Abstract
The recent drop in the price of natural gas has rekindled the interests in methanotrophs, the organisms capable of utilizing methane as the sole electron donor and carbon source, as biocatalysts for various industrial applications. As heterologous expression of the methane monooxygenases in more amenable hosts has been proven to be nearly impossible, future success in methanotroph biotechnology largely depends on securing phylogenetically and phenotypically diverse methanotrophs with relatively high growth rates. For long, isolation of methanotrophs have relied on repeated single colony picking after initial batch enrichment with methane, which is a very rigorous and time-consuming process. In this review, three unconventional isolation methods devised for facilitation of the isolation process, diversification of targeted methanotrophs, and/or screening of rapid growers are summarized. The soil substrate membrane method allowed for isolation of previously elusive methanotrophs and application of high-throughput extinction plating technique facilitated the isolation procedure. Use of a chemostat with gradually increased dilution rates proved effective in screening for the fastest-growing methanotrophs from environmental samples. Development of new isolation technologies incorporating microfluidics and single-cell techniques may lead to discovery of previously unculturable methanotrophs with unexpected metabolic potentials and thus, certainly warrant future investigation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Aerobic methanotrophs are a specialized group of organisms capable of utilizing methane as the sole source of energy and carbon (Hanson and Hanson 1996; Semrau et al. 2010). Until recently, only proteobacterial methanotrophs, belonging to either Alpha- or Gamma-proteobacteria class, were known; however, the discovery of verrucomicrobial methanotrophs has expanded the methanotroph diversity (Dunfield et al. 2007; Islam et al. 2008; Sharp et al. 2014). The list of microbial taxa (families) with physiologically confirmed methanotrophs now includes Methylococcaceae, Crenothrichaceae, Methylcystaceae, Beijerinckiaceae, and Methylacidiphilaceae (Semrau et al. 2010). Despite the broad phylogenetic and physiological diversity, these methanotrophs share the distinguishing physiological capability to catalyze the oxidation of methane (CH4) to methanol (CH3OH) using either particulate (pMMO) or soluble methane monooxygenases (sMMO).
These two MMOs have identical metabolic function in methanotrophs, but are structurally and evolutionarily distinct groups of metalloenzymes. The more prevalent form, pMMO, is a membrane-bound enzyme with copper in the active site, while sMMO, found only in selected groups of methanotrophs, is a cytoplasmic enzyme with a di-iron active center (Semrau et al. 2010, 2018). The pMMOs share evolutionary history with ammonia monooxygenases (AMO) and thus, are structurally similar to these cuproenzymes catalyzing ammonia (NH3) turnover to hydroxylamine (NH2OH) in ammonia oxidizing bacteria and archaea (Murrell et al. 2000a, b; Culpepper and Rosenzweig 2012; Hatzenpichler 2012). The sMMO are evolutionarily related to soluble di-iron monooxygenases and share modest structural similarity with toluene monooxygenases and phenol hydroxylases (Leahy et al. 2003; Coleman et al. 2006).
Methanotrophs harboring both pMMO(s) and sMMO(s) are not rare among cultured methanotrophic isolates; however, methanotrophs with only pMMO are the vast majority among methanotrophic isolates (Osborne and Haritos 2018). Methanotrophs possessing only sMMO fall within the Beijerinckiaceae family and recent researches have found these organisms dominating the methanotrophic populations in specialized ecological niches such as natural gas seeps and microbial mats of Movile Cave (Farhan Ul Haque et al. 2018; Kumaresan et al. 2018). Extrapolating the observations from the experiments with the methanotrophs possessing both pMMO and sMMO, it is now widely accepted that availability of bioavailable copper regulates the expression and activity of the MMOs (Semrau et al. 2010, 2018).
The methanotrophs, as the sole possessor of the MMOs, have drawn immense interests for their potential biotechnological applications; however, successful cases of industrial implementation have rarely been reported, due to several bottlenecks, one of them being the limited physiological diversity of isolated strains. In this review, the reasons why novel isolation techniques are essential for successful implementation of methanotroph biotechnology are explained and three novel approaches for methanotroph isolation are summarized.
The value of methanotrophs in biotechnology
The capability to wield the methane monooxygenases for conversion of CH4 to CH3OH are restricted exclusively to the aforementioned groups of methanotrophs. Several attempts to express sMMOs in heterologous hosts failed to reproduce the high CH4 turnover efficiency observed in methanotrophs, and no successful heterologous expression of pMMO has been reported to date (Jahng et al. 1996; Jahng and Wood 1994; Kalyuzhnaya et al. 2015; Murrell et al. 2000a; Strong et al. 2015). Due to insufficient knowledge on how active sMMO and pMMO complexes are formed in vivo, generating an artificial methanotroph from model laboratory organisms more amenable to manipulation (e.g., Escherichia coli, and Methylobacterium extorquens) remains elusive.
