Abstract
Polyunsaturated fatty acids (PUFAs) are essential lipids for cell function, normal growth, and development, serving as key structural components of biological membranes and modulating critical signal transduction events. Omega-3 (n-3) long chain PUFAs (LC-PUFAs) such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) have been shown to protect against inflammatory diseases and enhance brain development and function. This had led to a marked increase in demand for fish and fish oils in human diets, supplements, and aquaculture and created a need for new, sustainable n-3 LC-PUFA sources. We have studied for the first time homogenous preparations of the membrane-type ω6 and ω3 fatty acid desaturases from the fungus Mortierella alpina, as a model system to produce PUFAs. These desaturases possess a di-iron metal center and are selective for 18:1 n-9 and 18:2 n-6 acyl-CoA substrates, respectively. Sequence alignments and membrane topology predictions support that these enzymes have unique cap regions that may include the rearrangement and repositioning of the active site, especially when compared to the mammalian stearoyl–coenzyme A desaturase-1 (SCD1) and the related sphingolipid α-hydroxylase (Scs7p) that act upon different substrates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyunsaturated fatty acids (PUFAs) are essential for cell membrane function and cell signaling. Both omega-3 (n-3) and omega-6 (n-6) long chain (> 20 carbons) PUFAs (LC-PUFAs) such as arachidonic acid (ARA; 20:4 n-6), eicosapentaenoic acid (EPA; 20:5 n-3), and docosahexaenoic acid (DHA; 22:6 n-3) are precursors of bioactive metabolites, which have been implicated in several human inflammatory diseases including cardiovascular disease and cancer. Many studies support the beneficial effects of dietary n-3 PUFAs in the prevention of inflammatory diseases such as cancer and cardiovascular disease and normal brain function (Bazinet and Laye 2014; Gu et al. 2015; Weylandt et al. 2015). However, continued pressures on wild fish stocks together with dramatic increases in n-3 PUFA utilization by aquaculture as well as nutraceutical and pharmaceutical industries have place unsustainable requirements on the current global n-3 PUFA supply.
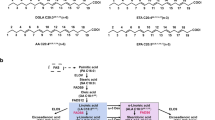
The formation of PUFAs (Fig. 1) is catalyzed by a variety of fatty acid desaturases and elongases (Shanklin et al. 2009; Wang et al. 2013). The regio- and stereo-selective introduction of an unsaturated bond by these enzymes is remarkable, given the lack of unique structural or chemical features along the aliphatic chain. The genes for the ω6 and ω3 desaturases have been lost in mammals during evolution. As such, the key products of this pathway, linoleic acid (LA; 18:2 n-6) and α-linolenic acid (ALA; 18:3 n-3), are essential fatty acids necessary for health. Their desaturation and elongation LC-PUFA products, ARA, EPA, and DHA, are often included in infant formulation and EPA and DHA are consumed as dietary supplements. Because of the increased demands for n-3 PUFA described earlier, there is a need for new, sustainable n-3 PUFA sources. Specifically, studies to determine the molecular mechanism and substrate specificity of ω6 and ω3 desaturases are needed to facilitate the engineering of microbes for the production of PUFAs and especially n-3 PUFAs.
Polyunsaturated fatty acid biosynthesis. a Nomenclature. The location of the site of unsaturation can be indicated either according to its relationship to the carboxylic acid moiety (Δx) or the terminal carbon atom (ωx). The fatty acid illustrated is stearidonic acid (18:4), with double bonds located at the Δ6,9,12,15 positions. b Overview of PUFA biosynthetic pathway in Mortierella alpina. The focus of this study is the ω6 and ω3 desaturases highlighted in bold. The following abbreviations are used: Des, desaturase; Elo, elongase. (ChemDraw and Adobe Illustrator were used to create this artwork)
Desaturases are found in two forms that recognize different substrate classes. The soluble acyl-ACP desaturases recognize lipids attached to the pantothenic linker arm of an acyl-carrier protein domain and are found in the plastids of higher plants (Shanklin et al. 2009). The crystal structures of the 18:0 Δ9-desaturase from Ricinus communis (castor) and a bifunctional desaturase from Hedera helix (ivy) reveal a di-iron metal center that is generated by two conserved His and four Glu residues (Guy et al. 2007; Moche et al. 2003). Depending on the oxidation state, the two iron atoms are ~ 3.2–4.2 Å apart with a μ-oxo bridge. In contrast, integral membrane desaturases act upon acyl-CoA-linked lipids and are found in endomembrane systems in prokaryotes and eukaryotes.
The recent crystal structures of the human and mouse SCD1 Δ9 membrane desaturases reveal a mushroom top-like cap that sits upon a 4-helical transmembrane (TM) bundle and other membrane-associated, amphipathic helices (AH) (Bai et al. 2015; Wang et al. 2015). The cap region juxtaposes four His-containing motifs to bind two metal ions and generates the substrate binding channels and interaction surface for the CoA moiety. Unfortunately, both the human and mouse SCD1 and the related yeast Scs7p sphingolipid α-hydroxylase are inactive. They are inactive since they contain two zinc atoms within the active site instead of the essential iron atoms (Zhu et al. 2015). While these structures do provide a rationale for the selective action at the ninth carbon atom, they do not provide insight into the substrate specificity of the ω6 and ω3 desaturases that act upon different carbon atom positions.
Work by our groups have focused on the desaturases from the oleaginous fungus Mortierella alpina, which can produce lipids up to 50% of its dry weight (Chen et al. 2013; Shi et al. 2015; Wang et al. 2011). M. alpina has a single copy of the ω3 and ω6 fatty acid desaturase genes and can generate EPA and ARA through the n-3 and the n-6 arms, respectively, of the PUFA biosynthetic pathway (Fig. 1), when cultured below 20 °C (Shimiziu et al. 1988; Shinmen et al. 1989). The data presented herein describes the first successful large-scale overexpression, purification, and characterization of the ω6 and ω3 desaturases, also known as the Δ12 and Δ15/17 desaturases, respectively. Both purified enzymes contain two molecules of Fe3+ per monomer with a unique UV-visible spectrum, are enzymatically active, and show marked preferences for the lipid chain length and site of unsaturation. Four His-containing motifs are conserved despite the low sequence homology to the Δ9-desaturase and other membrane desaturases. Transmembrane helix predictions and comparisons to the SCD1 and Scs7p topologies support that the ω6 and ω3 desaturases also contain a TM helical bundle. However, the cap region is most likely rearranged to yield substrate-binding pockets that tailor the unique specificity of these enzymes and may include repositioning of the metal center.
