer synthesized by numerous microorganisms under suitable cultivation conditions. Currently, the major problem in scaling up of this biopolymer is high substrate cost and the tedious production procedure using individual culture (Freches and Lemos 2017). Therefore, large-scale commercialization of biopolymer manufacture is limited compared with synthetic polymers. Biopolymers derived from microbial sources, however, have a favorable advantage due to their renewable resource sustainable production, biodegradability, and biocompatible properties. Hence, researchers and scientists alike have taken particular interest in synthesizing PHB through microbial fermentation of sugars or lipids (Arias et al. 2018; Dietrich et al. 2019)
PHB production process by using microbial consortia may be advantageous in terms of improving the yield within a short duration of the selected microbes that are mutually synergetic (Huang et al. 2017). Co-culture fermentation is a process in which the inoculum has two or more microorganisms. Co-culture strategy has the potential to carry out complex functions, very robust to the environmental changes, which result in decreased production costs. Theoretically, the co-culture strategy by consortia of microbes reduces capital and cost for operation due to the use of non-sterile equipment and adaptability to grow in waste effluents/raw materials (Dias et al. 2005). Some of the advantages of co-culture strategies are higher product yield, growth rate, and utilization capacity of the substrates. Another advantage of consortia is compartmentalization, where active/passive transport of the molecules in broth either substrate or intermediate moves across the cell membrane could be used to facilitate a decrease in undesired by-products (Shong et al. 2012)
Batch fermentation is the simplest to operate in the biological process, where all medium components added in bulk at the beginning of the fermentation process, and the batch runs until the carbon source is exhausted. Batch turnaround downtime for product formation is high. Although batch experiments is advantageous because of the ease of handling and maximum utilization of substrates, it is inefficient when the concentrations of substrates change and does not allow increasing the productivity. Carbon source limitation during production can lead to intracellular polymer degradation due to depolymerase activation (Clarke 2013). Fed-batch processes are applied in microbial culture fermentation through the supply of crucial nutrients at certain physiological conditions to drive the cell performance towards desired product production. In fed-batch mode, the initial batch phase loaded with bulk nutrients where initial concentrations optimized from batch conditions. In the fed-batch process, when the nutrients exhausted, required carbon and nitrogen concentrations were fed into the reactor to increase the growth of biomass and product, and also for cell maintenance (Clarke 2013). For intracellular polymer accumulation, it is vital to modulate the nutrients in the media, which triggers PHB productivity. Other reports, as well as one of our previous studies, revealed that for obtaining a high yield of PHB, carbon source should be in excess, and nitrogen source should be limited (Anusha et al. 2016; Khanna and Srivastava 2005).
High concentrations of intracellular material improved by modulating the initial C/N ratio. During the fed-batch period, optimizing and modulating C/N ratio in the bioreactor could increase the desired biomass (Tsouko et al. 2016). Many researchers have tried to optimize the feed by approaching either constant feed method (Mozumder et al. 2014a) or exponential feeding (Mozumder et al. 2014b). While most C/N ratio optimization was carried out in the feed (Suzuki et al. 1986), its modulation in the broth has not been attempted. Here we have optimized the C/N ratio in the culture broth to improve the PHB yield.
Aerobic bioprocess is highly exothermic. Heat released by living organisms during fermentation in a biocalorimeter is a universal phenomenon and throws information on kinetics and stoichiometry of cell growth and product formation (von Stockar et al. 2011). Overall, heat generated by a microbial system is a sum of enthalpy change due to all the metabolic reactions of the system. Since it indicates the metabolic status of cells immediately, it can be considered a useful control variable. As the volume of the reactor increases, the ratio of heat production to heat exchanging surface increases. Calorimetric methods are, therefore, promising to be also applied on an industrial scale (Voisard et al. 2002).
Control and monitoring the biological processes are vital factors for attaining high product yield as well as reproducibility. The use of process monitoring is exceptionally significant for improving the batch/fed-batch consistency, especially for the consortia of microbial strains. Several models, such as mechanistic (Mozumder et al. 2014b), metabolic (Špoljarić et al. 2013), and low structured (Pérez Rivero et al. 2016) models, were developed for PHB production in both batch as well as a fed-batch mode of operation for tracing the product yield. In the present study, we have modified already existing models by incorporating key parameters of online measurements of biocalorimetric data and validating the model for a very close prediction of the product concentration. This model predicts the variation of residual cell concentration and PHB with time and also predicts how the evolution of metabolic heat varies with these quantities.
In a biocalorimetry study by Schuler et al. (2012), with crabtree-negative yeast cells, based on heat profile measurement, growth rate and cell dry mass were estimated in fed-batch. Hrnčiřík et al. (2019) dealt with biocalorimetry for antibiotics Nystatin production by Streptomyces noursei, and the model which was developed by them dealt with the detection of biomass estimation. Most of the laboratory-scale works using biocalorimetry limited their models to biomass monitoring and did not deal with product formation kinetics. The main aim of the heat-based model approach is to optimize the feed parameters and predict the product (PHB) concentration data inline, which will help us understand the system better.
The objectives in this work were as follows: (i) first and foremost, to optimize and increase the yield and productivity of PHB by applying fed-batch strategy to maintain the C/N of the fermentation broth in a biocalorimeter (BioRC1e), (ii) to biochemically analyze the biocalorimetric measurements and online parameters determined during the fed-batch process of PHB production by microbial consortia, (iii) to establish relationship between the heat, product, and growth through modified Tessier model, (iv) to develop heat-based model for prediction of PHB and cell dry mass concentration through online approach, (v) to characterize the PHB obtained from fed-batch process by gas chromatography-mass spectrometry (GC-MS), Fourier transform infrared spectroscopy (FT-IR), and thermogravimetric analyzer (TGA).
Materials and methods
Seed medium and inoculum
Consortia of microbes having 2% (v/v) of Ralstonia eutropha MTCC 2487, 2% (v/v) of Pseudomonas putida MTCC 2475 and 2% (v/v) of Azotobacter vinelandii MTCC 2459 (totally 6% v/v of consortia) inoculated into the autoclaved seed medium (Anusha et al. 2016) consisting of peptone (8.5 g/L), ammonium sulphate (6 g/L), and meat extract (6 g/L) and yeast extract (8.5 g/L). The pH of the seed medium was 7. Each of the cultures in consortia was grown separately in the seed medium around 16 h in the 250-mL conical flask until it reaches the optical density (O.D) 1 (exponential stage cells) at 30 °C and 150 rpm and then inoculated into the production medium.
Initial production medium and culture conditions
The initial medium composition for fed-batch experiments consisted of glucose (carbon source), 25 g/L; ammonium sulphate (nitrogen source), 3 g/L; potassium dihydrogen orthophosphate, 12 g/L; citric acid, 2 g/L; magnesium sulphate, 1.5 g/L; and the trace element solution (TES) of 15 mL/L concentration (Anusha et al. 2016). The primary medium taken was reported in one of our previous works (Anusha et al. 2016). The magnesium sulphate and glucose were autoclaved in separate flasks. Before inoculation, the production medium was cooled down to room temperature. The initial pH of 7 was maintained in the production medium.