Due to this exclusive ownership of fully active MMOs, the aerobic methanotrophs have for long attracted interests for their potential utility in biotechnological applications. As both sMMO and pMMO are capable of co-oxidizing a broad range of xenobiotic organic compounds, including carcinogenic halogenated compounds and organic micropollutants, methanotrophs have been investigated for their potential application in in situ bioremediation of contaminated soils and groundwater (Benner et al. 2015; Little et al. 1988; Semrau 2011; Semrau et al. 2010). More recently, CH4 itself is considered as a target for bioremediation due to its high global warming potential (34 times that of CO2 in a 100-year scale), and various bioreactor configurations have been devised to reduce CH4 emissions from point sources, e.g., landfills and animal feeding operations, utilizing methanotrophic consortia (Ganendra et al. 2015; Limbri et al. 2014; Yoon et al. 2009). The ability of methanotrophs to mineralize and detoxify methyl-mercury has also been recently investigated, suggesting that methanotrophs may have value in detoxifying such toxic heavy metals (Lu et al. 2017). Production of various value-added chemical compounds using CH4 has also been a widely investigated research topic, as CH4 has been an attractive feedstock due to its low price and high availability, especially since the onset of the recent shale gas boom (Haynes and Gonzalez 2014; Strong et al. 2015). Single-cell protein for livestock feeds have been produced with gammaproteobacterial methanotrophs for decades, and utilization of alphaproteobacterial methanotrophs for polyhydroxy-alkanoate (PHA) production have been investigated as a biodegradable alternative for plastics (Kalyuzhnaya et al. 2015; Marshall et al. 2013; Pieja et al. 2017; Strong et al. 2016). Lately, genetic modifications of biosynthetic pathways in methanotrophs for conversion of CH4 to feedstock chemicals, e.g., CH3OH, organic acids, and fatty acids, or liquid fuels have been rigorously investigated in both academic and industrial sectors (Conrado and Gonzalez 2014; Hwang et al. 2018).
These potential industrial applications of methanotrophs capitalize on a variety of physiological traits that are not commonly shared by different methanotroph groups. For example, the capability to detoxify methyl mercury is limited to the selective group of methanobactin-producing alphaproteobacterial methanotrophs, while only fast-growing gammaproteobacterial strains, e.g., Methylococcus spp., proved profitable for single-cell protein production industry. As alpha- and gammaproteobacterial methanotrophs utilize distinct carbon assimilation and biosynthetic pathways, one pathway is favored over the other when a specific chemical is targeted as the product of metabolic engineering (Kalyuzhnaya et al. 2015). The ribulose monophosphate cycle in gammaproteobacterial methanotrophs concentrates the carbon flux to pyruvate synthesis via the Embden-Meyerhof-Parnas (EMP) pathway, thus favoring the production of chemical compounds branching from pyruvate, while the serine cycle in alphaproteobacterial methanotrophs is thought to be favorable for biofuel production due to the higher flux to acetyl-CoA. Theoretically, genetic manipulation would be able to fill such missing gaps in the pathways leading to desired processes or products once fast and efficient genetic tools are established in tractable methanotrophic strains; however, such approach has only been modestly successful (Henard et al. 2016).
Although synthetic biology approaches appear to be unlikely for biotechnological application of methanotrophs in the foreseeable future, the attractiveness of the aerobic methanotrophs as methane-transforming biocatalysts is difficult to ignore. Hence, the more realistic strategy for their biotechnological applications is to utilize methanotrophs without or with minimal genetic modifications. Therefore, having an inventory of methanotrophs with diverse traits and physiological properties (e.g., ownership of sMMO, resistance to change in pH and/or salinity, high/low carbon conversion efficiency and methanobactin production) and distinct biosynthetic metabolic pathways would be pertinent to meet biotechnological demands. General relevant traits necessary for potential industrial methanotrophs include rapid growth and high CH4 utilization rate, which would allow for faster population establishment and rapid metabolic turnover of CH4 and cometabolites, and facilitate genetic manipulation procedures should genetic manipulation be necessary (Jiang et al. 2010; Puri et al. 2015; Rocha-Rios et al. 2011; Rostkowski et al. 2013; Yan et al. 2016). Methanotrophs with relatively high growth rates, with specific growth rates higher than 0.1 h−1, have been reported (Table 1); however, the diversity of methanotrophs with such high growth rates under unrestricted access to methane is limited and most of these fast-growing methanotrophs are phylogenetically positioned within the family Methylococcaceae (Ho et al. 2013; Semrau et al. 2010). Therefore, an expansion in the library of fast-growing methanotrophs with a broader phylogenetic and metabolic spectrum would be necessary to facilitate implementation of methanotrophic biotechnology.