Materials and methods
Expression and purification of ω3 and ω6 desaturase
The desaturase-coding sequences were identified from M. alpina (no. 32222, American Type Culture Collection, Manassas, VA, USA), as previously described (Wang et al. 2011). The codon-optimized genes (GenScript) for the ω3 and ω6 desaturases were appended to a cassette containing the human rhinovirus 3C protease cleavage site, the IgG-specific ZZ-tag, and an RGS-His10-tag and cloned into the pPink-HC vector (Invitrogen) between the EcoRI and NaeI sites (Fig. S1) (Chen et al. 2006). Pichia pastoris strain 2 transformation and small-scale expression followed the general procedures described previously (Chen et al. 2013). The highest expressing clones were determined by Western blot using Anti-RGS-His10 HRP Conjugates (Qiagen).
For large-scale expression, uniform seed glycerol stocks were prepared as follows: a single colony was picked and inoculated in BMGY-buffered glycerol-complex medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate pH 6.0, 1.34% YNB-yeast nitrogen base (BD), 0.0004% biotin (Sigma), 1% glycerol) and shaken for 24 h at 28 °C and 250 rpm. Aliquots of glycerol stocks were then made by mixing the culture with sterile glycerol to a final concentration of 25% glycerol. Seed glycerol stocks (100 μL) were inoculated into 25 mL BMGY in 250-mL flask. The cells were grown for 24 h at 28 °C with shaking at 250 rpm to an OD600 of 10. Six 1-L BMGY cultures, in 2.8-L Fernbach flasks, were inoculated with 25-mL inoculum and grow at 28 °C for 30 h with vigorous shaking at 250 rpm to an OD600 of 100–120. Each culture was then centrifuged at 1500g for 10 min at room temperature, the cell pellets were resuspended in 200 mL of BMMY-buffered methanol-complex medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate pH 6.0, 1.34% YNB, 0.0004% biotin, 0.5% methanol) and cultured at 28 °C with shaking at 250 rpm to induce the expression. After induction for 24 h, the cells were harvested by centrifuging for 10 min at 1500g and stored frozen at − 80 °C.
A typical desaturase preparation was started by resuspending 100 g cells in lysis buffer (20 mM Hepes pH 7.5, 150 mM NaCl, 10% (w/v) glycerol, 100 μM PMSF, 100 μM, benzamidine) at a ratio of ~ 0.3 g (wet weight)/mL on ice. The cells were lysed by three passes at ~ 27,000 psi through an Avestin EmulsiFlex-C3 cell homogenizer with the outlet cooling coil immersed in an ice bath. The lysate was centrifuged at 1000g for 30 min at 4 °C. The pellets from this spin were resuspended in a total of 100 mL of lysis buffer, subjected to two additional passes through the homogenizer centrifuged again at 1000g for 30 min at 4 °C.
A membrane fraction was isolated by subjecting the supernatants from the post-lysis centrifugations to pelleting at 20,000g for 60 min at 4 °C. The pellet (~ 18 g) was resuspended in membrane buffer (50 mM Hepes pH 7.5, 150 mM NaCl, 15% (w/v) glycerol, 100 μM PMSF, 100 μM benzamidine) at 7 mL/g using a hand-held homogenizer (POLYTRON® PT 1200) set at the lowest power to prevent foaming.
For efficient extraction of desaturases, a wide variety of detergents with different head groups, chain lengths, and critical micelle concentration values (CMC) were tested: n-dodecyl-β-d-maltopyranoside (DDM), octaethylene glycol monododecyl ether (C12E8), n-octyl-β-d-glucoside (OG), Triton X-100, Tween-20, 5-cyclohexyl-1-pentyl-β-d-maltoside (CYMAL-5), n-decyl-β-d-maltoside (DM), n-octyl-β-d-thioglucopyranoside (OTG), N,N-dimethyldodecylamine N-oxide (LDAO), 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS), n-decylphosphocholine (Fos-10), and n-hexadecylphosphocholine (Fos-16) with SDS as a positive control. Then, 14 mL 10% solution of Fos-Choline-16 (w/v in water; Fos-16, Anatrace, Anagrade) was added to a final detergent concentration of 1% (w/v) and by incubation at 4 °C for 90 min, with agitation using a Rotisserie hybridization rotator (Labquake™, Thermo Scientific). Insoluble material was removed by centrifugation at 4000g for 30 min at 4 °C.
Affinity purification of desaturases was conducted using the C-terminally fused IgG-binding ZZ-tag. The solubilized membranes were incubated overnight at 4 °C on the rotator with 10 mL (packed resin) of IgG Sepharose 6 Fast Flow (GE Healthcare) that had been pre-equilibrated with buffer containing 20 mM Hepes pH 7.5, 150 mM NaCl, 15% (w/v) glycerol, and 0.002% (w/v) Fos-16. The IgG Sepharose was then washed three times at 4 °C with 30 resin volumes (each) of wash buffer consisting of 20 mM Hepes, pH 7.5, 150 mM NaCl, 15% (w/v) glycerol, 0.002% Fos-16, 100 μM PMSF, 100 μM benzamidine, and 1 mM DTT. Desaturases were released from the IgG Sepharose by treatment with rhinovirus 3C protease. One resin volume of the wash buffer containing 0.002% Fos-16 and rhinovirus 3C protease (HRV-3C) tagged with an N-terminal His6-tag (His6-HRV3C) at a ratio of 1:3 was incubated with the IgG Sepharose overnight at 4 °C. The released desaturases were separated from the IgG Sepharose resin at 4 °C by filtration using a 5.0 × 10 cm glass chromatography column (Bio-Rad) and the IgG Sepharose resin was washed three more times. The filtrate, containing the desaturase and protease, was collected in a tube and maintained on ice. The His-tagged protease was then removed by incubation of the filtrate for 2 h at 4 °C with 2 mL of HisPur Cobalt Superflow Agarose (Thermo Scientific) that had been pre-equilibrated with the wash buffer containing 0.002% Fos-16. The desaturases were separated from the bound protease using filtration as above; then the filtrate was concentrated ~ 3-fold to ~ 5 mL using a 50-kDa cutoff Vivaspin 20 concentrator (GE Healthcare).
