Abstract
Non-ribosomal peptide synthetases (NRPSs) are key enzymes in microorganisms for the assembly of peptide backbones of biologically and pharmacologically active natural products. The monomodular NRPS-like enzymes comprise often an adenylation (A), a thiolation (T), and a thioesterase (TE) domain. In contrast to the NRPSs, they do not contain any condensation domain and usually catalyze a dimerization of α-keto carboxylic acids and thereby provide diverse scaffolds for further modifications. In this study, we established an expression system for NRPS-like genes in Saccharomyces cerevisiae. By expression of four known genes from Aspergillus terreus, their predicted function was confirmed and product yields of up to 35 mg per liter culture were achieved. Furthermore, expression of ATEG_03090 from the same fungus, encoding for the last uncharacterized NRPS-like enzyme with an A-T-TE domain structure, led to the formation of the benzoquinone derivative atromentin. All the accumulated products were isolated and their structures were elucidated by NMR and MS analyses. This study provides a convenient system for proof of gene function as well as a basis for synthetic biology, since additional genes encoding modification enzymes can be introduced.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural products are fundamental to drug discovery and development (Newman and Cragg 2016). Like plants, microorganisms are important producers of biologically active natural products. The success rate for applying microbial drugs as pharmaceuticals is approximately 20-fold higher than those derived from plants (Bérdy 2012). Most of the natural products with promising biological activities are secondary metabolites produced by actinobacteria (Barka et al. 2016) or ascomycetous fungi, e.g., Penicillium and Aspergillus species (Bills and Gloer 2016).
Due to the ongoing development and improvement of sequencing technologies, the number of sequenced genomes has increased tremendously. Genome mining revealed the largely unexploited biosynthetic potential for natural products in many microorganisms (de Vries et al. 2017; Kolter and Van Wezel 2016). In the biosynthesis of most biologically active metabolites derived from microorganisms, multifunctional modular polyketide synthases and non-ribosomal peptide synthetases (NRPSs) assemble diverse skeletons (Bhetariya et al. 2016; Dejong et al. 2016; Süssmuth and Mainz 2017). NRPS modules, which consist at least of adenylation (A), thiolation (T), and condensation (C) domains, are responsible for the activation of amino acids and peptide formation. Usually only one A-T domain unit is essential for activation of one amino acid to be incorporated into the peptide chain. A thioesterase (TE) domain is responsible for release of the peptide residue from the enzyme template (Payne et al. 2017; Süssmuth and Mainz 2017). Further tailoring modifications like hydroxylation, prenylation, or cyclization lead to a vast number of diverse and biologically active products (Payne et al. 2017; Rudolf et al. 2017; Tang et al. 2017; Winkelblech et al. 2015).
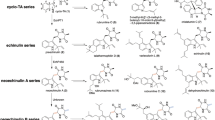
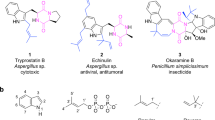
NRPS-like enzymes form a related enzyme group to NRPSs. These enzymes are also widely distributed in microorganisms. They consist of only one module with an A and a T domain, but lack the peptide-forming C domain. Instead, they possess a TE or reductase (R) domain at the C-terminus of the polypeptides (Guo and Wang 2014; Zhu et al. 2014). R-domains generate aryl aldehydes by reduction of carboxylic acids or thioesters (Wang et al. 2014; Wang and Zhao 2014). NRPS-like enzymes with an A-T-TE architecture use α-keto acids like phenyl pyruvic acid (1), 4-hydroxyphenyl pyruvic acid (2), or indolyl pyruvic acid (3) as substrates and catalyze their dimerization, resulting in the formation of lactones, ketals or benzoquinones (Fig. 1) (Braesel et al. 2015; Geib et al. 2016; Guo et al. 2013; Guo et al. 2015). The generated products serve as precursors for further modification, mostly prenylation and cyclization, leading to biologically active metabolites. For example, asterriquinones display diverse biological activities including antitumor and insulin mimic activities (Kim et al. 2007; Li et al. 2012). Prenylated butyrolactones show a wide range of biological activities such as antitumor, antiparasitic or anti-inflammatory activity (da Silva et al. 2017; Dewi et al. 2015; Guo et al. 2016).