Methanotroph isolation techniques
The traditional method for isolating microorganisms, including the methanotrophs, has remained largely unchanged for over 50 years. The majority of publicly available methanotroph cultures were isolated from enrichment of environmental samples in liquid medium supplemented with CH4 in the headspace gas, followed by repeated single-colony picking on agar plates (Dunfield et al. 2003; Kip et al. 2011; Lidstrom 1988; Wise et al. 1999; Zúñiga et al. 2011). Variation in medium compositions and incubation conditions enabled isolation of several unconventional methanotrophs (Dedysh 2011; Dunfield et al. 1999, 2003, 2007; Islam et al. 2008). The use of diluted media, e.g., diluted nitrate mineral salt medium (DNMS) with five-fold lower concentrations of salts, enabled isolation of acidophilic Beijerinckiaceae family of methanotrophs including Methylocella, Methylocapsa, and Methyloferula (Dunfield et al. 2003, 2010; Vorobev et al. 2011). Incubation at elevated temperatures and acidic pH and addition of CO2 to the headspace enabled isolation of the verrucomicrobial methanotrophs (Dunfield et al. 2007; Islam et al. 2008).
A major drawback of this isolation approach is extended time and rigorous effort needed for isolation, which limit the quantity and diversity of the strains isolated. Methanotrophs, even in cultures enriched with CH4 as the sole carbon substrate, tend to form aggregates with methylotrophs, as well as other heterotrophs living on the metabolites exuded by the methanotrophs (Ho et al. 2016; Kalyuzhnaya et al. 2013; Kim et al. 2018; Oshkin et al. 2015). In such cases, repetitions of spreading-and-single-colony-picking may prolong the isolation effort to yield cultures devoid of contaminant non-methanotrophic strains. The isolation procedure typically selects for abundant fast-growing methanotrophs, and rarely captures methanotrophs present in relatively low abundances (even if these are fast-growers) or slow-growers in environmental samples (Fig. 1). These inadequacies and isolation biases call for development of novel isolation techniques that can expedite the isolation procedure and target the organisms that were less likely to be isolated with the conventional isolation technique (Hoefman et al. 2012; Kim et al. 2018; Svenning et al. 2003).
The simulated growth curves of methanotrophs with varying growth rates and initial abundances, assuming incubation in 50-mL medium in 160-mL serum vial with limited amount of methane and air (20% v/v and 80% v/v, respectively, in the headspace). An identical growth yield of 0.94 (g dry cell weigh per g CH4) was assumed for all methanotrophs (Mahmoud 2017). The simulation result illustrates that initial enrichment with methane that precedes conventional isolation procedure selectively enriches only abundant and fast-growing methanotrophs. Neither minor fastest-growers nor slow-growers dominant in the original environmental sample is enriched, and thus, such organisms are likely to be excluded from isolation targets in the downstream procedures. Io: initial abundance (cells mL−1); Gr: growth rate (h−1). (A) Io: 8 × 107, Gr: 0.4; (B) Io: 8 × 107, Gr: 0.3; (C) Io: 4 × 108, Gr: 0.4; (D) Io: 8 × 108, Gr: 0.4; (E) Io: 4 × 108, Gr: 0.05; (F) Io: 1.6 × 109, Gr: 0.05; (G) Io: 8 × 106, Gr: 0.6
Increasing the throughput of the oft-used extinction culturing technique was suggested as a methodology to expedite methanotroph isolation procedures (Hoefman et al. 2012). Extinction culturing (i.e., isolation using serial dilution series) of 46 CH4-fed enrichments was performed simultaneously with multiple 96-well plates in a high-throughput manner, with 10−2-to-10−9 dilution series prepared for each enrichment (Fig. 2A). The most diluted samples exhibiting growth were then evaluated for CH4 oxidation capability and purity. With a single round of experiment, 14 monocultures of methanotrophs were obtained immediately following the extinction culturing and 8 more isolates were obtained after further purification steps. The initial enrichment step with 20% CH4 appeared to have selected towards isolation of rapid-growing methanotrophs; however, the methanotrophs targeted by this method were essentially the same as those targeted by the conventional isolation technique: fast-growing organisms that are relatively abundant in the environmental samples.
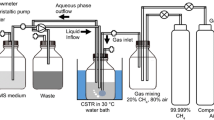
The schematic overview of the unconventional methods developed for the isolation of methanotrophs. (A) The high-throughput extinction cultivation; (B) the soil substrate membrane system; and (C) the continuously stirred tank reactor (CSTR) screening method for isolation of fast-growing methanotrophs. Further isolation from the resulting single colonies may be necessary to obtain pure cultures. The figures were adapted from the references cited in the text. aFirst with the dotted colonies transferred from 0.4-μm membranes and then with serial dilutions of the single colonies developed on the 0.2-μm membranes after the transfer
The isolation technique using soil substrate membrane system (SSMS), originally developed and applied for isolating methanotrophs (Svenning et al. 2003), shortened the time needed for isolation and yielded more diverse isolates than the conventional method. The initial processing of environmental samples in the SSMS procedure includes extraction of microbial consortium from an environmental sample and its serial dilution (Fig. 2B). The serially diluted samples were applied initially on 0.4-μm polycarbonate membranes, which were then inserted to petri dishes carrying soil slurry (soil suspended in MilliQ water) providing micronutrients to the microorganisms on the membranes. The colonies developed on the membranes were picked and dotted onto membranes with 0.2-μm pore size. The colonies on the 0.2-μm membrane were serially diluted and the dilutions were incubated on identically prepared soil membrane system. After the second membrane growth of the diluted colonies, the obtained cultures were confirmed against heterotroph contamination by floating the membrane on nutrient-rich medium. The SSMS technique enabled simultaneous isolation of gammaproteobacterial and alphaproteobacterial methanotrophs with remarkable diversity within a few months. The drawback of the method in isolating potential biotechnologically applicable methanotrophs, however, is that additional screening for fast-growers would be necessary as the method does not specifically select for fast-growing microorganisms. Although the SSMS technique was further developed into a more general technique for isolation of bacterial groups resistant to culturing, further improvement or application of this methodology for isolation of methanotrophs has not been reported (Ferrari et al. 2005, 2008).