The affinity-purified protein was subjected to size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare) that had been pre-equilibrated with 20 mM Hepes pH 7.5, 5% (w/v) glycerol, 150 mM NaCl, 0.002% Fos-16, 100 μM PMSF, 100 μM benzamidine). The column was eluted with the same buffer at a flow rate of 1 mL/min. Fractions containing the desaturase, based on SDS-PAGE analysis, were concentrated to 7–15 mg/mL, as determined by the Pierce BCA Protein Assay (Thermo Scientific) using bovine serum albumin as the reference standard. The iron content of the desaturases was determined by an Iron Assay Kit (Sigma) containing a calibrated standard (Fig. S2). The protein was incubated in the iron assay buffer for 2 h at 60 °C, in order to ensure the complete release of the iron from the protein and formation of the Fe2+-ferrozine complex. For the determination of the extinction coefficient, standard dilutions of the purified proteins were prepared and absorbances at 412 nm were monitored. According to Beer-Lambert law, the slope of the absorption spectrum is the extinction coefficient as the path length is fixed.
Expression and purification of human NADH cytochrome b5 reductase and human cytochrome b5
The expression plasmid for the soluble form of human NADH cytochrome b5 reductase (hCytb5R) was kindly provided by Dr. Lauren Trepanier (University of Wisconsin-Madison, USA) (Kurian et al. 2004). The codon-optimized gene for human cytochrome b5 (hCytb5) was generated by GenScript and subloned into the pET15b vector (Novagen). Protein expression was performed in BL21(DE3) Escherichia coli cells in six 1-L Fernbach flasks containing LB media and ampicillin (100 mg/L) at 37 °C and 250 rpm. Once an OD600 reached 0.6–0.8, the cells were cooled on ice, 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) added, and induction performed at room temperature for 4 h. The cell pellets were stored at − 80 °C. Both enzymes were purified using the following procedure. The cells were resuspended in 150 mL of buffer containing 20 mM Hepes pH 7.9, 500 mM KCl, 5 mM imidazole, 0.1% Triton X-100, 10% glycerol, 0.1 mM of the protease inhibitors PMSF and benzamidine, 1 mM MgCl2, and 40 μg/mL Dnase I. For hCytb5R, 0.2 mM FAD was also added. The cells were lysed using an Avestin EmulsiFlex-C3 cell homogenizer, and the supernatant loaded onto a 10-mL HisPur Cobalt Resin (Thermo Scientific) column. The column was washed with several column volumes of the same buffer without glycerol added and the protein eluted with a linear 5–500 mM imidazole gradient. The fractions containing the protein of interest were pooled and dialyzed against 20 mM Tris pH 8.0 overnight. The protein was purified from the dialysate using a Q-Sepharose FF column (GE Healthcare) and eluted with a linear gradient to 1 M NaCl. The final step of the purification involved separation on the HiLoad 16/60 Superdex 200 column equilibrated with 20 mM Hepes pH 7.5, 100 mM NaCl. The protein solution was concentrated using a Vivaspin concentrator. The His-tag was not removed from Cytb5R. The His-tag was removed from hCytb5 by the addition of 0.2 U/mg of biotinylated thrombin during the dialysis step; the thrombin was removed by incubation with streptavidin agarose beads. Removal of the His-tag was confirmed by MALDI-TOF MS. For hCytb5, a 3-fold M excess of hemin chloride, prepared by the published method (Goren and Fox 2008), was added and incubated for 1.5 h prior to injection of the Superdex 200 column, in order to ensure full loading of this critical cofactor.
Kinetic analysis of ω3 and ω6 desaturases
The enzymatic activities of ω3 and ω6 desaturases were determined by monitoring the reoxidation of hCytb5 in the presence and absence of fatty acid-CoA substrates (Oshino et al. 1971; Strittmatter et al. 1972). Assays were performed in quadruplicate on three separate days using 96-well UV-transparent half-area microplates (Corning) and a SpectraMax 340PC384 absorbance microplate reader. The 0.1-mL reaction mixtures contained 20 mM Hepes pH 7.5, 150 mM NaCl, 1 μM NADH, 0.002% Fos-16, different concentrations of fatty acid-CoA substrate (0–300 μM), 100 nM desaturase, 0.5 μM hCytb5, and 0.25 μM hCytb5R. Enzyme mixtures (100 nM desaturase, 0.5 μM hCytb5 and 0.25 μM hCytb5R, 20 mM Hepes pH 7.5, 150 mM NaCl, 0.002% Fos-16) and substrate/NADH mixtures (different concentrations of fatty acid-CoA substrate and 1 μM NADH) were incubated separately at 28 °C for 5 min. The incubated materials were then mixed and monitored at 422 nm and 28 °C. In this coupled assay, hCytb5 was instantaneously and completely reduced by NADH and hCytb5R. The hCytb5 remains reduced until all the NADH becomes consumed, and then hCytb5 becomes oxidized. The desaturase activity was calculated by determining the difference between the time for NADH oxidation in the absence and presence of fatty acid-CoA, indicated by the onset of hCytb5 reoxidation. And it was assumed that 1 mol of NADH is required for the formation of 1 mol of the unsaturated fatty acid-CoA product. The k cat, K m, and k cat/K m values were determined using nonlinear regression in the enzyme kinetics module of SigmaPlot 12.0 (Systat Software). Reaction products from the kinetic assays were verified by FAME (fatty acid methyl esters)-GC/MS analysis and the linolenate formed by M. alpina ω3 desaturase was also quantitated relative to added internal standards, as previously described (Chen et al. 2013). All acyl-CoA substrates were purchased from Avanti Polar Lipids (Alabaster, AL).