Genome mining of Aspergillus terreus (A. terreus) NIH 2624 for core genes of secondary metabolites revealed the presence of 16 putative NRPS-like genes (Guo and Wang 2014; Khaldi et al. 2010). Ten of the NRPS-like enzymes share a C-terminal R-domain and one of them, the product of ATEG_03630, was proven to be responsible for the aryl aldehyde formation (Wang et al. 2014). Six of the NRPS-like enzymes share an A-T-TE domain architecture. Five of them, encoded by ATEG_02004, ATEG_03563, ATEG_02815, ATEG_00700, and ATEG_08899, have been studied in the last years. By gene inactivation in the producer, heterologous expression in Aspergillus nidulans (A. nidulans) or in vitro assays, ApvA and MelA, encoded by ATEG_02004 and ATEG_03563, respectively, were proven to be responsible for the formation of aspulvinone E (4a) (Fig. 1) (Geib et al. 2016; Guo et al. 2013; Guo et al. 2015). It was proposed that ApvA is active in the mycelia and provides the backbone of aspulvinones (Guo et al. 2015), while MelA is active in conidia and produces 4a as a building block for the biosynthesis of the pigment Asp-melanin for protection from ultraviolet radiation (Geib et al. 2016). Heterologous expression of ATEG_02815 (btyA) in A. nidulans resulted in the formation of butyrolactone IIa (5) (van Dijk et al. 2016). Involvement of AtqA encoded by ATEG_00700 in the formation of asterriquinone CT5 was proven by a gene knockout study in its producer (Guo et al. 2013). PgnA by ATEG_08899 was identified as phenguignardic acid (6) synthetase after gene activation in A. terreus (Sun et al. 2016). The deduced product of the last uncharacterized NRPS-like gene ATEG_03090 was wrongly predicted to comprise only the sequence for an A and a T domain (Guo and Wang 2014) and its function remained unknown.
Functional proof of secondary metabolite genes is a prerequisite for using the genetic potential in biotechnology and synthetic biology. This is often challenging, as in the cases of the aforementioned examples for NRPS-like genes from A. terreus. Different strategies including gene inactivation in the producers (Guo et al. 2013), heterologous expression in A. nidulans (Guo et al. 2015; Sun et al. 2016) or Aspergillus niger (Geib et al. 2016) were applied for gene identification. Pre-genetic manipulation is usually required for the producers or heterologous Aspergillus hosts, before the inactivation of the genes of interest can be carried out. In some cases, the genes of interest are silent under laboratory conditions (Guo et al. 2013; Guo et al. 2015; Sun et al. 2016). Furthermore, many fungal strains are genetically difficult to be manipulated. Therefore, a convenient and producer-independent expression platform will be welcome for functional proof of metabolic genes including NRPS-like genes and for the production of secondary metabolites by synthetic biology via gene recombination.
Saccharomyces cerevisiae (S. cerevisiae) has been used as model organism for a long time and many tools for genetic modifications are available (Bond et al. 2016; Fletcher et al. 2016). Compared to the filamentous fungi, yeast grows faster and is genetically easy to handle. For our purpose, we used the S. cerevisiae strain BJ5464-NpgA, which carries the phosphopantetheinyl transferase gene npgA from A. nidulans. NpgA is required for post-translational modification of NRPSs to their active holo forms and is commonly used for heterologous expression of NRPS or PKS genes (Haynes et al. 2013; Ishiuchi et al. 2012; Lee et al. 2009). In this study, we took the four known genes ATEG_02004, ATEG_03563, ATEG_02815 and ATEG_08899 to test their expression in the yeast strain. The function of the uncharacterized NRPS-like gene ATEG_03090 was afterwards identified by expression and identification of the accumulated product.
Materials and methods
Computer-assisted sequence analysis
Alignments of amino acid sequences and domain structure prediction were carried out by using the programs BLAST (http://blast.ncbi.nlm.nih.gov) and CD-Search (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), respectively.
Strains, plasmids, and culture conditions
Escherichia coli XL1 blue MRF’ (Stratagene) was used for plasmid construction. Cultivation was carried out at 37 °C in liquid or on solid lysogeny broth (LB) medium, supplemented with carbenicillin at a final concentration of 50 μg/ml for selection. A. terreus FGSC A1156 (identical to the genome reference strain NIH 2624) was grown in YME medium containing 0.4% yeast extract, 1.0% malt extract and 0.4% glucose. Yeast strain BJ5464-NpgA (Haynes et al. 2013) was cultivated at 30 °C on yeast medium containing 1% yeast extract, 2.0% peptone and 2.0% glucose or synthetic complete (SC) medium without uracil (SC-U) consisting of 0.67% yeast nitrogen base with ammonium sulfate, 0.065% CSM-his-leu-ura (MP Biomedicals), histidine and leucine as described by Sherman (2002). For induction of gene expression, 2.0% galactose instead glucose were used as carbon source.
mRNA isolation and cDNA synthesis
A. terreus FGSC A1156 was grown for 3 days in a 250-ml cylindrical flask containing 50 ml YME medium at 30 °C and 200 rpm in the dark. The mycelium was filtrated, washed with water and grinded to a fine powder. Total mRNA was extracted by using the E.Z.N.A® Fungal RNA Kit (Omega bio-tec) as described in the manual. The mRNA was directly applied for cDNA synthesis with the ProtoScript® First Strand cDNA Synthesis Kit (New England BioLabs) using oligo-dT primers as well as the Random Primer Mix.