In contrast to the SSMS method which accentuates the diversity of acquired isolates, the recently demonstrated isolation technique utilizing a continuous cultivation system aimed to select the fastest-growing methanotroph from an environmental sample (Kim et al. 2018). This isolation method was based on the fundamental chemostat principle, that is, only organisms with growth rates higher than the dilution rates would be retained in the chemostat at steady-state (Jannasch 1969). After initial batch incubation of an environmental sample with CH4 as the sole source of carbon and energy, the enriched methane-oxidizing consortium was incubated in a fed-batch reactor with continuous flow of the gas phase (80:20 mixture of air and CH4 provided at 1.0 mL min−1) for further enrichment of fast-growing methanotrophs (Fig. 2C). The reactor was eventually converted to a chemostat with the initial dilution rate of 0.1 h−1 and the dilution rate was gradually increased with 0.05 h−1 increments to screen out methanotrophs with a slower growth rate or contaminating heterotrophs and methylotrophs. The reactor sample collected from the highest dilution rate before washout was selected for further purification by repeated single colony selections. In less than 2 months, the method successfully isolated a methanotrophic strain affiliated to the genus Methylomonas (originating from a stream sediment) with a specific growth rate of 0.4 h−1, a remarkably high value for growth on methane (Kim et al. 2018).
The feasibility of this isolation method has only been tested under prototypical incubation conditions for optimal methanotrophic growth, i.e., neutral pH, optimal CH4:air ratio (20:80), 10 mM NO3− as the nitrogen source, and sufficient copper (10 μM). Incubation conditions modified to exert selection pressures towards specific phenotypes, e.g., the absence of copper endowing selective advantage to sMMO-expressing organisms or the absence of dissolved N source selecting for nitrogen-fixing methanotrophs, as well as use of diverse environmental samples as the initial inocula, may allow for diversification of target isolates (Auman et al. 2001; Murrell et al. 2000b). Lanthanides (e.g., cerium, lanthanum, and praseodymium) were found to promote methanol oxidation and growth by the methanotrophs with Xox-type methanol dehydrogenases (Xox-MeDH) and thus, variation of lanthanide compositions in the media may also provide additional selection for methanotrophs relying exclusively on Xox-MeDH (Semrau et al. 2018). This enrichment/screening methodology would also be suitable for enriching methanotrophs capable of utilizing low methane concentrations (e.g., < 100 ppmv). Such methanotrophs have potential applicability in CH4 removal systems for landfills and cattle barns, as CH4 emitted from such sources rarely exceed 100 ppmv in concentration (Joo et al. 2015; Ngwabie et al. 2009; Yoon et al. 2009).
Conclusion and outlook
Methanotrophs have recently regained immense interests as the biocatalysts for industrial applications. Although securing of fast-growing isolates with diverse metabolic pathways and physiological properties is a prerequisite for successful biotechnological applications of methanotrophs, surprisingly few advances have been made to improve isolation techniques in the past decades. The unique isolation approaches reviewed here, may not be sufficiently optimized, although these methods have proved their potential. Development of novel isolation techniques for methanotrophs may also take advantage of the recent scientific advances in microfluidics, cell imaging, and Raman microspectroscopy techniques (Hol and Dekker 2014; Ishii et al. 2010; Ma et al. 2014; Song et al. 2016). Several cases of successful implementation of microfluidics-based single-cell techniques to isolate as yet “uncultivable” microorganisms have been recently reported (Jiang et al. 2016; Zhang et al. 2014). Applying these state-of-the-art techniques to methanotroph isolation may yield a substantial expansion of the current methanotroph inventory. Crenothrix spp., the enigmatic filamentous gammaproteobacterial methanotrophs, recently confirmed as major CH4 sinks in stratified lakes, have evaded isolation for more than a century since its initial discovery (Oswald et al. 2017; Stoecker et al. 2006). As these methanotrophs were observed to actively incorporate 13C-labeled CH4 to their biomass, these evasive organisms may be able to be isolated with help of Raman microspectroscopy and microfluidics (and/or optical tweezer) techniques (Wang et al. 2016).