Preliminary fed-batch experiments
The preliminary experiments for fed-batch were performed in a shake flask (working volume of 1 L) at 30 °C at 150 rpm using an orbital shaker (model: Scigenics LE). Experiments were performed to understand the behavior of the feed method. Three fed-batch strategies (nitrogen feeding (N), carbon feeding (C), and carbon-nitrogen feeding (C & N) were designed to understand which strategy gives optimal PHB. The initial production medium condition for fed-batch remains the same. These study facilities to understand which feeding method is best suitable for the high yield of PHB. Feeding strategy started from 10 to 26 h of fermentation to maintain uniform consistency of the experiments, and the 10th h was the period where the substrate was exhausted to half of its initial concentration. Nitrogen source feeding method consisted of 0.2 g/(L.h) of ammonium sulphate (concentration, 0.66 g/mL), carbon source feed consisted of 2 g/(L h) (concentration of glucose feed, 0.9 g/mL), and carbon and nitrogen source feeding consisted of feeding of both glucose and ammonium sulphate in the rate 2 g/(L h) of glucose and 0.2 g/(L h) of ammonium sulphate (glucose, 0.9 g/mL and ammonium sulphate, 0.66 g/mL). Seed culture of 6% v/v having microbes (R. eutropha, P. putida, and A. vinelandii in ratio of 1:1:1 each of 2%) used as inoculum for fed-batch studies. The absorbance of the seed medium was constituted to be 0.2 g/L (1.2 optical densities) of cell dry mass (CDM). The initial pH of the medium was at 7, for all the fed-batch experiments. At regular time intervals, samples were taken for analysis. Experiments were performed twice, and the average of two runs was reported in this study.
Fed-batch shake flask experiments
Fed-batch strategies in shake flasks were performed to analyze the C/N ratio effects for improving the productivity and yield of PHB. Initial conditions of production medium and the inoculum percentage remain the same as mentioned above. C/N ratios were optimized in the production medium from the 10th to 26th h of bioprocess through feeding of glucose and ammonium sulphate. It was understood from the BioRC1e batch experiments (data not shown) that 1.2 g/L of ammonium concentration is crucial concentration in the medium for yield improvement. Hence, the concentration to be there in the production medium broth in fed-batch phase is fixed as 1.2 g/L. The details of the feed strategies are given in Table 1. Carbon source (glucose) plays absolutely necessary for feeding; it is the one getting converted into product. Ammonium sulphate role in the production medium is to synthesis the DNA material and protein synthesis, thereby increasing the cell turnover rate. The C/N ratio here represented in the form of (moles) of glucose/(moles) of ammonium sulphate. For converting glucose in moles: g of glucose taken/180, ammonium sulphate moles: g of ammonium sulphate/132.13. The ammonium sulphate and glucose were added as per the requirements of the cells to maintain the ratio inside the production medium broth during fed-batch phase.
Analytical methods
The cell dry mass (CDM) was determined for the samples withdrawn at a regular interval of time. CDM was measured after centrifugation of the sample at 8000 rpm at 4 °C for 10 min. The pellet was washed with distilled water and again centrifuged, and the pellet dried for 4 h at 85 °C using an oven.
The glucose concentration was estimated by di-nitro-salicylic acid (DNS) for the supernatant and was analyzed using a spectrometer (model: Shimadzu UV-2600) at 510 nm (Miller 1959). The nitrogen source—ammonium sulphate—was estimated by the Solórzano method (Solórzano 1969).
PHB concentration was estimated for the samples collected at regular interval of time; the harvested cells were centrifuged at 8000 rpm at 4 °C for 10 min. The pellet obtained was treated with an equal proportion of 6% sodium hypochlorite at ambient temperature for 1 h. Centrifugation was performed at 8000 rpm for 12 min (model: Sigma 3-18 K refrigerated centrifuges). The pellet washed twice with methanol and water, and centrifuged at 10,000 rpm at 4 °C for 5 min, and the pellet obtained was dried and further purified by the addition of boiling chloroform. The chloroform in excess was removed through evaporation. For estimation of PHB concentration, 10 mL of sulfuric acid was added to the purified PHB powder, and the sample was heated at 100 °C for 10 min in a water bath for the conversion of PHB polymer into its monomer—crotonic acid (Law and Slepecky 1961). Crotonic acid was measured at 235 nm using a UV spectrometer (model: Shimadzu UV-2600) and compared with a standard graph.
Biocalorimeter experiments for fed-batch
Biocalorimeter (BioRC1e) experiments (make: BioRC1e, Mettler-Toledo, Switzerland) performed by maintaining the isothermal condition throughout the process by the temperature maintenance circulation system (models: Julabo FL601 and Julabo FP51). BioRC1e is a 2.2-L double jacketed reactor vessel with a working volume of 1.5 L. Online probes such as turbidity, dissolved oxygen (DO) (model: Mettler–Toledo M700), and pH sensors have installed for monitoring the bioprocess. The software iControl RC1e measures the heat generated from the experiment. The inside reactant temperature (Tr) was maintained continuously by controlling the jacket temperature (Tj), which indeed aids in maintaining isothermal condition by circulating silicone oil in the jacket region. A four-bladed Rushton turbine impeller acts as an agitator for proper mixing. Through a sparger, oxygen was supplied and the flow rate was adjusted using a rotameter. A 0.2-μm membrane filter was used to sterilize the oxygen bubbles, and it is sparged exactly below the impeller. The measurement and principle of biocalorimeter were reported previously in one of our papers (Anusha et al. 2016). The heat is measured by Eq. 1.
where qr is the heat evolved during the process (W) and Tr and Tj represent the temperature of reactor content and jacket (°C). U is the overall heat transfer coefficient (W/m2 K), and A is area of heat exchange (m2).
Baseline experiments nullified heat produced from the production medium without inoculum and also due to aeration and agitation effects. The heat generated in the baseline experiment is represented as qb, heat generated inside the reactor is qr, and exact heat during metabolic process during fermentation is represented by Eq. 2.
The initial concentrations of glucose and ammonium sulphate for fed-batch runs were 25 g/L and 3 g/L. Experiments were performed for the C/N ratio of 10 and 15 for fed-batch processes during the interval 10–26 h of growth. For C/N 10 studies, 16.36 g/L of glucose and 1.2 g/L of ammonium sulphate concentrations were maintained. For C/N 15 studies, 24.54 g/L glucose and 1.2 g/L ammonium sulphate concentrations were maintained. Required feed for maintaining the ratio in the broth was added accordingly. The initial pH of the medium was at 7. For all fed batch experiments, inoculum was 6% (v/v) (consisting of co-culture strains: R. eutropha, P. putida, and A. vinelandii), 150 rotations per minute (rpm) of agitation, and 1 liter per minute (lpm) of aeration was maintained. At regular interval of time, the samples were taken out and the offline analysis such as cell dry mass (CDM), glucose, ammonium sulphate, and PHB was measured.
Oxygen uptake rate and carbon dioxide evolution rate estimation
The exit gas analyzer (model: Siemens—Ultramat 23) monitored the oxygen and carbon dioxide from the reactor. Carbon dioxide evolution rate (CER) and oxygen uptake rate (OUR) data were estimated by the gas balance method (Doran 1995).
Yield calculations during the fed-batch process
The bioenergetics studies state the metabolic process underway and the effective use of the substrate. Heat yield coefficient due to glucose consumption estimated as YQ/S (kJ/g of substrate—glucose), yield coefficient of heat due to CDM (CDM represented as X) formation: YQ/X (kJ/g), and heat yield due to oxygen consumption quantified as YQ/O2 (kJ/mol of O2). These yield coefficients were calculated from the slope of cumulative heat production during the batch phase/fed-batch phase with the carbon source—glucose—consumed, CDM formed, and the amount of oxygen consumed (Battley 1998).
Calculation of productivity (qp) for PHB experimental data is given in Eq. 4.
YX/S calculated by considering (highest cell dry mass (CDM) achieved during fed-batch process) − initial X concentration (g/L)) to the total glucose consumed (Initial glucose + feed added − unused glucose) (g/L). Product (PHB) yield coefficient due to glucose (YP/S) was calculated by considering the highest PHB concentration obtained (g/L) in fed-batch phase to the amount of total glucose consumed (initial glucose + feed added − unused glucose) (g/L). Product yield coefficient due to CDM (YP/X) is calculated by considering (highest concentration of PHB formed during fed-batch (g/L) − initial PHB (g/L)) to total CDM formed (g/L).
Bioprocess models
Here, the modeling of bioprocess for understanding and prediction of the product (PHB) are based on the modified Tessier model and a heat-based model. The purpose of developing a model here is to predict product concentration with time and also to predict how the evolution of metabolic heat varies with these quantities.