Construction of expression plasmids
The five NRPS-like genes were amplified from cDNA. Primers for the amplification are listed in Table S1. The pESC-URA vector was cut with suitable restriction enzymes in the MCS1 and the fragments were inserted into the vector by homologous recombination according to the protocol described by Jacobus and Gross (2015).
Cultivation of the transformants for metabolite production
For quantification of the produced metabolites, 5 ml SC-U with 2.0% glucose were inoculated with a single colony of the transformant of interest. The expression was started by inoculating 50 ml SC-U medium with 2% galactose with the pre-culture to an absorption of 0.1 at 600 nm. 0.5 ml samples were taken at different time points and the same volume methanol was added. The samples were stored at − 20 °C till analysis. Transformants producing aspulvinone E were cultivated in the dark.
For product isolation, pre-precultures were cultivated for 40 h. 50 ml SC-U medium was inoculated with 1 ml of these cultures. After 24 h, these cultures were pelleted at 3000 rpm for 5 min and then washed with 25 ml water. The cells were used to inoculate 2 l of SC medium with 2% galactose as carbon source. After 48 h, the cultures were extracted three times with the same volume of ethyl acetate, dried over Na2SO4 and evaporated to dryness at 35 °C.
HPLC analysis
For determination of the product formation, the yeast extracts were analyzed on an Agilent 1200 series HPLC system (Böblingen, Germany) by using a Multospher 120 RP-18 column (250 × 4 mm, 5 μm, CS-Chromatographie Service, Langerwehe, Germany) with a flow rate of 1 ml/min. Water (A) and acetonitrile (B), both containing 0.05% trifluoroacetic acid, were used as mobile phases. A linear gradient from 10 to 40% B in 15 min and a second one from 40 to 45% B in 25 min were used for analysis of 4a, 5 and 8. 6 was eluted with a gradient from 10 to 60% B in 10 min, followed by 60 to 80% B in another 10 min. The column was then washed for 5 min with 95% B and equilibrated with 10% B for 5 min.
For isolation, a Multospher 120 RP-18 column (250 × 4 mm, 5 μm, CS-Chromatographie Service, Langerwehe, Germany) was used with the same solvents and similar gradients, but with a flow rate of 3 ml/min.
LC-MS analysis
For LC-MS analysis, an Agilent 1260 series HPLC system equipped with a photodiode array detector and a Bruker micrOTOF-Q III mass spectrometer was used. The samples were separated on a Multospher 120 RP-18 column (250 × 4 mm, 5 μm, CS- Chromatographie Service, Langerwehe, Germany). A flow rate at 0.5 ml/min and a linear gradient of 5–100% acetonitrile in water, both containing 0.1% formic acid, in 10 min was used for separation. Subsequently the column was washed with 100% acetonitrile with 0.1% formic acid before it was equilibrated to the starting conditions for 5 min.
For detection of the conversion from 4a to 4b, the same column and solvents were used, but a linear gradient of 5–100% acetonitrile in water in 40 min and a flow rate of 0.25 ml/min was applied. The column was washed for 5 min followed by an equilibration to 5% for 10 min.
NMR analysis
1H–NMR spectra were taken on an ECA500 spectrometer (JEOL, Tokyo, Japan). Chemical shifts were referenced to the respective solvent signal. The spectra were processed with MestReNova 6.0.2.2–5475.
Accession numbers
The revised coding regions have been deposited in the GenBank database under accession numbers MG384312 for ATEG_02004, MG384313 for ATEG_03563, MG384314 for ATEG_02815, MG384316 for ATEG_08899 and MG384315 for ATEG_03090.
Results
Proof of the coding sequences and plasmid construction for yeast expression
For preparation of expression in S. cerevisiae, we confirmed the coding sequences of the NRPS-like genes by PCR amplification. For this purpose, mRNA was isolated from A. terreus A1156 and converted to cDNA. The coding sequences of the genes of interest were amplified from the obtained cDNA by using primers listed in Table S1. The PCR products were cloned via the pGEM®-T easy vector system or directly via homologous recombination into the yeast expression vector pESC-URA (Table 1). In the expression constructs, the NRPS-like genes are under control of a galactose-inducible promoter.