Accumulating evidence suggests a synergistic effect of methanotroph and non-methanotroph interaction in modulating methane oxidation (Ho et al. 2014). To this end, the concerted effort of a methanotroph paired to non-methanotrophic microorganisms have been shown to significantly stimulate methane oxidation rates (Ho et al. 2014). Given that many methanotrophs resist cultivation, but could be highly enriched with specific accompanying community members (Oshkin et al. 2015) indicate an intricate relationship likely driven by metabolic exchange (Oshkin et al. 2015; Krause et al. 2017). This begs the question whether it is necessary to isolate individual methanotrophs, or capitalize on naturally occurring enriched methane-oxidizing consortia for biotechnological purposes. However, previous lessons from successful industrial implementations of microbial biotechnology, e.g., bioremediation of chlorinated ethene-contaminated soils with Dehalococcoides consortia, suggest that the discovery of new microorganisms may lead to unexpected breakthroughs, resolving long-standing dilemmas (He et al. 2003; Löffler et al. 2013). Thus, the development of isolation and culturing techniques targeting elusive methanotrophs warrant continued attention and efforts for successful implementation of methanotroph biotechnology.
References
Auman AJ, Speake CC, Lidstrom ME (2001) nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl Environ Microbiol 67:4009–4016. https://doi.org/10.1128/aem.67.9.4009-4016.2001
Benner J, de Smet D, Ho A, Kerckhof F, Vanhaecke L, Heylen K, Boon N (2015) Exploring methane-oxidizing communities for the co-metabolic degradation of organic micropollutants. Appl Microbiol Biotechnol 99:3609–3618. https://doi.org/10.1007/s00253-014-6226-1
Coleman NV, Bui NB, Holmes AJ (2006) Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol 8:1228–1239. https://doi.org/10.1111/j.1462-2920.2006.01015.x
Conrado R, Gonzalez R (2014) Envisioning the bioconversion of methane to liquid fuels. Science 343:621–623. https://doi.org/10.1126/science.1246929
Culpepper MA, Rosenzweig AC (2012) Architecture and active site of particulate methane monooxygenase. Crit Rev Biochem Mol Biol 47:483–492. https://doi.org/10.3109/10409238.2012.697865
Dedysh S (2011) Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front Microbiol 2:184. https://doi.org/10.3389/fmicb.2011.00184
Dunfield PF, Liesack W, Henckel T, Knowles R, Conrad R (1999) High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl Environ Microbiol 65:1009–1014
Dunfield PF, Khmelenina VN, Suzina NE, Trotsenko YA, Dedysh SN (2003) Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int J Syst Evol Microbiol 53:1231–1239. https://doi.org/10.1099/ijs.0.02481-0
Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M (2007) Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882. https://doi.org/10.1038/nature06411
Dunfield PF, Belova SE, Vorobev AV, Cornish SL, Dedysh SN (2010) Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int J Syst Evol Microbiol 60:2659–2664. https://doi.org/10.1099/ijs.0.020149-0
Farhan Ul Haque M, Crombie AT, Ensminger SA, Baciu C, Murrell JC (2018) Facultative methanotrophs are abundant at terrestrial natural gas seeps. Microbiome 6:118. https://doi.org/10.1186/s40168-018-0500-x
Ferrari BC, Binnerup SJ, Gillings M (2005) Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol 71:8714–8720. https://doi.org/10.1128/aem.71.12.8714-8720.2005
Ferrari BC, Winsley T, Gillings M, Binnerup S (2008) Cultivating previously uncultured soil bacteria using a soil substrate membrane system. Nat Protoc 3:1261–1269. https://doi.org/10.1038/nprot.2008.102
Ganendra G, Mercado-Garcia D, Hernandez-Sanabria E, Peiren N, De Campeneere S, Ho A, Boon N (2015) Biofiltration of methane from ruminants gas effluent using autoclaved aerated concrete as the carrier material. Chem Eng J 277:318–323. https://doi.org/10.1016/j.cej.2015.04.128
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 60:439–471
Hatzenpichler R (2012) Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol 78:7501–7510. https://doi.org/10.1128/AEM.01960-12
Haynes CA, Gonzalez R (2014) Rethinking biological activation of methane and conversion to liquid fuels. Nat Chem Biol 10:331–339. https://doi.org/10.1038/nchembio.1509
He J, Ritalahti KM, Yang K-L, Koenigsberg SS, Löffler FE (2003) Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65. https://doi.org/10.1038/nature01717