Phylogenetic analysis and transmembrane topology predictions
Selected membrane fatty acid desaturase amino acid sequences were aligned with ClustalW in the MEGA, ver. 6 (Tamura et al. 2013) using default parameters, including the Gonnet scoring matrix, a gap penalty of 10, and a gap extension penalty of 0.1. The resulting alignment was used to generate a distance-based unrooted phylogram using the neighbor-joining method performed under the Phylogeny module. Parameters for developing neighbor-joining tree included the use of the Poisson substitution model and a pairwise deletion method to treat gaps in the program. The transmembrane helices of the M. alpina ω3 and ω6 desaturases, mouse stearoyl-CoA desaturase, and yeast fatty acid 2-hydrolase Scs7p were predicted using the PolyPhobius program (http://phobius.sbc.su.se/poly.htmL) (Käll et al. 2005).
Nucleotide sequence and amino acid sequence accession number
The natural nucleotide sequence of M. alpina ω3 desaturase and M. alpina ω6 desaturase gene was deposited in the GenBank database under the accession number KF433065 and KF433064. The codon-optimized nucleotide sequence of M. alpina ω3 desaturase, M. alpina ω6 desaturase, and human cytochrome b5 gene was deposited in the GenBank database under the accession number MF101847, MF101848, and MF101850, respectively. The amino acid sequence accession numbers used: human SCD1 (GI: 53759151), mouse SCD1 (GI: 31543675), zebrafish SCD1 (GI: 28394115), Acheta domesticus Δ9 desaturase (GI: 13430287), Saccharomyces cerevisiae Δ9 desaturase (GI: 172064), M. alpina Δ9 desaturase (GI: 556911674), M. alpina ω6 desaturase (GI: 556911676), and M. alpina ω3 desaturase (GI: 556911678). Kluyveromyces lactis CBS 2359 Δ12 desaturase (KLLA0F07095g), Cyberlindnera fabianii YJS4271 Δ12 desaturase (CYFA0S32e00958g), Candida albicans SC5314 (C6_01110W_A) Δ12 desaturase, Lachancea cidri CBS 2950 Δ12 desaturase (LACI0G04676g), K. lactis CBS 2359 Δ15 desaturase (KLLA0B00473g), C. fabianii YJS4271 Δ15 desaturase (CYFA0S01e00826g), C. albicans SC5314 Δ15 desaturase (C1_13070C_A), L. cidri CBS 2950 Δ15 desaturase (LACI0G19350g).
Results
M. alpina ω3 and ω6 desaturases possess a di-iron center
The ω3 and ω6 desaturase genes from M. alpina were expressed as a C-terminal fusion to an HRV-3C protease recognition site-ZZ-His-tag, using the P. pastoris PichiaPink system (Fig. S1). This particular heterologous expression system has been used for a variety of soluble and membrane proteins and allows for the rapid identification and selection of expression clones. Transformants having the highest expression level, as determined by Western blot, were selected for large-scale expression and purification. To extract the recombinant desaturases from the cell membrane, the membrane isolation protocol was optimized and a variety of detergents with different head groups, chain lengths, and critical micelle concentration (CMC) values were tested (DDM, C12E8, OG, Triton X-100, Tween-20, CYMAL-5, DM, OTG, LDAO, CHAPS, Fos-Choline-10 and Fos-Choline-16 (Fos-16), and SDS as a positive control; see the “Materials and Methods” section). The optimal protocol involved removing cellular debris by centrifugation at 1000g for 30 min and then isolating the membranes by centrifugation at 20,000g for 1 h. Only Fos-16 effectively extracted the desaturases from the membrane, when compared to the efficiency of SDS. The solubilized proteins were then purified by IgG affinity chromatography and released from the IgG beads by treatment with HRV-3C protease and dithiothrietol. The proteins were further purified and buffer exchanged by passage over a size-exclusion column.
The resulting proteins were homogenous and consistent with their theoretical molecular weights (ω6 46,836 Da; ω3 desaturase 47,426 Da (Fig. 2a). Upon concentration, the purified ω3 and ω6 desaturases were noticeably yellow in color. The UV-visible spectra of these non-heme-containing desaturases (Fig. 2b, c) revealed a peak of absorption at 412 nm (inset), indicating the presence of bound iron. Quantitation of the iron resulted in values of 1.9 ± 0.2 and 2.0 ± 0.2 mol of iron per mol of protein for the ω6 and ω3 desaturase, respectively. The extinction coefficients were determined to be 5210 ± 184 and 5865 ± 298 M−1 cm−1 for the ω6 and ω3 desaturase, respectively. This work represents the first homogenous preparation of these important ω3/ω6 desaturase representatives. Moreover, as described in more detail in the “Discussion” section, the presence of a di-iron metal center contrasts with the recent crystal structures of the mouse and human SCD1 desaturases and the Scs7p hydroxylase. Both of these enzymes were inactive since they contained two zinc atoms in the active site instead of two iron atoms (Bai et al. 2015; Wang et al. 2015; Zhu et al. 2015).