DNA sequencing of the obtained constructs revealed that all five genes consist merely of one exon and the gene products comprise 925 to 946 amino acids sharing a clear A-T-TE architecture and the annotation of the genome sequence was incorrect for ATEG_02004, ATEG_03563, ATEG_02815 and ATEG_03090. The false annotation of ATEG_02004, ATEG_03563 and ATEG_03090 was caused by insertion of one base pair, resulting in the prediction of more exons for the first two genes. In case of ATEG_03090, only the coding sequence of the A and T domain with 844 amino acids was included in the previous prediction (Table S2) (Guo and Wang 2014). The predicted exon length of ATEG_08899 in the NCBI database was confirmed by sequencing the expression construct. However, differences in two base pairs were revealed, leading to one mutation N410D in the amino acid sequence.
Expression of known NRPS-like genes in yeast
The constructs pEH10, pEH13, pEH29 and pEH27, carrying ATEG_02004, ATEG_03563, ATEG_02815 and ATEG_08899 (Table 1), respectively, were introduced into the S. cerevisiae strain BJ5464-NpgA. Gene expression was induced by addition of galactose and the cultures were incubated for 48 h. The strain with the empty vector pESC-URA was used as a negative control. Already 24 h after induction, the culture expressing ATEG_02004 encoding ApvA showed a bright yellow color, indicating the formation of the expected aspulvinone E (4a). To prove the product formation in the transformants, the complete 48 h old cultures of the four transformants were extracted with ethyl acetate. The organic phases were evaporated to dryness, dissolved in methanol and analyzed on LC-MS.
In comparison to the negative control (Fig. 2a), additional peaks were clearly detected in the transformants with the constructs pEH10, pEH29 and pEH27 (Fig. 2b, d and e). LC-MS analysis of these peaks showed the expected [M + H]+ ions for 4a, 5 and 6 as the products (Table S3). They also represent predominant products in the chromatograms with UV detection at 296 nm (Fig. 2b, d, and e). The presence of two peaks in the transformant with pEH10 could indicate the light-induced isomerization of aspulvinone E (4a) to isoaspulvinone E (4b) (Geib et al. 2016).
LC-MS analysis of the metabolite profiles of S. cerevisiae cultures expressing NRPS-like genes 48 h after induction. Products were detected with a photo array detector and illustrated for absorption at 296 nm (shown in black). The extracted ion chromatograms with the respective [M + H]+ ions are highlighted in color
To provide evidence that 4a is the product of the NRPS-like enzyme ApvA and 4b the isomerization product upon light exposition, yeast cells carrying pEH10 were grown in the dark and extracted with ethyl acetate. For better separation of 4a from 4b, we prolonged the elution time for LC-MS analysis. One predominant product peak with the same retention time as 4a was detected on the LC-MS chromatogram of the ethyl acetate extract (Fig. 3a). This extract was then exposed to sun light at room temperature and its conversion was monitored by LC-MS analysis (Fig. 3b). Conversion of 4a to 4b can already be clearly detected after 30 min and increased with the exposing time. After 4 h, the ratio of 4b to 4a nearly reached the one observed in the culture maintained under normal conditions (Fig. 2b). This experiment proved unequivocally the light-induced conversion of 4a to 4b.
To confirm their structures, 4a, 5 and 6 were isolated on HPLC from the cultures with pEH10, pEH29, and pEH27, respectively, and subjected to NMR and MS analyses. Interpretation of the obtained NMR spectra (Fig. S1-S4) and comparison of these data (Table 2) with those of the expected products proved unequivocally 4a, 5 and 6 to be aspulvinone E (Dewi et al. 2015), butyrolactone IIa (van Dijk et al. 2016) and phenguignardic acid (Sun et al. 2016), respectively. These results were also confirmed by MS data (Table S3).
Monitoring the concentration of 4a (kept in the dark), 5 and 6 in the yeast cultures revealed an exponential increase of product formation from 15 to 28 h and nearly reached steady state after 40 h (Fig. 4). Product yields of 13, 15 and 35 mg per liter culture are calculated for 4a, 5 and 6 after 100 h, respectively.