Henard CA, Smith H, Dowe N, Kalyuzhnaya MG, Pienkos PT, Guarnieri MT (2016) Bioconversion of methane to lactate by an obligate methanotrophic bacterium. Sci Rep 6:21585. https://doi.org/10.1038/srep21585
Hirayama H, Suzuki Y, Abe M, Miyazaki M, Makita H, Inagaki F, Uematsu K, Takai K (2011) Methylothermus subterraneus sp. nov., a moderately thermophilic methanotroph isolated from a terrestrial subsurface hot aquifer. Int J Syst Evol Microbiol 61:2646–2653. https://doi.org/10.1099/ijs.0.028092-0
Hirayama H, Fuse H, Abe M, Miyazaki M, Nakamura T, Nunoura T, Furushima Y, Yamamoto H, Takai K (2013) Methylomarinum vadi gen. nov., sp. nov., a methanotroph isolated from two distinct marine environments. Int J Syst Evol Microbiol 63:1073–1082. https://doi.org/10.1099/ijs.0.040568-0
Ho A, Kerckhof F, Luke C, Reim A, Krause S, Boon N, Bodelier PLE (2013) Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ Microbiol Rep 5:335–345. https://doi.org/10.1111/j.1758-2229.2012.00370.x
Ho A, de Roy K, Thas O, De Neve J, Hoefman S, Vandamme P, Heylen K, Boon N (2014) The more, the merrier: heterotroph richness stimulates methanotrophic activity. ISME J 8:1945–1948. https://doi.org/10.1038/ismej.2014.74
Ho A, Angel R, Veraart AJ, Daebeler A, Jia Z, Kim SY, Kerckhof F, Boon N, Bodelier PLE (2016) Biotic interactions in microbial communities as modulators of biogeochemical processes: methanotrophy as a model system. Front Microbiol 7:1285. https://doi.org/10.3389/fmicb.2016.01285
Hoefman S, van der Ha D, De Vos P, Boon N, Heylen K (2012) Miniaturized extinction culturing is the preferred strategy for rapid isolation of fast-growing methane-oxidizing bacteria. Microb Biotechnol 5:368–378. https://doi.org/10.1111/j.1751-7915.2011.00314.x
Hol FJH, Dekker C (2014) Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346:6208. https://doi.org/10.1126/science.1251821
Hwang IY, Nguyen AD, Nguyen TT, Nguyen LT, Lee OK, Lee EY (2018) Biological conversion of methane to chemicals and fuels: technical challenges and issues. Appl Microbiol Biotechnol 102:3071–3080. https://doi.org/10.1007/s00253-018-8842-7
Ishii S, Tago K, Senoo K (2010) Single-cell analysis and isolation for microbiology and biotechnology: methods and applications. Appl Microbiol Biotechnol 86:1281–1292. https://doi.org/10.1007/s00253-010-2524-4
Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland N-K (2008) Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci 105:300–304. https://doi.org/10.1073/pnas.0704162105
Jahng D, Wood TK (1994) Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl Environ Microbiol 60:2473–2482
Jahng D, Kim CS, Hanson RS, Wood TK (1996) Optimization of trichloroethylene degradation using soluble methane monooxygenase of Methylosinus trichosporium OB3b expressed in recombinant bacteria. Biotechnol Bioeng 51:349–359. https://doi.org/10.1002/(SICI)1097-0290(19960805)51:3%3C349::AID-BIT10%3E3.0.CO;2-H
Jannasch HW (1969) Estimations of bacterial growth rates in natural waters. J Bacteriol 99:156–160
Jiang H, Chen Y, Jiang P, Zhang C, Smith TJ, Murrell JC, Xing X-H (2010) Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering. Biochem Eng J 49:277–288. https://doi.org/10.1016/j.bej.2010.01.003
Jiang C-Y, Dong L, Zhao J-K, Hu X, Shen C, Qiao Y, Zhang X, Wang Y, Ismagilov RF, Liu S-J, Du W (2016) High-throughput single-cell cultivation on microfluidic streak plates. Appl Environ Microbiol 82:2210–2218. https://doi.org/10.1128/aem.03588-15
Joo HS, Ndegwa PM, Heber AJ, Ni JQ, Bogan BW, Ramirez-Dorronsoro JC, Cortus E (2015) Greenhouse gas emissions from naturally ventilated freestall dairy barns. Atmos Environ 102:384–392. https://doi.org/10.1016/j.atmosenv.2014.11.067
Kalyuzhnaya MG, Yang S, Rozova ON, Bringel F, Smalley NE, Clubb J, Konopka M, Orphan VJ, Beck D, Trotsenko YA, Vuilleumier S, Khmelenina VN, Lidstrom ME (2013) Highly efficient methane biocatalysis revealed in methanotrophic bacterium. Nat Commun 4:2785. https://doi.org/10.1038/ncomms3785
Kalyuzhnaya MG, Puri AW, Lidstrom ME (2015) Metabolic engineering in methanotrophic bacteria. Metab Eng 29:142–152. https://doi.org/10.1016/j.ymben.2015.03.010
Kim J, Kim DD, Yoon S (2018) Rapid isolation of fast-growing methanotrophs from environmental samples using continuous cultivation with gradually increased dilution rates. Appl Microbiol Biotechnol 102:5707–5715. https://doi.org/10.1007/s00253-018-8978-5