Biochemical characterization of M. alpina ω3 and ω6 desaturase. a SDS-PAGE analysis of 5 μg of each FOS16-solubilized protein. M, maker; lane 1, ω6 desaturase; lane 2, ω3 desaturase. b UV-visible spectrum of ω6 desaturase. Inset, 350–500-nm region illustrating the iron charge transfer bands. The sample contained 14-μM protein, 20 mM Hepes pH 7.5, 150 mM NaCl, 5% glycerol. c UV-visible spectrum of ω3 desaturase. The same parameters used as in panel b. d Coupled reaction scheme to monitor the desaturase activity. NADH and human cytochrome b5 reductase (hCytb5R) are used to rapidly reduce human cytochrome b5 (hCytb5) and in turn the desaturase. The addition of lipid CoA substrate results in the reoxidation of hCytb5, as the reaction is setup so that NADH (0.1 nmol) is limiting. e Representative progress curves for the reaction of the ω3 desaturase with 18:2-CoA. Note the rapid reoxidation of hCytb5 by the addition of lipid substrate versus air oxidation in the blank. Reaction conditions: 20 mM Hepes pH 7.5, 150 mM NaCl, 1 μM NADH, 0.002% Fos16, 100 nM desaturase, 0.5 μM hCytb5, and 0.25 μM hCytb5R. f Michaelis-Menten analysis of the reaction between the ω3 desaturase and 18:2-CoA. See Table 2 for kinetic values determined for this substrate and others. (SigmaPlot, ChemDraw, and Adobe Illustrator were used to create this artwork)
M. alpina ω6 and ω3 desaturases are selective for 18:1 and 18:2 acyl-CoA substrates
The ω6 and ω3 desaturases are located at key branch points of the PUFA biosynthetic pathway (Fig. 1). Previous studies have analyzed the change in whole-cell lipid profiles of S. cerevisiae that overexpresses these and other fungal desaturases (Kainou et al. 2006; Sakuradani et al. 2005; Sakuradani et al. 1999). The lack of purified enzyme has precluded detailed kinetic studies. It is not clear, for example, whether the ω6 desaturase enzymes can work efficiently on substrates of different chain lengths (16:1 and 18:1), and how efficient can the ω3 desaturase work on different ω6 fatty acid substrates.
The desaturation of fatty acids by integral membrane desaturases requires the coupling of the electron-transport proteins NADH-cytochrome b5 reductase and cytochrome b5 (Enoch et al. 1976). In order to determine the activity of the M. alpina desaturases against a variety of fatty acid-CoA substrates, a continuous assay was used that contains the soluble forms of human cytochrome b5 reductase (hCytb5R) and human cytochrome b5 (hCytb5) (Fig. 2d) (Goren and Fox 2008; Oshino et al. 1971; Strittmatter et al. 1972). This assay, developed by Strittmatter et al. (Strittmatter et al. 1972), monitors the reduction and reoxidation of hCytb5 at 422 nm to indicate the NADH oxidation in the absence and presence of fatty acid-CoA, as the hCytb5R absorbance interferes at 340 nm. Upon the addition of NADH, the hCytb5 was reduced rapidly (occurs within the mixing time of the samples) and completely, remained in the reduced state until all the NADH was consumed, and was then auto-oxidized at a measurable rate. The presence of substrate greatly reduces the time required for NADH oxidation, enabling the calculation of the rate of reaction. Figure 2e illustrates the principle of the assay using the ω3 desaturase with 18:2 n-6 substrate. The rate at different acyl-CoA substrate concentrations (Fig. 2f) was used to determine the k cat, K m, and k cat/K m values for each substrate.
The mobilities of the reaction products on GC and the double bond position of different reaction products were verified by FAME-GC/MS analysis (Fig. S3 and S4). The α-linolenic acid (18:3 n-3) formed by M. alpina ω3 desaturase activity using 20 μM linoleoyl (18:2 n-6)-CoA as substrate was also quantitated relative to added internal standards. A comparison of the spectrophotometric assay and FAME analysis was shown in Table 1. The linolenate formation calculated from the rates of NADH oxidation was comparable to the amount of linolenate quantitated by FAME analysis. This shows that the simple and rapid spectrophotometric procedure agrees well with results obtained by the usual FAME-GC/MS analysis.
The M. alpina ω6 desaturase exhibited a ~ 2-fold greater k cat/K m value for oleoyl (18:1 n-9)-CoA over pamitoleoyl (16:1 n-7)-CoA (Table 2), supporting that the preferred substrate is oleoyl-CoA. This effect appears to be predominantly due to the 3-fold lower k cat value for 16:1 n-7. This enzyme was not able to act upon saturated fatty acids. The ω3 desaturase showed marked differences for linoleoyl (18:2 n-6)-, γ-linolenoyl (18:3 n-6)-, and arachidonoyl (20:4 n-6)-CoA substrates and was inactive towards 18:0-, 20:0-, and 18:1-CoA (data not shown). While the γ-linolenoyl-CoA substrate was turned over with a k cat value ~ 2-fold higher than that for linoleoyl-CoA, the K m value for the linoleoyl-CoA was ~ 6-fold lower. The arachidonoyl-CoA substrate had the lowest apparent affinity with a K m value of 157 μM, a value ~ 10-fold higher than linoleoyl-CoA. Altogether, these data support that linoleoyl-CoA is the best substrate for the ω3 desaturase, they also suggest that γ-linolenoyl-CoA can also be efficiently utilized.
M. alpina ω3 and ω6 desaturases are distantly related to the ∆9 desaturases
Three conserved histidine-containing motifs (one HX4H and two HX2HH) have been described for integral membrane desaturases, alkane hydroxylases (e.g., yeast Scs7p) and xylene monooxygenases (Shanklin and Whittle 2003; Shanklin et al. 1994). In the SCD1 and Scs7p structures, a fourth motif ((N/H)X3H) was identified (Bai et al. 2015; Wang et al. 2015; Zhu et al. 2015). The location of these motifs is illustrated in the sequence alignments comparing M. alpina ω3 and ω6 desaturases with human/mouse SCD1 and more closely related yeast Δ12 and Δ15 desaturases (Santomartino et al. 2017) (Fig. 3a). The four histidine-containing motifs are found within the M. alpina ω3 and ω6 desaturases and yeast Δ12 and Δ15 desaturases, but their sequence identities to the SCD1 and Scs7p proteins are all very low (< 20%) and several key differences exist. For example, the first motif is HX3H instead of HX4H and the (N/H)X3H motif appears to be TX3H, and 7- and 10-residue inserts flank the second HX2HH motif. Thus, the close juxtaposition of all four His motifs in the crystal structures suggests that significant structural rearrangements of the active site region must occur in order to accommodate the unique sequence features of the ω3 and ω6 desaturases. Interestingly, the M. alpina ω6 and ω3 desaturases and yeast Δ12 and Δ15 desaturases, except for the Δ12 enzyme from C. fabianii, contain additional HX2H or HXHH sequences in an insertion adjacent to the HX2HH motif. These additional sequences suggest the possibility of a unique metal center coordination sphere for the non-SCD-like desatures. This notion is further supported by the clear separation of the ∆9, ω6/ω3, and Δ12/Δ15 branches identified by a phylogenetic tree analysis (Fig. 3b).