In the LC-MS chromatogram of the yeast extract with pEH13 (ATEG_03563), two product peaks were detected with the extracted [M + H]+ ions for 4a and 4b, which also share the same retention times with the products of ApvA. Due to the low productivity, no significant color change was observed during the cultivation and 4a and 4b are nearly not detectable by UV absorption at 296 nm. Nevertheless, 4a and 4b can be unequivocally identified as aspulvinone E and isoaspulvinone E, respectively, by comparison of their UV spectra and exact [M + H]+ ions with those of the ApvA products. The low activity of MelA was also observed in a previous study (Guo et al. 2015).
Functional proof of the last uncharacterized NRPS-like enzyme with an A-T-TE architecture from A. terreus
After the successful establishment of the expression system for NRPS-like genes in yeast, the uncharacterized NRPS-like gene ATEG_03090 from A. terreus became subject of our investigation. As aforementioned, the coding region of this gene was corrected from 2535 to 2763 bps. The deduced product EAU36364 comprises 920 amino acid residues with an A-T-TE domain structure instead of 844 amino acids comprising only an A and a T domain. A Blastp search with the revised sequence of EAU36364 led to the identification of the best hit for ApvA from A. terreus, with sequence identity of 57% on the amino acid level. Furthermore, EAU36364 shares identities of 43 and 44% with the two didemethylasterriquinone synthetases TdiA from A. nidulans and AtqA from A. terreus, respectively. Its function cannot be predicted by sequence and domain structure analysis (Table S4).
The revised full length coding sequence of ATEG_03090 was cloned into the pESC-URA vector under the control of a galactose inducible promoter. The transformant was cultivated as described above and the metabolite profile was analyzed 48 h after induction by LC-MS. As shown in Fig. 2f, an additional peak (8) at 7.5 min was clearly detected in the HPLC chromatogram with UV absorption at 296 nm. This peak was subsequently isolated on HPLC from the yeast culture and subjected to NMR and MS analyses. In the NMR spectrum in (CD3)2SO (Fig. S4), signals of two sets of coupling aromatic protons (1:1) were detected at 7.21 and 6.78 ppm, respectively. In addition, two (broad) singlets for phenolic protons with similar integral intensity were observed at 9.49 and 10.76 ppm. Comparison of the NMR data (Table 2) with those in literature led to the identification of 8 as atromentin (Schneider et al. 2008; Wackler et al. 2012), which was also confirmed by its [M + H]+ ion at m/z 325.0707.
Discussion
In this study, five NRPS-like genes from A. terreus encoding enzymes with an A-T-TE architecture have been successfully expressed in S. cerevisiae. Product formation in substantial amounts was detected in transformants of four genes and the identities of these products were confirmed by NMR and MS analyses after isolation from the yeast cultures. These compounds are precursors of biologically active metabolites (da Silva et al. 2017; Gao et al. 2013; Kim et al. 2007) and could be further modified by tailoring enzymes. Therefore, this study provides a convenient platform for functional proof of NRPS-like genes and for their application in the synthetic biology. We also demonstrated that the product of ApvA and MelA, aspulvinone E, was rapidly converted to its isomer isoaspulvine E by sun light. Both compounds are detected in cultures without light protection.
The uncharacterized NRPS-like gene ATEG_03090 was proven to encode an enzyme with an A-T-TE domain structure being responsible for the formation of atromentin. To the best of our knowledge, this is the first report on an atromentin synthetase in ascomycetes. Formation of atromentin by NRPS-like enzyme has been demonstrated in basidiomycetes, e.g., by GreA from Suillus grevillei with a polypeptide chain of 958 amino acids and AtrA form Tapinella panuoides with 957 amino acids (Schneider et al. 2008; Wackler et al. 2012). To distinguish AtrA from the basidiomycete, we termed the product of ATEG_03090 AtrAAt. In Streptomyces rapamycinicus, the NRPS-like enzyme EchA catalyzes the dimerization of two phenylpyruvic acid molecules to form polyporic acid, which contains the same benzoquinone core as atromentin (Table S4) (Zhu et al. 2014). A sequence identity of 39% was calculated for AtrAAt and EchA (Table S4), being comparable to those of AtrAAt with different atromentin synthetases from basidiomycetes (Table S4). Formation of other products than atromentin with a quinone core is also known in ascomycota. Even A. terreus has a NRPS-like enzyme (AtqA), which catalyzes the formation of the benzoquinone derivative didemethylasterriquinone D (Fig. 1). The sequence identity of AtrAAt and AtqA amounts to 44%, higher than those of AtrAAt with the enzymes mentioned above. Even this value is still much lower than the 56% of AtrAAt with ApvA, being responsible for the formation of products with different structure cores. This provides an excellent example that gene function should be proven experimentally. It would be interesting to compare the structures and binding motifs of the NRPS-like enzymes form different species.