Modified Tessier model
The modeling process starts with expressing the entire system in terms of differential equations because of the simplicity in relating the rates of the different quantities. These equations were integrated and the biokinetic parameters estimated by fitting the theoretical heat data with the experimental heat data using non-linear regression techniques available in the python programming language (SciPy).
Two different stages are having batch and fed-batch processes in the same experiment, which have modeled separately. The initial conditions for the batch are already known, and that will be used to predict the quantities until the 10th h. After the 10th h, the feeding of the substrate started, and thus, the equations used for the batch stage were modified to account for the change in volume. The same biokinetic parameters predicted in the batch phase (0th–10th h) were used for predicting the quantities in the fed-batch stage.
Formulating the batch stage equations for modified Tessier model
From the experimental data, during the fed-batch stage, the substrate limitations due to glucose or nitrogen were absent for optimized C/N ratio of 10. High glucose concentrations were assumed not to play any inhibitory role in product formation. Various growth models were considered for fitting the data such as the Monod model (Gerson et al. 1988), Tessier model (Beyenal et al. 2003), and Mulchandani model (Mulchandani et al. 1989). Finally, the modified Tessier which incorporated the Luedeking and Piret model (Luedeking and Piret (1959)) and Mou and Cooney (1976) expressions was chosen as it best described the metabolic process during the fed-batch stage, which relied on the strategy of maintaining an optimum concentration of glucose throughout the fed-batch stage. The chosen model also fitted the experimental data with higher accuracy when compared with the other models. In the Tessier equation (Beyenal et al. 2003) (Eq. 5), the residual cell concentration was taken for consideration.
The consumption of glucose is assumed to be solely dependent on the growth (Eq. 8) of residual cell concentration and formation of the PHB. Hence, a relation was formed linearly relating the rate of consumption of glucose with the rates of formation of the residual cell and product.
Similarly, the consumption of nitrogen (Eq. 9) is assumed to be dependent only on the growth of the active residual cells and not due to PHB formation.
The rate of formation of PHB (Eq. 10) was determined by Luedeking and Piret (1959) which is based on the assumption of PHB formation being dependent on both the growth and non-growth phases.
Heat plays a significant role in metabolic processes of the culture, and it can be used as an indicator of these metabolic changes described by Mou and Cooney (1976). Even while observing the trends of both the metabolic heat and biomass, it was seen that the metabolic heat follows a pattern similar to the rate of change of biomass growth and product formation (depicted from Fig. 4)—values of PHB, cell dry mass, and residual cell concentrations data given (rate of changes can be derived from Fig. 4). In addition to this, the heat released or consumed will be due to the maintenance of the cell also. Hence, the above expression (Eq. 11) stated by Mou and Cooney (1976) was chosen for the modified Tessier-based model for understanding the biokinetics of the process. The expression was differentiated for predicting the parameters.
Equations 12 and 13 are used in the batch model (0–10 h). The parameters were estimated by fitting the experimental data of instantaneous metabolic heat values to the system of equations by using the least-squares fit algorithm available in the python programming language library (Scipy), which was optimized by reducing the difference between the squares of the theoretical and experimental quantities. This model is the modified form of Tessier model which incorporates valuable expressions from Leudeking and Piret (1959) and Mou and Cooney (1976) expressions which describe the biokinetics of the process.
Formulating fed-batch equations for modified Tessier model
As mentioned previously in the modeling approach, the parameters (Table 2) for residual cell concentration and PHB prediction remain the same for both the batch and fed-batch phases of the experiment. However, the heat yield due to maintenance is different during the batch phase and fed-batch phase; hence, the model was also split into two stages to calculate the two different heat yields.
Attempts were also made to include the dilution effects caused by the changes in volume in the fed-batch stage. Although the feed rates of the substrates varied at different stages of the experiment for maintaining the ratio of glucose to nitrogen, the changes were minor when the volume changes due to sample collection. Hence, it was assumed that the feed rates were constant and fixed at 5.893 mL/h for glucose and 0.7935 mL/h for ammonium sulphate. The concentrations of the feed were known and fixed at 0.9 g/mL for glucose and 0.66 g/mL for ammonium sulphate. The sample collection rate (Sc) was at 5 mL/h.
To appropriately portray the experimental conditions during the fed-batch stage, the expressions for substrate consumption rate were modified to account for the maintenance of concentrations of glucose and ammonium sulphate. During C/N ratio of 10 maintenance of the culture broth, the rates of change of glucose and ammonium sulphate in the medium were zero.
The following Eqs. 14–21 are the modified equations used to predict the various quantities during the fed-batch process.
Since we have maintained glucose and ammonium sulphate concentration in the broth from 10 to 26 h, the change in the glucose concentration during that phase was zero (Eqs. 15 and 16).
\( \mu ={\mu}_{R-\mathit{\max}}\left(1-{e}^{\frac{-{S}_g}{K_s}}\right) \) recalling Eq. 5.
Substituting the μ calculated using Tessier model into the growth Eq. 17, we get the change of residual cell concentration.
Change of product concentration was estimated using modified Luedeking-Piret equation (Luedeking and Piret 1959) represented in Eq. 18, which takes into consideration the changes in volume.
Differentiating Eq. 5 to estimate the change in μ with time (t),
The evolution of instantaneous heat depends on the growth of cells (in this case, for Tesssier-based model, residual cell concentration (R)) and PHB and also due to cell maintenance.
Differentiating Eq. 20 and substituting Eq. 17–19, we get Eq. 21
where R represents residual cell concentration (g/L), μ specific growth rate of residual cells (h−1), μR-max maximum specific growth rate of residual cell concentration (h−1), Sg glucose concentration (g/L), Sn ammonium sulphate concentration (g/L), P PHB product concentration (g/L), Ks half velocity constant (g/L), q metabolic heat (kJ), V active volume of the reaction (L), F1 glucose flow rate (L/h), F2 ammonium sulphate flow rate (L/h), Sc sample collection rate (L/h), k1 residual cell concentration yield on heat (g L−1 W−1), k2 PHB yield on heat (g L−1 W−1), k3 non-growth-associated constant, k4 growth-associated constant, k5 glucose utilized per residual cells formed (g/g), k6 glucose utilized per PHB formed (g/g), k7 ammonium sulphate utilized per residual cells formed (g/g), and k8 heat yield during maintenance (W g−1 L−1).
The formulated Eqs. 5–13 represent batch and fed-batch region expressions. These equations are time derivatives and describe the product, biomass, glucose, nitrogen source, and heat as a function of time.
These differential equations are simultaneous and have been integrated numerically using the 4th-order Runge-Kutta method and were then curve fitted to the experimental data of C/N of 10 to get the optimum values of the parameters k1 to k8. Once the best fit curve was found and the value of these parameters was computed, these parameters were then put back into the equations corresponding to the product (PHB), residual cell concentration (R), glucose, nitrogen source, and heat and their corresponding values were predicted by feeding in various time values to the equations. The accuracy of these predicted values was compared with the independent validation experimental data to establish the relationship of the biokinetic parameters.
Heat-based model
Santharam et al. (2017) predicted product through heat data by incorporating the Luedeking-Piret equation (Eq. 24). The equations proposed by Santharam et al. (2017) are as follows:
where V0 is the initial working volume of the reactor, Vt is the instantaneous volume during the fed-batch process which was assumed to be constant due to the extremely minute volume changes, qr represents the instantaneous heat generated during the process, X is the cell dry mass (CDM) (g/L), Qt is the cumulative heat generated at particular time (kJ/L), and Q0 is the initial heat which is taken to be 0 for the process. YQ/X for the batch process is the slope of the plot between cumulative heat generated and the biomass formed (g/L), X0 is the initial biomass concentration, and Xt is CDM at time t.
where μ(estimated,t) (h−1) is the growth rate determined based on heat data. Qp is the specific rate of PHB production (h−1), and P is the product—PHB concentration (g/L). In this heat-based model, the growth- and non-growth-associated constants are represented as α and β, for better clarity. Equations 24 and 25 are modified from the Luedeking-Piret equation (Luedeking and Piret 1959) (Eq. 10).