Primary structure analysis of M. alpina ω6 and ω3 desaturases surrounding the conserved His-box motifs. a Partial sequence alignment of M. alpina ω3 and ω6 desaturase with integral stearoyl-coenzyme A desaturases and yeast Δ12 and Δ15 desaturases. Conserved histidine residues in the primary coordination sphere of the di-iron unit are highlighted by hollow box. Additional His residues that could contribute to the metal center are highlighted by dashed box. Accession numbers used: human SCD1 (GI: 53759151), mouse SCD1 (GI: 31543675), zebrafish SCD1 (GI: 28394115), Acheta domesticus Δ9 desaturase (GI: 13430287), Saccharomyces cerevisiae Δ9 desaturase (GI: 172064), Mortierella alpina Δ9 desaturase (GI: 556911674), Mortierella alpina ω6 desaturase (GI: 556911676) and Mortierella alpina ω3 desaturase (GI: 556911678). Kluyveromyces lactis CBS 2359 Δ12 desaturase (KLLA0F07095g), Cyberlindnera fabianii YJS4271 Δ12 desaturase (CYFA0S32e00958g), Candida albicans SC5314 (C6_01110W_A) Δ12 desaturase, Lachancea cidri CBS 2950 Δ12 desaturase (LACI0G04676g), Kluyveromyces lactis CBS 2359 Δ15 desaturase (KLLA0B00473g), Cyberlindnera fabianii YJS4271 Δ15 desaturase (CYFA0S01e00826g), Candida albicans SC5314 Δ15 desaturase (C1_13070C_A), Lachancea cidri CBS 2950 Δ15 desaturase (LACI0G19350g). b Phylogenetic relationships between M. alpina ω3 and ω6 desaturases, yeast Δ12 and Δ15 desaturases, and other lipid-coenzyme A desaturases. The neighbor-joining tree was developed using a Poisson substitution model and gaps were treated using the method of pairwise deletion in the program MEGA ver. 6 (Tamura et al. 2013). The lengths of the branches and the numbers next to each node represent the amount of change in evolutionary lineages over time and the probability of cluster, respectively. (Microsoft Word, MEGA, and Adobe Illustrator were used to create this artwork)
ω3 and ω6 desaturases have the same membrane topology as ∆9 desaturases
The recent crystal structures provide a new framework to predict the membrane topology of the M. alpina desaturases. The crystal structures all exhibit a transmembrane (TM) four helical bundle and a mushroom top-like domain that contains the metal-binding His motifs and substrate-binding surfaces and channels (Bai et al. 2015; Wang et al. 2015; Zhu et al. 2015). This region also contains several membrane-associated amphipathic helices (AH). A recent evaluation of 12 programs that predict TM helices found that PolyPhobius outperformed all others (Reeb et al. 2015). We used this program to predict the helices of the MmSCD1 and Scs7p sequences and the M. alpina ω3 and ω6 desaturases. The former was a key validation step, as the SCD1 and Scs7p structures represent a new class of membrane proteins not used in the program development.
PolyPhobius was readily able to identify multiple helices for MmSCD1 and Scs7p (Fig. 4a, top 2 panels). For MmSCD1 and Scs7p, the transmembrane helices found in the crystal structure have the highest probability. Interestingly, the AHs are also predicted, but with lower probability. It is important to note here that the Scs7p protein also contains an N-terminal CytB5 domain, consistent with inability of the program to identify TM helices in this region. The prediction for these two proteins correlates well with the crystal structures (Fig. 4b) and places all His motifs on the same side of the membrane.
Topology predictions for M. alpina ω6 and ω3 desaturases. a Comparison of the sequence-based transmembrane predictions for MmSCD1, Scs7p to those of the M. alpina ω6 and ω3 desaturases. Plots generated by PolyPhobius (Käll et al. 2005). The probability of residues being on one side of the membrane or another is indicated in blue and green. Putative helices are highlighted in gray. For MmSCD1 and Scs7p, the transmembrane (TM) and membrane-associated, amphipathic helices (AH), identified from their crystal structures, are indicated. b Topology diagrams. The structures of MmSCD1 and Scs7p both contain four TM, multiple AH, and other cap helices. This arrangement places all four His-box motifs (His residues shown as green spheres) on the same side of the membrane. Using this key criterion, the identified helices in panel b, and the conservation of the His-box motifs in Fig. 3, the predicted topologies for the M. alpina ω6 and ω3 desaturases are shown. (PolyPhobius, Microsoft Powerpoint, and Adobe Illustrator were used to create this artwork)
In contrast to the MmSCD1 and Scs7p, the predictions for the M. alpina ω3 and ω6 desaturases and closely related yeasts Δ12 and Δ15 desaturases from K. lactis CBS 2359, C. fabianii YJS4271, C. albicans SC5314, and L. cidri CBS 2950 all identify six helices with high probability; the topology models of M. alpina ω3 and ω6 desaturases are illustrated in Fig. 4a, bottom two panels. However, if one carefully traces the path through the membrane, the predictions would place the HX2HH motif on the opposite side of the membrane from the other His motifs, an impossibility to maintain the formation of the metal center. If one considers the sequence alignments, however, the two central helices are most likely AH helices, like AH1 in MmSCD1 and AH7 in Scs7p (Fig. 4b). Based on this analysis and the conservation of histidine motifs, we propose that the ω3 and ω6 desaturases of M. alpina and other closely related yeasts, Δ12 and Δ15 desaturases contain a 4-TM helical bundle and two intervening AH helices. This topology contrasts with the arrangement seen in the SCD1 and alkyl hydrolase proteins. As will be discussed more in the next section, the presence of a unique cap region is consistent with the different substrate specificities of the enzymes while maintaining a similar di-metal center for catalysis.