Previous studies showed that most of the NRPS-like enzymes are involved in the biosynthesis of secondary metabolites and catalyze the formation of different scaffolds (Fig. 1) (Guo et al. 2013; Guo and Wang 2014). These backbones can be modified by introducing genes encoding tailoring enzymes like prenyltransferases (Winkelblech et al. 2015) into the S. cerevisiae transformants carrying NRPS-like genes. Therefore, undesired modification of the backbones by host enzymes should be avoided. Our results in this study showed that the products of the four known enzymes ApvA, MelA, BtyA and PgnA are identical to those detected in the naïve producer or in the heterologous Aspergillus hosts (Geib et al. 2016; Guo et al. 2013; Guo et al. 2015; Sun et al. 2016), indicating that products of these NRPS-like enzymes were not further converted by the yeast enzymes. Combinational expression of the NRPS-like and prenyltransferase genes in yeast strains are now under investigation.
References
Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clément C, Ouhdouch Y, Van Wezel GP (2016) Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev 80(1):1–43. https://doi.org/10.1128/MMBR.00019-15
Bérdy J (2012) Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot 65(8):441. https://doi.org/10.1038/ja.2012.54
Bhetariya PJ, Prajapati M, Bhaduri A, Mandal RS, Varma A, Madan T, Singh Y, Sarma PU (2016) Phylogenetic and structural analysis of polyketide synthases in Aspergilli. Evol Bioinformatics Online 12:109–119
Bills GF, Gloer JB (2016) Biologically active secondary metabolites from the fungi. Microbiol Spectr 4:1–32
Bond C, Tang Y, Li L (2016) Saccharomyces cerevisiae as a tool for mining, studying and engineering fungal polyketide synthases. Fungal Genet Biol 89:52–61. https://doi.org/10.1016/j.fgb.2016.01.005
Braesel J, Gotze S, Shah F, Heine D, Tauber J, Hertweck C, Tunlid A, Stallforth P, Hoffmeister D (2015) Three redundant synthetases secure redox-active pigment production in the basidiomycete Paxillus involutus. Chem Biol 22(10):1325–1334. https://doi.org/10.1016/j.chembiol.2015.08.016
da Silva IP, Brissow E, Kellner Filho LC, Senabio J, de Siqueira KA, Vandresen FS, Damasceno JL, Mendes SA, Tavares DC, Magalhaes LG, Junior PA, Januario AH, Soares MA (2017) Bioactive compounds of Aspergillus terreus-F7, an endophytic fungus from Hyptis suaveolens (L.) Poit. World J Microbiol Biotechnol 33(3):62. https://doi.org/10.1007/s11274-017-2228-3
de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, Anderluh G, Asadollahi M, Askin M, Barry K, Battaglia E, Bayram O, Benocci T, Braus-Stromeyer SA, Caldana C, Canovas D, Cerqueira GC, Chen F, Chen W, Choi C, Clum A, Dos Santos RA, Damasio AR, Diallinas G, Emri T, Fekete E, Flipphi M, Freyberg S, Gallo A, Gournas C, Habgood R, Hainaut M, Harispe ML, Henrissat B, Hilden KS, Hope R, Hossain A, Karabika E, Karaffa L, Karanyi Z, Krasevec N, Kuo A, Kusch H, LaButti K, Lagendijk EL, Lapidus A, Levasseur A, Lindquist E, Lipzen A, Logrieco AF, MacCabe A, Makela MR, Malavazi I, Melin P, Meyer V, Mielnichuk N, Miskei M, Molnar AP, Mule G, Ngan CY, Orejas M, Orosz E, Ouedraogo JP, Overkamp KM, Park HS, Perrone G, Piumi F, Punt PJ, Ram AF, Ramon A, Rauscher S, Record E, Riano-Pachon DM, Robert V, Rohrig J, Ruller R, Salamov A, Salih NS, Samson RA, Sandor E, Sanguinetti M, Schutze T, Sepcic K, Shelest E, Sherlock G, Sophianopoulou V, Squina FM, Sun H, Susca A, Todd RB, Tsang A, Unkles SE, van de Wiele N, van Rossen-Uffink D, Oliveira JV, Vesth TC, Visser J, Yu JH, Zhou M, Andersen MR, Archer DB, Baker SE, Benoit I, Brakhage AA, Braus GH, Fischer R, Frisvad JC, Goldman GH, Houbraken J, Oakley B, Pocsi I, Scazzocchio C, Seiboth B, vanKuyk PA, Wortman J, Dyer PS, Grigoriev IV (2017) Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol 18(1):28. https://doi.org/10.1186/s13059-017-1151-0