Kip N, Ouyang W, van Winden J, Raghoebarsing A, van Niftrik L, Pol A, Pan Y, Bodrossy L, van Donselaar EG, Reichart G-J, Jetten MSM, Sinninghe Damsté JS, Op den Camp HJM (2011) Detection, isolation, and characterization of acidophilic methanotrophs from sphagnum mosses. Appl Environ Microbiol 77:5643–5654. https://doi.org/10.1128/aem.05017-11
Krause SMB, Johnson T, Karunaratne YS, Fu Y, Beck DAC, Chistoserdova L, Lidstrom ME (2017) Lanthanide-dependent cross-feeding of methane-derived carbon is linked by microbial community interactions. Proc Natl Acad Sci 114:358–363. https://doi.org/10.1073/pnas.1619871114
Kumaresan D, Stephenson J, Doxey AC, Bandukwala H, Brooks E, Hillebrand-Voiculescu A, Whiteley AS, Murrell JC (2018) Aerobic proteobacterial methylotrophs in Movile Cave: genomic and metagenomic analyses. Microbiome 6:1. https://doi.org/10.1186/s40168-017-0383-2
Leahy JG, Batchelor PJ, Morcomb SM (2003) Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev 27:449–479. https://doi.org/10.1016/S0168-6445(03)00023-8
Lidstrom ME (1988) Isolation and characterization of marine methanotrophs. Antonie Leeuwenhoek 54:189–199. https://doi.org/10.1007/bf00443577
Limbri H, Gunawan C, Thomas T, Smith A, Scott J, Rosche B (2014) Coal-packed methane biofilter for mitigation of greenhouse gas emissions from coal mine ventilation air. PLoS One 9:e94641. https://doi.org/10.1371/journal.pone.0094641
Little CD, Palumbo AV, Herbes SE, Lidstrom ME, Tyndall RL, Gilmer PJ (1988) Trichloroethylene biodegradation by a methane-oxidizing bacterium. Appl Environ Microbiol 54:951–956
Löffler FE, Ritalahti KM, Zinder SH (2013) Dehalococcoides and reductive dechlorination of chlorinated solvents. In: Stroo HF, Leeson A, Ward CH (eds) Bioaugmentation for groundwater remediation. Springer New York, New York, pp 39–88
Lu X, Gu W, Zhao L, Farhan Ul Haque M, DiSpirito AA, Semrau JD, Gu B (2017) Methylmercury uptake and degradation by methanotrophs. Sci Adv 3:e1700041. https://doi.org/10.1126/sciadv.1700041
Ma L, Kim J, Hatzenpichler R, Karymov MA, Hubert N, Hanan IM, Chang EB, Ismagilov RF (2014) Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in human microbiome project's most wanted taxa. Proc Natl Acad Sci 111:9768–9773. https://doi.org/10.1073/pnas.1404753111
Mahmoud AMA (2017) Biological conversion process of methane into methanol using mixed culture methanotrophic bacteria enriched from activated sludge system. Dissertation, York university
Marshall CW, LaBelle EV, May HD (2013) Production of fuels and chemicals from waste by microbiomes. Curr Opin Biotechnol 24:391–397. https://doi.org/10.1016/j.copbio.2013.03.016
Murrell JC, Gilbert B, McDonald IR (2000a) Molecular biology and regulation of methane monooxygenase. Arch Microbiol 173:325–332. https://doi.org/10.1007/s002030000158
Murrell JC, McDonald IR, Gilbert B (2000b) Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol 8:221–225. https://doi.org/10.1016/S0966-842X(00)01739-X
Ngwabie NM, Jeppsson KH, Nimmermark S, Swensson C, Gustafsson G (2009) Multi-location measurements of greenhouse gases and emission rates of methane and ammonia from a naturally-ventilated barn for dairy cows. Biosyst Eng 103:68–77. https://doi.org/10.1016/j.biosystemseng.2009.02.004
Osborne CD, Haritos VS (2018) Horizontal gene transfer of three co-inherited methane monooxygenase systems gave rise to methanotrophy in the Proteobacteria. Mol Phylogenet Evol 129:171–181. https://doi.org/10.1016/j.ympev.2018.08.010
Oshkin IY, Beck DAC, Lamb AE, Tchesnokova V, Benuska G, McTaggart TL, Kalyuzhnaya MG, Dedysh SN, Lidstrom ME, Chistoserdova L (2015) Methane-fed microbial microcosms show differential community dynamics and pinpoint taxa involved in communal response. ISME J 9:1119–1129. https://doi.org/10.1038/ismej.2014.203
Oswald K, Graf JS, Littmann S, Tienken D, Brand A, Wehrli B, Albertsen M, Daims H, Wagner M, Kuypers MMM, Schubert CJ, Milucka J (2017) Crenothrix are major methane consumers in stratified lakes. ISME J 11:2124–2140. https://doi.org/10.1038/ismej.2017.77
Pieja AJ, Morse MC, Cal AJ (2017) Methane to bioproducts: the future of the bioeconomy? Curr Opin Chem Biol 41:123–131. https://doi.org/10.1016/j.cbpa.2017.10.024
Puri AW, Owen S, Chu F, Chavkin T, Beck DAC, Kalyuzhnaya MG, Lidstrom ME (2015) Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense. Appl Environ Microbiol 81:1775–1781. https://doi.org/10.1128/aem.03795-14