Discussion
Integral membrane fatty acid desaturases are widely distributed in nature and desaturate bonds at a variety of different positions along the aliphatic chain of acyl-CoA substrates (Fig. 1). This ability suggests that the structure of each enzyme is tailored to first recognize a substrate of a particular chain length(s) and then to perform the desaturase reaction at a specific position. Previous studies of the membrane desaturases have been limited to either studying the effects of the heterologous expression on the lipid profiles of a host organism such as S. cerevisiae or monitoring the activity of the enzyme reconstituted in complex proteoliposomes (Enoch et al. 1976; Sakuradani et al. 2005; Sakuradani et al. 1999). While the results from these studies are confounded by the presence of other lipid and protein mixtures, they have played a key role in enzyme identification and classification. The present study has produced homogeneous preparations of the ω3 and ω6 desaturases from M. alpina reconstituted in detergent micelles (Fig. 2a), verified the presence of a metal center containing two iron atoms, and determined the kinetic parameters for a panel of substrates for the first time. Comparisons of sequence alignments, crystal structures and predictions of membrane topology all support that the ω3 and ω6 desaturases are unique.
The quantitation of iron content and the spectra of the M. alpina ω3 and ω6 desaturases are consistent with the presence of a di-iron, non-heme metal center (Fig. 2b, c). As mentioned earlier, the presence of zinc in the active sites of the SCD1 and Scs7p proteins prevents a direct comparison of spectra. One would expect that the spectra of the SCD1 proteins to be similar, since the sequence alignments (Fig. 3a) support the conservation of a 9-His metal center. The differences of the histidine-containing motifs and the presence of additional HX2H and HXHH sequences in the ω6 and ω3 desaturases suggest the possibility of a unique metal center coordination sphere.
A survey of the literature finds that the absorbance maximum of 412 nm is similar to a reaction intermediate of the metal center of methane monooxygenase from Methylococcus capsulatus (Bath) (λ max = 420 nm), which contains a 2-His/4-Glu di-iron metal center (Liu et al. 1995). The di-iron center of toluene-4-monooxygenase also contains an acid-rich motif (λ max = 458 nm) (Pikus et al. 1996). These absorption maxima differ significantly from those found in the soluble desaturases that contain 2-His/4-Glu motifs (λ max = 345–375 nm) (Fox et al. 1993; Shanklin et al. 2009; Whittle et al. 2005). Nonetheless, detailed spectral analyses of each of these proteins and others support the presence of an oxygen atom bridging between the metal ions. Additional spectral and structural studies are clearly needed for the membrane desaturases to clarify the details of the metal center and its oxidation state during catalysis, but this will have to await the optimization of iron and iron-57 incorporation into these proteins on a much larger preparative scale.
With the presence of iron in our preparations, we could access the activity of the ω6 and ω3 desaturases against a panel of substrates. Using a coupled assay system containing the soluble forms of human Cytb5R and Cytb5 (Fig. 2d–f and Table 2), the specificity activity for the optimal substrates is on par with historical preparations of the rat liver microsomal stearoyl-CoA desaturase (235–464 versus 240–360 nmol mg−1 min−1protein, respectively) (Goren and Fox 2008; Oshino et al. 1971; Strittmatter et al. 1972; Strittmatter et al. 1974). The low μM K m values are also consistent with these studies and those of the soluble ∆9 desaturases (Whittle et al. 2005). The k cat values are, however, ~ 10-fold lower than those of the soluble ∆9 desaturases, suggesting that the coupling of the human Cytb5R-Cytb5 is not as efficient as the endogenous, membrane-associated reduction system. In the FAME-GC/MS analysis of reaction products (Fig. S3 and S4), fatty acids 18:2 n-6 and 18:3 n-3 were detected in the blank sample. Control experiments involving lipid extraction from the purified proteins revealed that the purified ω3 and ω6 desaturases contain fatty acids 16:0, 16:1 n-7, 18:0, 18:1 n-9, 18:2 n-6, and 18:3 n-3, some of which are also substrates for these enzymes (Fig. S5 and Table S1). We tried adding excess NADH without the presence of exogenous substrates to stimulate catalysis and modification of these lipids. The amount of 18:3 n-3 and 18:2 n-6 did not vary with time (data not shown). These observations lead us to think that the co-purified fatty acids do not occupy the substrate-binding tunnel near the di-metal center but may block other portions of the active site, which might also explain the lower turnover rates.
Based on the k cat/K m values, the preferred acyl-CoA substrate for the ω6 desaturase was 18:1 n-9 (Table 2). The optimal substrate for the ω3 desaturase was linoleoyl-CoA with conversion to α-linolenic acid (18:3 n-3). Interestingly, the ω3 desaturase also converted γ-linolenoyl-CoA to stearidonic acid (18:4 n-3). These data are consistent with the change in lipid profiles, when the proteins are heterologously expressed (Kainou et al. 2006; Sakuradani et al. 2005; Sakuradani et al. 1999). The fact that the enzyme can utilize both linoleoyl-CoA and γ-linolenoyl-CoA but arachidonoyl-CoA is a poor substrate in M. alpina has implications for the engineering of this organism for the industrial production of n-3 LC-PUFAs such as EPA and DHA. The data implies: (i) that this organism has the capacity to convert both linoleic acid and γ-linolenoyl-CoA to α-linolenic acid and stearidonic acid, respectively, thereby converting eighteen-carbon n-6 to n-3 PUFAs for further metabolism to n-3 LC-PUFAs (Fig. 1) and (ii) that the ω3 desaturase does not efficiently convert n-6 LC-PUFAs such as ARA to n-3 LC-PUFAs such as EPA. Consequently, fermentation conditions or variant strains that promote conversion of eighteen-carbon n-6 into n-3 PUFAs should enhance the concentrations of n-3 LC-PUFAs and be advantageous.