Dejong CA, Chen GM, Li H, Johnston CW, Edwards MR, Rees PN, Skinnider MA, Webster AL, Magarvey NA (2016) Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat Chem Biol 12(12):1007–1014. https://doi.org/10.1038/nchembio.2188
Dewi RT, Tachibana S, Fajriah S, Hanafi M (2015) α-Glucosidase inhibitor compounds from Aspregillus terreus RCC1 and their antioxidant activity. Med Chem Res 24(2):737–743. https://doi.org/10.1007/s00044-014-1164-0
Fletcher E, Krivoruchko A, Nielsen J (2016) Industrial systems biology and its impact on synthetic biology of yeast cell factories. Biotechnol Bioeng 113(6):1164–1170. https://doi.org/10.1002/bit.25870
Gao H, Guo W, Wang Q, Zhang L, Zhu M, Zhu T, Gu Q, Wang W, Li D (2013) Aspulvinones from a mangrove rhizosphere soil-derived fungus Aspergillus terreus Gwq-48 with anti-influenza A viral (H1N1) activity. Bioorg Med Chem Lett 23(6):1776–1778. https://doi.org/10.1016/j.bmcl.2013.01.051
Geib E, Gressler M, Viediernikova I, Hillmann F, Jacobsen ID, Nietzsche S, Hertweck C, Brock M (2016) A non-canonical melanin biosynthesis pathway protects Aspergillus terreus conidia from environmental stress. Cell Chem Biol 23(5):587–597. https://doi.org/10.1016/j.chembiol.2016.03.014
Guo CJ, Knox BP, Sanchez JF, Chiang YM, Bruno KS, Wang CCC (2013) Application of an efficient gene targeting system linking secondary metabolites to their biosynthetic genes in Aspergillus terreus. Org Lett 15(14):3562–3565. https://doi.org/10.1021/ol401384v
Guo C-J, Sun W-W, Bruno KS, Oakley BR, Keller NP, Wang CCC (2015) Spatial regulation of a common precursor from two distinct genes generates metabolite diversity. Chem Sci 6(10):5913–5921. https://doi.org/10.1039/C5SC01058F
Guo C-J, Wang CCC (2014) Recent advances in genome mining of secondary metabolites in Aspergillus terreus. Front Microbiol 5:717
Guo F, Li Z, Xu X, Wang K, Shao M, Zhao F, Wang H, Hua H, Pei Y, Bai J (2016) Butenolide derivatives from the plant endophytic fungus Aspergillus terreus. Fitoterapia 113:44–50. https://doi.org/10.1016/j.fitote.2016.06.014
Haynes SW, Gao X, Tang Y, Walsh CT (2013) Complexity generation in fungal peptidyl alkaloid biosynthesis: a two-enzyme pathway to the hexacyclic MDR export pump inhibitor ardeemin. ACS Chem Biol 8(4):741–748. https://doi.org/10.1021/cb3006787
Ishiuchi K, Nakazawa T, Ookuma T, Sugimoto S, Sato M, Tsunematsu Y, Ishikawa N, Noguchi H, Hotta K, Moriya H, Watanabe K (2012) Establishing a new methodology for genome mining and biosynthesis of polyketides and peptides through yeast molecular genetics. Chembiochem 13(6):846–854. https://doi.org/10.1002/cbic.201100798
Jacobus AP, Gross J (2015) Optimal cloning of PCR fragments by homologous recombination in Escherichia coli. PLoS One 10(3):e0119221. https://doi.org/10.1371/journal.pone.0119221
Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND (2010) SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47(9):736–741. https://doi.org/10.1016/j.fgb.2010.06.003
Kim H, Deng L, Xiong X, Hunter WD, Long MC, Pirrung MC (2007) Glyceraldehyde 3-phosphate dehydrogenase is a cellular target of the insulin mimic demethylasterriquinone B1. J Med Chem 50(15):3423–3426. https://doi.org/10.1021/jm070437i
Kolter R, Van Wezel GP (2016) Goodbye to brute force in antibiotic discovery? Nat Microbiol 1(2):15020. https://doi.org/10.1038/nmicrobiol.2015.20
Lee KKM, Da Silva NA, Kealey JT (2009) Determination of the extent of phosphopantetheinylation of polyketide synthases expressed in Escherichia coli and Saccharomyces cerevisiae. Anal Biochem 394(1):75–80. https://doi.org/10.1016/j.ab.2009.07.010