The process was divided into three different phases: batch, fed-batch, and decline, and each phase modeled separately. The data, too, were split into three parts according to the cumulative values of heat, which corresponding to the batch phase (0–10 h), fed-batch (10–32 h), and decline (32–36 h) phases of the process. For each part of process, the slope of the graph plotted between biomass (g/L) and cumulative heat (kJ) was calculated using linear regression to find YQ/X. The values of YQ/X for batch phase were 17.17 kJ/g, fed-batch phase 6.16 kJ/g, and decline phase − 14.22 kJ/g, respectively. The YQ/X found for each stage was put into Eq. 23, along with the corresponding instantaneous heat and cumulative heat data, to compute the cell dry mass values. The predicted CDM values were used in Eq. (27), along with the instantaneous heat and cumulative heat to forecast the values of α and β. These parameters were estimated by first solving the differential equation and then curve fitting the non-linear equation to the experimental PHB data. The estimated heat-based α and β values were 1.699 and 0.07 for the batch stage, 1.7 and 0.02 for the fed-batch stage, and 1.69967 and 0.02 for the decline process. The instantaneous heat and the offline collected data (biomass) from the process of production of PHB were used as input for the model. These computations were achieved by using functions available in the numerical computing library of python called SciPy.
These predicted values of α and β were substituted back into Eq. (27) and were used to predict the PHB production values, which compared with the experimental values obtained in the C/N of 10 validation experiment.
Model calibration and model validation
Here the function yi refers to experimental values and the function f(xi, E) refers to the predicted values from the model using the parameter E. This function is also known as the loss function used as part of the Levenberg-Marquardt algorithm (Kanzow et al. 2004) (Eq. 28) used in the least-squares optimization module of the SciPy library in python. When using the least-squares optimization technique, the value of the above function is minimized by varying the parameter value E for successive iterations. In reality, however, the amounts of parameters to be varied and determined are too many. A trust region method was used by introducing boundary conditions for the parameters. The boundary conditions for the parameters chosen from its known ranges were observed and calculated in previous experiments.
For model validation purposes the experimental values were compared with the predicted values from the model. For comparison purposes, Nash-Sutcliffe model (Eq. 29) efficiency coefficient was used to evaluate the accuracy of the model predictions (Moriasi et al. 2007). As the coefficient is close to 1, it points to the fact that the prediction is accurate for the validation experiments.
Characterization of PHB
Fourier transform infrared spectroscopy and gas chromatograph-mass spectrometry
For Fourier transform infrared spectroscopy (FT-IR) analysis, the PHB powder extracted from fed-batch experiment was characterized using FT-IR spectroscopy. Spectral readings (model: PerkinElmer) were taken in the range of 400–4000 cm−1 for functional groups’ confirmation of the PHB.
Gas chromatograph-mass spectrometry (GC-MS) analysis was carried out using 5 mg of PHB extracted from fed-batch experiment into a glass tube; 1 mL of chloroform, 0.15 mL of sulfuric acid, and 0.85 mL of methanol were added. The mixture was heated at 100 °C for 2 h. The mixture undergoes methanolysis. Mixture was allowed to cool and equal amount of water added for the separation of aqueous and organic phases. Organic layer was carefully removed and the resulted methyl esters analyzed by GC-MS (model: Agilent). This method characterizes the monomeric composition of PHB (Braunegg et al. 1978).
Thermogravimetric analysis
The PHB obtained from the fed-batch process was characterized for thermal stability by thermal gravimetric analysis (TGA) (model: TA Instrument). The thermal degradation study of PHB was characterized from 30 to 400 °C.
Results
Preliminary studies for C & N feeding
The results for carbon and nitrogen mixed feeding for preliminary fed-batch experiments are shown in Fig. 1. Among the feeding strategies adopted, the maximum PHB productivity was seen for mixed feeding (C & N feeding together) at 28 h of fed-batch process with maximum PHB concentration of 17.13 ± 0.84 g/L inside 27.88 ± 1.39 g/L CDM. Ammonium sulphate (nitrogen source) feeding alone did not improve the productivity and yield of PHB. Nitrogen source only aids in DNA synthesis and protein production in the cells (Kampen 2014). In the case of carbon source feeding, it directly involves in PHB formation and improves the yield of PHB. Approximately 2.3 times increase in concentration of PHB was obtained with carbon feeding alone.
The productivity of PHB obtained for ammonium sulphate, glucose, and glucose-ammonium sulphate feeding was 0.22 ± 0.011, 0.53 ± 0.02, and 0.61 ± 0.03 g L−1 h−1. Approximately 2.8 times increase in productivity was observed in mixed feeding (C and N feeding).
For studying the influence of the concentration of C/N in the production broth, one has to optimize the ratios. Further experiments were performed to choose the C (mole)/N (mole) maintenance in the fed-batch for improving the yield thereon.
Effect of C/N ratio feeding strategy in shake flask
In the shake flask experiment, C (moles) of glucose/N (moles) of ammonium sulphate optimized varied from 5 to 20. To maintain the ratio, feed glucose and ammonium sulphate were supplied according to the requirements represented in Table 1. The supernatant was analyzed offline for glucose and ammonium sulphate. A comparison of cell dry mass (CDM), PHB, and residual cell concentration during the fed-batch process in shake flask is represented in Fig. 2. It was observed from the fed-batch studies that for C/N ratio of 10, maximum CDM of 43.26 ± 2.16 g/L was achieved with PHB concentration of 28.5 ± 1.42 g/L. Thus from Table 1, the ratio C/N of 10 maintenance in the culture broth resulted in maximum PHB productivity.
For maintaining the appropriate ratio, glucose and nitrogen feeding has to be supplied throughout the fed-batch stage and the details are given in Table 1 and feed added for C/N maintenance in the shake flask is given as (supplementary material Fig. S1 and Fig. S2). The repression of the product in the C/N ratio of 20 is due to glucose concentration above the threshold limit. This repression leads to the carbon source being diverted to the by-product production.
According to Verlinden et al.’s (2007) studies, single fed-batch fermentation that is limited nitrogen leads to a low amount of PHB due to declining biomass. Therefore, in this present fed-batch design of PHB production, an attempt has been made to maintain certain threshold concentration of carbon and nitrogen such that the growth metabolism, biomass turnover, and PHB accumulation were sustained without any disruption in these distinct metabolic pathways.
The bacterial consortia require optimal carbon and nitrogen availability in the media to support all three aspects of PHB biosynthesis growth, biomass, and carbon/energy reserve biopolymer. We focussed on verifying the relative “C & N” levels that will result in maximal PHB accumulation. After optimization, we found the C/N ratio of 10 to be favorable for better yield of PHB accumulation. Therefore, further experiments were performed using an optimum C/N ratio of 10.
Fed-batch analysis in BioRC1e
Although BioRC1e results for C/N 10 and 15 were carried out here, the result of C/N 10 alone was discussed here. Interestingly, PHB and biomass yield for C/N 10 and 15 were similar. However, for C/N 15, the glucose added was more (Table 1), i.e., 107 g/L required for achieving 50 ± 1.5 g/L of CDM and 30.2 ± 1.5 g/L of PHB, while the C/N of 10 used only 87.8 g/L glucose for producing 30.3 ± 1.4 g/L of PHB. Hence, further experimental discussion is based on C/N 10 fed-batch observations. The result of C/N of 15 is given in the supplementary material Fig. S3.