Rocha-Rios J, Quijano G, Thalasso F, Revah S, Muñoz R (2011) Methane biodegradation in a two-phase partition internal loop airlift reactor with gas recirculation. J Chem Technol Biotechnol 86:353–360. https://doi.org/10.1002/jctb.2523
Rostkowski KH, Pfluger AR, Criddle CS (2013) Stoichiometry and kinetics of the PHB-producing type II methanotrophs Methylosinus trichosporium OB3b and Methylocystis parvus OBBP. Bioresour Technol 132:71–77. https://doi.org/10.1016/j.biortech.2012.12.129
Semrau JD (2011) Bioremediation via methanotrophy: overview of recent findings and suggestions for future research. Front Microbiol 2:209. https://doi.org/10.3389/fmicb.2011.00209
Semrau JD, DiSpirito A, Yoon S (2010) Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. https://doi.org/10.1111/j.1574-6976.2010.00212.x
Semrau JD, DiSpirito AA, Gu W, Yoon S (2018) Metals and methanotrophy. Appl Environ Microbiol 84:e02289. https://doi.org/10.1128/AEM.02289-17
Sharp CE, Smirnova AV, Graham JM, Stott MB, Khadka R, Moore TR, Grasby SE, Strack M, Dunfield PF (2014) Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ Microbiol 16:1867–1878. https://doi.org/10.1111/1462-2920.12454
Song Y, Yin H, Huang WE (2016) Raman activated cell sorting. Curr Opin Chem Biol 33:1–8. https://doi.org/10.1016/j.cbpa.2016.04.002
Stoecker K, Bendinger B, Schoning B, Nielsen PH, Nielsen JL, Baranyi C, Toenshoff ER, Daims H, Wagner M (2006) Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci 103:2363–2367. https://doi.org/10.1073/pnas.0506361103
Strong PG, Xie S, Clarke WP (2015) Methane as a resource: can the methanotrophs add value? Environ Sci Technol 49:4001–4018. https://doi.org/10.1021/es504242n
Strong PJ, Kalyuzhnaya M, Silverman J, Clarke WP (2016) A methanotroph-based biorefinery: potential scenarios for generating multiple products from a single fermentation. Bioresour Technol 215:314–323. https://doi.org/10.1016/j.biortech.2016.04.099
Svenning MM, Wartiainen I, Hestnes AG, Binnerup SJ (2003) Isolation of methane oxidising bacteria from soil by use of a soil substrate membrane system. FEMS Microbiol Ecol 44:347–354. https://doi.org/10.1016/S0168-6496(03)00073-4
Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN (2011) Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int J Syst Evol Microbiol 61:2456–2463. https://doi.org/10.1099/ijs.0.028118-0
Wang Y, Huang WE, Cui L, Wagner M (2016) Single cell stable isotope probing in microbiology using raman microspectroscopy. Curr Opin Biotechnol 41:34–42. https://doi.org/10.1016/j.copbio.2016.04.018
Wise MG, McArthur JV, Shimkets LJ (1999) Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis. Appl Environ Microbiol 65:4887–4897
Yan X, Chu F, Puri AW, Fu Y, Lidstrom ME (2016) Electroporation-based genetic manipulation in type I methanotrophs. Appl Environ Microbiol 82:2062–2069. https://doi.org/10.1128/aem.03724-15
Yoon S, Carey J, Semrau J (2009) Feasibility of atmospheric methane removal using methanotrophic biotrickling filters. Appl Microbiol Biotechnol 83:949–956. https://doi.org/10.1007/s00253-009-1977-9
Zhang D, Berry JP, Zhu D, Wang Y, Chen Y, Jiang B, Huang S, Langford H, Li G, Davison PA, Xu J, Aries E, Huang WE (2014) Magnetic nanoparticle-mediated isolation of functional bacteria in a complex microbial community. ISME J 9:603–614. https://doi.org/10.1038/ismej.2014.161
Zúñiga C, Morales M, Le Borgne S, Revah S (2011) Production of poly-β-hydroxybutyrate (PHB) by Methylobacterium organophilum isolated from a methanotrophic consortium in a two-phase partition bioreactor. J Hazard Mater 190:876–882. https://doi.org/10.1016/j.jhazmat.2011.04.011
Funding
This research was funded by the National Research Foundation of Korea (NRF) (grant number 2015M3D3A1A01064881 and 2016K2A9A2A06004870). The authors were also financially supported by the Deutsche Forschungsgemeinschaft (grant number HO6234/1-1) and the Leibniz Universität Hannover.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors (Dunfield et al. 1999)
Rights and permissions
About this article
Cite this article
Kwon, M., Ho, A. & Yoon, S. Novel approaches and reasons to isolate methanotrophic bacteria with biotechnological potentials: recent achievements and perspectives. Appl Microbiol Biotechnol 103, 1–8 (2019). https://doi.org/10.1007/s00253-018-9435-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9435-1