Li X, Zheng SL, Li X, Li JL, Qiang O, Liu R, He L (2012) Synthesis and anti-breast cancer activity of new indolylquinone derivatives. Eur J Med Chem 54:42–48. https://doi.org/10.1016/j.ejmech.2012.04.019
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79(3):629–661. https://doi.org/10.1021/acs.jnatprod.5b01055
Payne JA, Schoppet M, Hansen MH, Cryle MJ (2017) Diversity of nature's assembly lines—recent discoveries in non-ribosomal peptide synthesis. Mol BioSyst 13(1):9–22. https://doi.org/10.1039/C6MB00675B
Rudolf JD, Chang CY, Ma M, Shen B (2017) Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function. Nat Prod Rep 34(9):1141–1172. https://doi.org/10.1039/C7NP00034K
Schneider P, Bouhired S, Hoffmeister D (2008) Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides. Fungal Genet Biol 45(11):1487–1496. https://doi.org/10.1016/j.fgb.2008.08.009
Sherman F (2002) Getting started with yeast. Methods Enzymol 350:3–41. https://doi.org/10.1016/S0076-6879(02)50954-X
Sun WW, Guo CJ, Wang CC (2016) Characterization of the product of a nonribosomal peptide synthetase-like (NRPS-like) gene using the doxycycline dependent Tet-ON system in Aspergillus terreus. Fungal Genet Biol 89:84–88. https://doi.org/10.1016/j.fgb.2016.01.014
Süssmuth RD, Mainz A (2017) Nonribosomal peptide synthesis—principles and prospects. Angew Chem Int Ed Engl 56(14):3770–3821. https://doi.org/10.1002/anie.201609079
Tang MC, Zou Y, Watanabe K, Walsh CT, Tang Y (2017) Oxidative cyclization in natural product biosynthesis. Chem Rev 117(8):5226–5333. https://doi.org/10.1021/acs.chemrev.6b00478
van Dijk JW, Guo CJ, Wang CC (2016) Engineering fungal nonribosomal peptide synthetase-like enzymes by heterologous expression and domain swapping. Org Lett 18(24):6236–6239. https://doi.org/10.1021/acs.orglett.6b02821
Wackler B, Lackner G, Chooi YH, Hoffmeister D (2012) Characterization of the Suillus grevillei quinone synthetase GreA supports a nonribosomal code for aromatic alpha-keto acids. Chembiochem 13(12):1798–1804. https://doi.org/10.1002/cbic.201200187
Wang M, Beissner M, Zhao H (2014) Aryl-aldehyde formation in fungal polyketides: discovery and characterization of a distinct biosynthetic mechanism. Chem Biol 21(2):257–263. https://doi.org/10.1016/j.chembiol.2013.12.005
Wang M, Zhao H (2014) Characterization and engineering of the adenylation domain of a NRPS-like protein: a potential biocatalyst for aldehyde generation. ACS Catal 4(4):1219–1225. https://doi.org/10.1021/cs500039v
Winkelblech J, Fan A, Li S-M (2015) Prenyltransferases as key enzymes in primary and secondary metabolism. Appl Microbiol Biotechnol 99(18):7379–7397. https://doi.org/10.1007/s00253-015-6811-y
Zhu J, Chen W, Li YY, Deng JJ, Zhu DY, Duan J, Liu Y, Shi GY, Xie C, Wang HX, Shen YM (2014) Identification and catalytic characterization of a nonribosomal peptide synthetase-like (NRPS-like) enzyme involved in the biosynthesis of echosides from Streptomyces sp. LZ35. Gene 546(2):352–358. https://doi.org/10.1016/j.gene.2014.05.053
Acknowledgements
A. terreus FGSC A1156 was kindly provided by Mathias Brock (HKI Jena, Germany). We also thank Stefan Newel from Philipps-Universität Marburg for taking NMR spectra.
Funding
The Bruker micrOTOF QIII mass spectrometer was financially supported in part by a grant from the Deutsche Forschungsgemeinschaft (INST 160/620-1 to S.-M. L.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 322 kb)
Rights and permissions
About this article
Cite this article
Hühner, E., Backhaus, K., Kraut, R. et al. Production of α-keto carboxylic acid dimers in yeast by overexpression of NRPS-like genes from Aspergillus terreus. Appl Microbiol Biotechnol 102, 1663–1672 (2018). https://doi.org/10.1007/s00253-017-8719-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8719-1