The biocalorimeteric (BioRC1e) online measured parameters such as heat (Q), turbidity, pH, and dissolved oxygen are presented in Fig. 3. The heat curve indicates three separate stages of the microbial culture, i.e., log, stationary, and stage of death. The lack of the lag phase shows a robust approach for optimizing and adapting the culture to the setting of the medium and reactor. In the fed-batch period, cells avoid carbon source waste and maximize PHB accumulation. This was achieved by applying a composition and rate of feed that keeps a desired concentration of glucose in the reactor, i.e., 16.36 g/L glucose concentration in the production medium (C feed concentration) (glucose concentration of 0.9 g/mL). Ammonium sulphate was maintained at 1.2 g/L throughout the fed-batch stage in the production medium.
Heat peak value attains to 3.98 ± 0.2 W during the growth phase (0th–14th h, this is the phase where accelerated growth of the microbial cells takes place). Until 10 h, the experiment was run as a batch process; from the 10th to 26th h, feeding of both carbon and nitrogen substrates was began. Cumulative heat in the batch stage (0th–10th h) was 70 ± 2.5 kJ/L. During the fed-batch phase (10–26 h), the cumulative heat production was 410 kJ/L ± 18.5. In fed-batch, the rate of cell growth during the initial period was maximized in a rich nutrient medium; once an acceptable concentration of cells reached (until batch stage), the change to optimum product formation was initiated by feeding required carbon and nitrogen source for proper threshold maintenance C/N ratio of 10.
A drop in pH was observed from 7 to 6 during the initial 0th–10th h, due to utilization of glucose. In the fed-batch process, feed glucose and ammonium sulphate were buffered to pH 7. The addition of feed increased pH of the reaction (12th–14th h) due to feed pH interference. Again pH dropped from 7 to 5.6 due to the possible production of organic acids. Glucose utilization by the Entner-Doudoroff pathway releases organic acids during pyruvate production (Richhardt et al. 2013).
The dissolved oxygen (DO) trend depicted in Fig. 3 reveals that from the 0th to 6th h, it decreases because the cells start to consume oxygen in the dissolved state and contribute to overall growth; this phase is called an accelerated growth phase. From 6 to 36 h, demand for oxygen is more by the growing cells, and hence, DO concentration decreases. Both heat and DO portrait the same information about the state of culture growth. It was seen that at 36 h, deceleration in the growth takes place. Hence, the dissolved oxygen is not required by the cells and DO peaks up.
The turbidity profile from the initial stage starts to increase from 0 h and reaches a maximum at 28 h, where it touches the highest turbidity value of 3000 ± 150 nephelometric turbidity units (NTU); at the period of decline growth phase, the turbidity remains same. Turbidity is the marker of the cell density of the fermentation process.
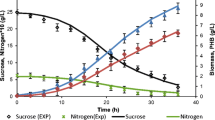
The offline analysis during fed-batch production in BioRC1e for PHB biosynthesis is depicted in Fig. 4. Until the batch experiment mode (0th–10th h), before feed addition about 6.2 ± 0.31 g/L of CDM formed with a PHB concentration of 4.3 ± 0.21 g/L, glucose was well utilized by the culture during batch phase, and at the end of batch, 11.4 ± 0.57 g/L of glucose and 0.9 g/L ± 0.045 of ammonium sulphate were found until the 10th h (batch stage). From the 10th–28th h, the highest PHB concentration of 30.3 ± 1.5 g was obtained for a CDM of 45.5 ± 1.8 g/L (Fig. 4). Glucose and ammonium sulphate addition profiles during fed-batch mode by the co-culture are depicted in Fig. 5. After 28 h, PHB starts to decline as the cells begin to lyse which is well portrayed in the heat data (Fig. 3). In the famine phase, PHB decrease was observed probably due to its depolymerization for sustaining the energetics of the culture.
At the end of the experiment, about 6 g/L ± 0.3 of glucose and 0.5 ± 0.02 g/l of ammonium sulphate remained unutilized in the culture broth. The post 26 h is like the famine phase where “carbon” addition stopped (Fig. 5). In the intermediate fed-batch phase, we included this novel C/N optimization strategy. By this, we were able to achieve the favorable metabolic milieu for enhanced PHB production.
The maximum height and duration of the exothermic peak indicated the overall biomass activity, whereas the kinetics of the initial thermal response to the addition of substrates may be related to the degree of biomass adaptation to the substrates added (Redl and Tiefenbrunner 1981).
Oxygen uptake rate and carbon dioxide evolution rate during fed-batch
Calorimetric and respirometric measurements have been successfully applied to monitor the aerobic process of the co-culture during a fed-batch stage and due to the addition of feed and to evaluate and compare biomass activities. Oxygen uptake rate (OUR) and carbon dioxide evolution rate (CER) profiles during fed-batch process are represented in Fig. 6. In the batch stage tenure (0–10 h), 2.8 ± 0.14 g L−1 h−1 of CER and 26 ± 0.9 g L−1 h−1 of OUR were observed. In the fed-batch stage, 3.4 ± 0.17 g L−1 h−1 CER and 32 ± 1.4 g L−1 h−1 OUR were maintained throughout in the feeding stage, which reiterates the respiratory metabolism of culture.
Yield coefficients for C/N ratio of 10 during the fed-batch process in BioRC1e
Heat yield due to cell dry mass (CDM): YQ/X relates the energy dissipation in microbial growth and has direct influence on the expected biomass yield. Heat yield due to CDM formed was 11.96 ± 0.45 kJ/g until the batch phase (0th–10th h). The yield of heat due to CDM formation in the period 10th–26th h (fed-batch phase) was 5.76 ± 0.22 kJ/g. In the fed-batch stage, less energy dissipation occurred, and more biomass building took place. The more the energy dissipation occurs, the less the energy will cling to biomass growth (von Stockar et al. 2011).
Heat generated due to the total amount of oxygen consumed (YQ/O2) was calculated from the slope of total cumulative heat dissipated in the total period (0–28 h) to the total oxygen consumed in moles. YQ/O2 was found to be 394.24 ± 18.71 kJ/O2 consumed. Biokinetic parameters for the C/N ratio of 10 from BioRC1e run are represented in Table 1. YP/X was calculated as 0.67 attributes to the accumulation capacity of PHB of co-culture during fed-batch mode. Substrate utilization capacity during the fed-batch stage (10th–20th h), the biomass yield coefficient due to substrate (YX/S) was found to be 0.42. It links the glucose utilization capacity for total biomass formation.
Bioprocess modeling
Here, we have carried out two types of modeling: (i) modified Tessier model for evaluation of product concentration in offline mode, basically to correlate heat with product and cell growth; (ii) the second approach is on the prediction of PHB based on instantaneous heat signals online.
Modified Tessier model for prediction of PHB and residual cell concentration and establishment of biokinetic relationship
The model development focuses on the prediction of intracellular PHB and the use of metabolic yield of heat as measured by BioRC1e as the nodal parameter around which the production estimate was made.
In addition to biomass growth, the production of metabolites must also be monitored and controlled if biocalorimetry is to be applied successfully as a process analytical technology (PAT) tool. In its attempt to quantify a metabolite (PHB) by linking it to metabolic heat data, the work is therefore significant.
The estimated parameters generated from the modified Tessier model are listed in Table 2. These parameters as mentioned previously was determined by non-linear regression techniques available in the python library (Scipy. Optimize), which use the Runge-Kutta 4th-order methods. This was achieved by identifying the model parameters using Eqs. 5–13, and the initial experimental data sets. The estimated parameter values (Table 2) were determined in this way, and the simulated data for PHB, residual cell concentration, heat obtained from the validation experiment, and the model simulated values were compared. The growth-associated constant (k4) was much greater than the non-growth-associated constant (k3) (Table 2) which describes that the bioprocess behind PHB mostly relies on the growth-associated factor. Using estimated parameters and the time derivatives of Eqs. 5–13 results in model-predicted values. It was observed from the biokinetic studies from Table 2 that, for 1 g of PHB production, 2.5 g of glucose was used. The independent validation experimental results and the model simulated data for C/N of 10 are compared in Fig. 7. There is a good match between the experimental and predicted trends of metabolic heat (Fig. 7a), which reiterates the validity of the established model equations in this work. This link describes that heat linked to growth, product, and cell maintenance and thus gives us insight about the efficiency of instantaneous experimental heat data to predict product concentration online. To further, the model equations generated in this work were used to predict the concentration of PHB and the residual cell concentration for validation experiments (Fig. 7b, c) at any stage of the bioprocess. The goodness of the model is well established by looking at the Nash-Sutcliffe efficiency. The Nash-Sutcliffe efficiency for PHB prediction was found to be 0.94, stating the very closer correlation between model and the experimental data from which we can infer the high accuracy of the modified Tessier model (Eq. 21) for understanding the biokinetic behavior behind the bioprocess of PHB production by offline approach.
Heat model-based prediction of PHB
The modified equations put forward by Santharam et al. (2017) were used in the model, with online parameter heat as the input, to predict the biomass values and to compute the optimized values of the growth- and non-growth-associated factors. The calculated growth (α) and non-growth (β) parameters for the C/N 10 experiment were as follows: α (1.699) and β (0.07) for the batch stage, α (1.7) and β (0.02) for the fed-batch stage, and α (1.69967) and β (0.02) for the decline process. The parameters α and β for the C/N 15 experiment were found to be 1.7 and 0.0817 for the batch process, 1.7 and 0.018 for the fed-batch process, and 1.699 and 0.011 for the decline phase respectively. An independent experiment for validation of the model was carried using C/N 10 α and β for predicting the PHB concentration. For the prediction of CDM, the following parameters in Eq. 23 were substituted; the initial biomass concentration (for all experiments, initial biomass was fixed, i.e., 0.2 g/L, cumulative heat data (Qt), and the slope values (YQ/X). Using the computed parameters:α and β and the instantaneous heat data (qr), cumulative heat data (Qt) and the slope prefixed values (YQ/X) were substituted in Eq. 27, and then, Eq. 27 integrated values resulted in model-predicted PHB data. Further, the values predicted by the model are in good agreement with the C/N 10 validation experimental values, which affirms the accuracy of the model. The model assumed that the physiological conditions like aeration, agitation, inoculum size, temperature, and production medium are maintained constant across all experiments. As the volume change during the fed-batch process was minimal, it was assumed to be constant.
Finally, the efficiency of CDM and PHB prediction was evaluated using the Nash-Sutcliffe model efficiency coefficient. The model predicted the CDM and PHB production values with a Nash-Sutcliffe efficiency coefficient of 0.9630 and 0.9758 on validating it on a new set of experimental data, which reaffirms the accuracy of the model (Fig. 8) for prediction of CDM and PHB.
Characterization of PHB obtained from fed-batch process
Fourier transform infrared spectroscopy for PHB from fed-batch experiment
The Fourier transform infrared spectroscopy (FT-IR) spectra of the PHB sample extracted from BioRC1e experiment of the fed-batch process for a C/N ratio of 10 are given as supplementary material Fig. S4. The peaks at 1727 and 1288 cm−1 depict the specific rotation around the carbon atom. The peak at 1727 cm−1 represents the ester group C=O stretching exhibited by the crystalline nature of PHB. The 1288 cm−1 corresponds to the C–O stretching. The peak at 3440 cm−1 is due to the group -OH, because of moisture in the sample. The spectra were compared with the standard graph of the PHB (Supplementary material Fig. S4).
Gas chromatography-mass spectrometry of PHB monomer
PHB is a biopolymer; exact m/z in the gas chromatography-mass spectrometry (GC-MS) is not possible. Nevertheless, the characterization of monomers can be characterized using GC-MS (methods for analysis of poly (3-hydroxy alkanoates) (PHA) monomer composition). Methanolyzed PHB (Supplementary material Fig. S5) revealed the retention time and m/z fragmentation pattern using GC-MS. Prominent peaks with retention time values of 3.993, 4.419, and 7.653 using GC and corresponding derivatized products having m/z signals at base peak 86 when compared with NIST library show the presence of butenoic acid (another name—crotonic acid) confirming the presence of monomers of poly (3-hydroxybutyrate).
Thermogravimetric analysis of PHB obtained from fed-batch process
Thermogravimetric analysis (TGA) plot (Supplementary material Fig. S4) shows thermal degradation onset at 228 °C. TGA plots also reveal that the absence of weight loss in TGA up to 270 °C. From initial weight % of 2.42 mg, residue formed is 0.34 mg (14.41% residual weight) which corresponds to ash content in the sample. The extracted PHB is thermally stable till 270 °C. TGA presented here corroborates well with the study of Kunasundari et al. (2017).
Discussion
The carbon and nitrogen sources feeding together signify the importance of regulation of glucose and ammonium sulphate role in either biomass turnover or productivity. There are also interactions between the regulation of carbon and nitrogen inside the microbial cells. The Entner-Doudoroff pathway was induced by glucose, but full induction occurs if amino acids are also available (Richhardt et al. 2013). Thus, from the preliminary shake flask studies, it can be concluded that the carbon and nitrogen feeding together has a more significant influence on product formation.
Fed-batch operation may also be useful if the concentration of the substrate is so high that the microorganism is affected by detrimental osmotic effects. Many researchers developed control strategies for fed-batch culture to optimize the feed rate of specified nutrient concentrations either by constant feed (Mozumder et al. 2014a) or by exponential feeding method (Mozumder et al. 2014b), also with C/N feeding (Suzuki et al. 1986). However, they tried to optimize the feed concentration, not the concentration of C/N in the production broth during the fed-batch stage. Why optimization of the C/N ratio? The right proportion of carbon source to sustain energy metabolism for growth and product formation enabled higher productivity of PHB. However, carbon input should be adequate to maintain the PHB accumulation and not allow the PHB to be broken down (PHB depolymerase) to sustain energy metabolism.
Generally speaking, fed-batch culture is useful if prolonging a particular phase of batch culture favors optimal product formation. The threshold limit of substrate concentrations above or below which the formation of a product is inhibited or repressed. From this study, both the C/N ratio of 5 and C/N of 20 are not suitable for high PHB productivity. The highest concentration of PHB was obtained for C/N of 10. Hence, through shake flask studies, it was observed that lower threshold glucose concentration and higher glucose concentration did not favor increase in PHB concentration. Also, nitrogen source (1.2 g/L for ratio maintenance) (ammonium sulphate) was required for biomass. And nitrogen depletion at the cost of biomass turnover is not favorable for PHB production. This appears like a paradox, wherein it is well known that nitrogen depletion leads to PHB accumulation.
Experimental results for PHB production using feeding strategy in BioRC1e portrayed that C/N ratio 10 was crucial for the maximum productivity of PHB. The heat (Q) too clearly depicts the metabolic growth state of the culture during the fed-batch mode. In the growth phase, BioRC1e depicted an increase in instantaneous heat values exponentially. Instantaneous heat values were constant over the period in the fed-batch stage which described the energetics of the co-culture during fed-batch mode which was highly energy conserving and directly related to the energy storage product PHB. Heat depleted from 28 h of growth indicated that the growth stage follows decline phase. This actual metabolic state fingerprinting through heat using a biocalorimeter makes them useful for high-throughput monitoring and control of the bioprocesses. Thus, heat measurements can be used as an effective tool for in situ monitoring of the biological process and make them inevitable even at the industrial level. OUR and CER data reiterates the microbial viability of co-culture aerobic behavior during fed-batch process.
Recent researches using co-culture suggested that the yield of PHB improved when the fed-batch strategy was adapted. Colombo et al. (2017) researched polyhydroxyalkanoates (PHA) accumulation using consortia of microbial strains in a 100-L anaerobic percolated bio-cell reactor with organic fraction of municipal solid waste (OFMSW) as substrate. PHA product yield is 114.4 ± 14.5 g/kg of OFMSW (PHB yield, 60.63 g/kg since PHB/PHV were in the ratio 53/47). Pepè Sciarria et al. (2018) obtained polyhydroxyalkanoates (PHA) yield of 0.77 ± 0.18 mmol CPHA mmol−1 Cfed using mixed population of microbes by aerobic feeding process of recalcitrant of CO2. The CO2 in the process of microbial electro-synthesis was converted to acetate and butyrate by microbial sources; finally, the feed CO2 was converted into bio-plastic PHB. When compared with reported literature, Dias et al. (2005) found PHB productivity to be 1.29 g L−1 h−1. In our study, we achieved 1.89 ± 0.09 g L−1 h−1 PHB productivity during fed-batch stage (10th–26th h) and overall productivity for entire period (0–28 h) achieved was 1.1 ± 0.05 g L−1 h−1. Ahn et al. (2001) achieved the highest fed-batch productivity of 4.6 g L−1 h−1 by using whey as the substrate. Industrially it is not a cost-effective process to use whey as the carbon source. The drawback in their study is that natural carbon sources cannot be reliably replicated.
In the modified Tessier model, the experimental PHB concentration matches close to the model-predicted PHB concentration. We can arrive at further insights into the changes occurring in the metabolic processes. The estimated growth-associated kinetic constant (k4) indicates that the product formation was predominantly influenced by the rate of increase of cell concentration (Table 2). It was also observed that the heat yield coefficient for the maintenance of active residual cells during the fed-batch stage is negative. The modified Tessier equations incorporating the Luedeking-Piret equation (Luedeking and Piret 1959) and Mou and Cooney (1976) expressions and a better understanding of the relationship between the heat to the growth, PHB, and the cell maintenance were established. Thus, it established the fact that heat signals provide greater insight into the bioprocess happening of the system during PHB production when monitored through a biocalorimeter.
Türker (2004) tried to estimate biomass concentration using elemental, electron, and heat balances, and he found that heat balance gave a good accuracy in determining biomass concentration compared with elemental and electron balances. Santharam et al. (2017) predicted the product—inulinase (extracellular enzyme)—through heat data by incorporating the Luedeking-Piret equation (Luedeking and Piret 1959). The advantages of using a heat-based model over other conventional models like Tessier are that the heat-based model derives data from instantaneous heat measurements and through non-invasive methods.
In the heat-based model, from the instantaneous metabolic heat signals of the experiment along with the known parameters (α, β, YQ/X), online PHB prediction was established in this study. This method does not alter the reaction environment and hence leads to greater accuracy. Heat-based model prevents human error. The iControl software did the online data collection, thereby decreasing the human intervention in data collection. Also, heat-based model provides computational simplicity. Other models require the solving of several complex simultaneous differential equations, and these equations have to be curve fitted to experimental data to find α and β (heat-based), whereas in the case of the heat-based model, it is required only to solve a single differential equation, thus retaining the accuracy.
The mathematical modeling approach dealt with in this study could successfully predict the PHB concentrations both in offline and online modes. The offline mode employs a modified Tessier model. The heat-based model describes the PHB prediction online. The heat-based model prediction of PHB online using Eq. 27, suggested that the PHB formation in the culture is the growth-associated process. This model validated with new experimental data. Through the modeling exercise, our studies have demonstrated the use of the online instantaneous metabolic heat alone to monitor the bioprocess and control the reaction. Metabolic heat is the net result of biomass growth, product formation, and cell maintenance, as described in our model Eq. 20. The model Eq. 27 can be used to predict the PHB concentration online. Based on the instantaneous calorimetric response, it is possible to feed the required nutrients for a constant C/N. If we do not maintain the C/N, we have observed a drop in heat and the PHB concentration. Thus, the reactor controlled for optimal production of PHB.
To improve the yield of a PHB, which is sensitive to nutrient factors (excess or deficiency), it is necessary to optimize the concentration of the most critical nutrients. Here, we optimized the carbon and nitrogen ratio in the broth and achieved high PHB yield by the specific microbial consortia. Further, the model developed in this study could help predict PHB product concentration. The Nash-Sutcliffe efficiency described the goodness of the model. Here, we have compared our experimental results with the model-predicted concentration of PHB and cell dry mass to prove that heat-based correlations is a feasible one. The heat-based model demonstrated a significantly shorter execution time and better conformity because of the consideration of the online parameter heat.
References
Ahn WS, Park SJ, Lee SY (2001) Production of poly (3-hydroxybutyrate) from whey by cell recycle fed-batch culture of recombinant Escherichia coli. Biotechnol Lett 23:235–240. https://doi.org/10.1023/A:1005633418161
Anusha SM, Leelaram S, Surianarayanan M (2016) Production of poly (3-hydroxybutyric acid) by Ralstonia eutropha in a biocalorimeter and its thermokinetic studies. Appl Biochem Biotechnol 179:1–19. https://doi.org/10.1007/s12010-016-2049-0
Arias DM, Uggetti E, García-Galán MJ, García J (2018) Production of polyhydroxybutyrates and carbohydrates in a mixed cyanobacterial culture: effect of nutrients limitation and photoperiods. New Biotechnol 42:1–11. https://doi.org/10.1016/j.nbt.2018.01.001
Battley EH (1998) The development of direct and indirect methods for the study of the thermodynamics of microbial growth. Thermochim Acta 309:17–37. https://doi.org/10.1016/S0040-6031(97)00357-2
Beyenal H, Chen SN, Lewandowski Z (2003) The double substrate growth kinetics of Pseudomonas aeruginosa. Enzym Microb Technol 32:92–98. https://doi.org/10.1016/S0141-0229(02)00246-6
Braunegg G, Sonnleitner B, Lafferty RM (1978) A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol 6:29–37. https://doi.org/10.1007/BF00500854
Clarke KG (2013) Microbial kinetics during batch, continuous and fed-batch processes, Bioprocess engineering, an introductory engineering and life science approach, Woodhead Publishing, pp. 97–146. doi:https://doi.org/10.1533/9781782421689.97
Colombo B, Favini F, Scaglia B, Sciarria TP, D’Imporzano G, Pognani M, Alekseeva A, Eisele G, Cosentino C, Adani F (2017) Enhanced polyhydroxyalkanoate (PHA) production from the organic fraction of municipal solid waste by using mixed microbial culture. Biotechnol Biofuels 10:1–15. https://doi.org/10.1186/s13068-017-0888-8
Dias JML, Serafim LS, Lemos PC, Reis MAM, Oliveira R (2005) Mathematical modelling of a mixed culture cultivation process for the production of polyhydroxybutyrate. Biotechnol Bioeng 92:209–222. https://doi.org/10.1002/bit.20598
Dietrich K, Dumont M, Del LF, Orsat V (2019) Sustainable PHA production in integrated lignocellulose biorefineries. New Biotechnol 49:161–168. https://doi.org/10.1016/j.nbt.2018.11.004
Doran PM (1995) Bioprocess engineering principles. Academic Press, London, p 439
Freches A, Lemos PC (2017) Microbial selection strategies for polyhydroxyalkanoates production from crude glycerol : effect of OLR and cycle length. New Biotechnol 39:22–28. https://doi.org/10.1016/j.nbt.2017.05.011
Gerson DF, Kole MM, Ozum B (1988) Substrate concentration control in bioreactors. Biotechnol Genet Eng Rev 6:67–105. https://doi.org/10.1080/02648725.1988.10647846
Hrnčiřík P, Moucha T, Mareš J, Náhlík J, Janáčová D (2019) Software sensors for biomass concentration estimation in filamentous microorganism cultivation process. Chem Biochem Eng Q 33:141–151. https://doi.org/10.15255/cabeq.2018.1387
Huang L, Chen Z, Wen Q, Lee D (2017) Enhanced polyhydroxyalkanoate production by mixed microbial culture with extended cultivation strategy. Bioresour Technol 241:802–811. https://doi.org/10.1016/j.biortech.2017.05.192
Kampen WH (2014) Nutritional requirements in fermentation processes. In: Vogel HC, Todaro CM (eds) Fermentation and biochemical engineering handbook (Third Edition). William Andrew Publishing, Boston, pp 37–57. https://doi.org/10.1016/B978-1-4557-2553-3.00004-0
Kanzow C, Yamashita N, Fukushima M (2004) Levenberg-Marquardt methods with strong local convergence properties for solving nonlinear equations with convex constraints. J Comput Appl Math 172:375–397. https://doi.org/10.1016/j.cam.2004.02.013
Khanna S, Srivastava AK (2005) Statistical media optimization studies for growth and PHB production by Ralstonia eutropha. Process Biochem 40:2173–2182. https://doi.org/10.1016/j.procbio.2004.08.011
Kunasundari B, Arza CR, Maurer FHJ, Murugaiyah V, Kaur G, Sudesh K (2017) Biological recovery and properties of poly(3-hydroxybutyrate) from Cupriavidus necator H16. Sep Purif Technol 172:1–6. https://doi.org/10.1016/j.seppur.2016.07.043
Law JH, Slepecky RA (1961) Assay of poly-β-hydroxybutyric acid. J Bacteriol 82:33–36
Luedeking R, Piret EL (1959) A kinetic study of the lactic acid fermentation. Batch process at controllet pH. J Biochem Microbiol Technol Eng 1:393–412. doi: https://doi.org/10.1002/jbmte.390010406
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. https://doi.org/10.1021/ac60147a030
Moriasi DN, Arnold JG, Van Liew MW, Bingner RL, Harmel RD, Veith TL (2007) Model evaluation guidelines for systematic quantification of accuracy in watershed simulations. Transactions of the ASABE 50:885–900. https://doi.org/10.13031/2013.23153
Mou DG, Cooney CL (1976) Application of dynamic calorimetry for monitoring fermentation processes. Biotechnol Bioeng 18:1371–1392. https://doi.org/10.1002/bit.260181005
Mozumder MSI, De Wever H, Volcke EIP, Garcia-Gonzalez L (2014a) A robust fed-batch feeding strategy independent of the carbon source for optimal polyhydroxybutyrate production. Process Biochem 49:365–373. https://doi.org/10.1016/j.procbio.2013.12.004
Mozumder MSI, Goormachtigh L, Garcia-Gonzalez L, De Wever H, Volcke EIP (2014b) Modeling pure culture heterotrophic production of polyhydroxybutyrate (PHB). Bioresour Technol 155:272–280. https://doi.org/10.1016/j.biortech.2013.12.103
Mulchandani A, Luong JHT, Groom C (1989) Applied microbiology biotechnology substrate inhibition kinetics for microbial growth and synthesis of poly-β-hydroxybutyric acid by Alcaligenes eutrophus ATCC 17697. Appl Microbiol Biotechnol 30:11–17. https://doi.org/10.1007/bf00255990
Pepè Sciarria T, Batlle-Vilanova P, Colombo B, Scaglia B, Balaguer MD, Colprim J, Puig S, Adani F (2018) Bio-electrorecycling of carbon dioxide into bioplastics. Green Chem 20:4058–4066. https://doi.org/10.1039/c8gc01771a
Pérez Rivero C, Sun C, Theodoropoulos C, Webb C (2016) Building a predictive model for PHB production from glycerol. Biochem Eng J 116:113–121. https://doi.org/10.1016/j.bej.2016.04.016
Redl B, Tiefenbrunner F (1981) Determination of hydrolytic activities in wastewater systems by microcalorimetry. Water Res 15:87–90. https://doi.org/10.1016/0043-1354(81)90187-1
Richhardt J, Bringer S, Bott M (2013) Role of the pentose phosphate pathway and the Entner-Doudoroff pathway in glucose metabolism of Gluconobacter oxydans 621H. Appl Microbiol Biotechnol 97:4315–4323. https://doi.org/10.1007/s00253-013-4707-2
Santharam L, Samuthirapandi AB, Easwaran SN, Mahadevan S (2017) Modeling of exo-inulinase biosynthesis by Kluyveromyces marxianus in fed-batch mode: correlating production kinetics and metabolic heat fluxes. Appl Microbiol Biotechnol 101:1877–1887. https://doi.org/10.1007/s00253-016-7971-0
Schuler MM, Sivaprakasam S, Freeland B, Hama A, Hughes K-M, Marison IW (2012) Investigation of the potential of biocalorimetry as a process analytical technology (PAT) tool for monitoring and control of Crabtree-negative yeast cultures. Appl Microbiol Biotechnol 93:575–584. https://doi.org/10.1007/s00253-011-3507-9
Shong J, Rafael M, Diaz J, Collins CH (2012) Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol 23:798–802. https://doi.org/10.1016/j.copbio.2012.02.001
Solórzano L (1969) Determination of ammonia in natural waters by the phenol hypochlorite method. Limnol Oceanogr 14:799–801. https://doi.org/10.4319/lo.1969.14.5.0799
Špoljarić IV, Lopar M, Koller M, Muhr A, Salerno A, Reiterer A, Malli K, Angerer H, Strohmeier K, Schober S, Mittelbach M, Horvat P (2013) Mathematical modeling of poly[(R)-3-hydroxyalkanoate] synthesis by Cupriavidus necator DSM 545 on substrates stemming from biodiesel production. Bioresour Technol 133:482–494. https://doi.org/10.1016/j.biortech.2013.01.126
Suzuki T, Yamane T, Shimizu S (1986) Mass production of poly-β-hydroxybutyric acid by fed-batch culture with controlled carbon/nitrogen feeding. Appl Microbiol Biotechnol 24:370–374. https://doi.org/10.1007/BF00294592
Tsouko E, Papanikolaou S, Koutinas AA (2016) Production of fuels from microbial oil using oleaginous microorganisms. In: Luque R, Lin CSK, Wilson K, Clark J (eds) Handbook of Biofuels Production (Second Edition). Woodhead Publishing, pp 201–236. https://doi.org/10.1016/B978-0-08-100455-5.00008-4
Türker M (2004) Development of biocalorimetry as a technique for process monitoring and control in technical scale fermentations. Thermochim Acta 419:73–81. https://doi.org/10.1016/j.tca.2004.01.036
Verlinden RAJ, Hill DJ, Kenward MA, Williams CD, Radecka I (2007) Bacterial synthesis of biodegradable polyhydroxyalkanoates. J Appl Microbiol 102:1437–1449. https://doi.org/10.1111/j.1365-2672.2007.03335.x
Voisard D, von Stockar U, Marison IW (2002) Quantitative calorimetric investigation of fed-batch cultures of Bacillus sphaericus 1593M. Thermochim Acta 394:99–111. https://doi.org/10.1016/S0040-6031(02)00243-5
von Stockar U, Marison I, Janssen M, Patiño R (2011) Calorimetry and thermodynamic aspects of heterotrophic, mixotrophic, and phototrophic growth. J Therm Anal Calorim 104:45–52. https://doi.org/10.1007/s10973-010-1278-7
Acknowledgments
One of the authors (Anusha SM) wishes to acknowledge the Council of Scientific & Industrial Research (CSIR), New Delhi, for the CSIR-GATE fellowship. The authors express their gratitude to Prof. NR Rajagopal for continuous support. The authors thank Mr. Saravana Raj Adimoolam for valuable and helpful scientific discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 919 kb)
Rights and permissions
About this article
Cite this article
Subramanian, A.M., Nanjan, S.E., Prakash, H. et al. Biokinetics of fed-batch production of poly (3-hydroxybutyrate) using microbial co-culture. Appl Microbiol Biotechnol 104, 1077–1095 (2020). https://doi.org/10.1007/s00253-019-10274-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10274-7