The TM and AH profiles from the MmSCD1 and Scs7p crystal structures are in excellent agreement with those predicted by PolyPhobius program (Fig. 4), even though this membrane protein fold was not included in the development of this algorithm. Both proteins contain a four-TM helix bundle with a cap that contains either three or four membrane-associated AH helices. The predictions for the ω6 and ω3 desaturases include a four-TM helix bundle but only two AH helices. Given the latter difference and the sequence changes in and around the four His-containing motifs of the di-iron metal center, one could easily envision that the cap structure of these enzymes must rearrange. It is also intriguing that the ω6 and ω3 desaturases have additional His residues near one of the HX2HH motifs. Thus, it is entirely possible that the composition and location of the metal center has changed. Further support for this notion comes from the conservation of the additional His-rich motif in several yeast Δ12 and Δ15 desaturases (Fig. 3).
A distinct cap for each desaturase subclass would be consistent with the need for a binding surface or pocket that would position the optimal substrate (18:1 n-9 or 18:2 n-6) with the appropriate register to the metal center. As observed in the ligand complexes of the MmSCD1 and HsSCD1 structures, the aliphatic portion of the substrate packs within clearly defined channels that help to “kink” the substrate and to position carbon atoms 9 and 10 adjacent to the di-zinc-containing metal center (Bai et al. 2015; Wang et al. 2015). The lack of conservation of the CoA-binding residues, between SCD1 and the ω6 and ω3 desaturases, further supports the presence of a distinct cap region for each enzyme. The future determination of the crystal structure of these enzymes will be invaluable to visualize the proposed topology and active site and to rationalize the regio- and stereo-specificity of these important enzymes for the production of PUFAs for dietary supplementation and the prevention and treatment of numerous diseases.
References
Bai Y, McCoy JG, Levin EJ, Sobrado P, Rajashankar KR, Fox BG, Zhou M (2015) X-ray structure of a mammalian stearoyl-CoA desaturase. Nature 524(7564):252–256. https://doi.org/10.1038/nature14549
Bazinet RP, Laye S (2014) Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat Rev Neurosci 15(12):771–785. https://doi.org/10.1038/nrn3820
Chen C, Huang Q-L, Jiang S-H, Pan X, Hua Z-C (2006) Immobilized protein ZZ, an affinity tool for immunoglobulin isolation and immunological experimentation. Biotechnol Appl Biochem 45(2):87–92. https://doi.org/10.1042/BA20060055
Chen H, Gu Z, Zhang H, Wang M, Chen W, Lowther WT, Chen YQ (2013) Expression and purification of integral membrane fatty acid desaturases. PLoS One 8(3):e58139. https://doi.org/10.1371/journal.pone.0058139
Enoch HG, Catalá A, Strittmatter P (1976) Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J Biol Chem 251(16):5095–5103
Fox BG, Shanklin J, Somerville C, Munck E (1993) Stearoyl-acyl carrier protein delta 9 desaturase from Ricinus communis is a diiron-oxo protein. Proc Natl Acad Sci U S A 90(6):2486–2490. https://doi.org/10.1073/pnas.90.6.2486
Goren MA, Fox BG (2008) Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr Purif 62(2):171–178. https://doi.org/10.1016/j.pep.2008.08.002
Gu Z, Shan K, Chen H, Chen YQ (2015) n-3 polyunsaturated fatty acids and their role in cancer chemoprevention. Curr Pharmacol Rep 1(5):283–294. https://doi.org/10.1007/s40495-015-0043-9
Guy JE, Whittle E, Kumaran D, Lindqvist Y, Shanklin J (2007) The crystal structure of the ivy Δ4-16:0-ACP desaturase reveals structural details of the oxidized active site and potential determinants of regioselectivity. J Biol Chem 282(27):19863–19871. https://doi.org/10.1074/jbc.M702520200
Käll L, Krogh A, Sonnhammer EL (2005) An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(Suppl 1):i251–i257. https://doi.org/10.1093/bioinformatics/bti1014
Kainou K, Kamisaka Y, Kimura K, Uemura H (2006) Isolation of delta12 and omega3-fatty acid desaturase genes from the yeast Kluyveromyces lactis and their heterologous expression to produce linoleic and alpha-linolenic acids in Saccharomyces cerevisiae. Yeast 23(8):605–612. https://doi.org/10.1002/yea.1378
Kurian JR, Bajad SU, Miller JL, Chin NA, Trepanier LA (2004) NADH cytochrome b5 reductase and cytochrome b5 catalyze the microsomal reduction of xenobiotic hydroxylamines and amidoximes in humans. J Pharmacol Exp Ther 311(3):1171–1178. https://doi.org/10.1124/jpet.104.072389
Liu KE, Valentine AM, Wang DL, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ (1995) Kinetic and spectroscopic characterization of intermediates and component interactions in reactions of methane monooxygenase from Methylococcus capsulatus (Bath). J Am Chem Soc 117(41):10174–10185. https://doi.org/10.1021/ja00146a002
Moche M, Shanklin J, Ghoshal A, Lindqvist Y (2003) Azide and acetate complexes plus two iron-depleted crystal structures of the di-iron enzyme delta9 stearoyl-acyl carrier protein desaturase. Implications for oxygen activation and catalytic intermediates. J Biol Chem 278(27):25072–25080. https://doi.org/10.1074/jbc.M301662200
Oshino N, Imai Y, Sato R (1971) A function of cytochrome b5 in fatty acid desaturation by rat liver microsomes. J Biochem 69(1):155–167
Pikus JD, Studts JM, Achim C, Kauffmann KE, Munck E, Steffan RJ, McClay K, Fox BG (1996) Recombinant toluene-4-monooxygenase: catalytic and Mössbauer studies of the purified diiron and rieske components of a four-protein complex. Biochemistry 35(28):9106–9119. https://doi.org/10.1021/bi960456m
Reeb J, Kloppmann E, Bernhofer M, Rost B (2015) Evaluation of transmembrane helix predictions in 2014. Proteins 83(3):473–484. https://doi.org/10.1002/prot.24749
Sakuradani E, Abe T, Iguchi K, Shimizu S (2005) A novel fungal ω3-desaturase with wide substrate specificity from arachidonic acid-producing Mortierella alpina 1S-4. Appl Microbiol Biotechnol 66(6):648–654. https://doi.org/10.1007/s00253-004-1760-x
Sakuradani E, Kobayashi M, Ashikari T, Shimizu S (1999) Identification of Delta12-fatty acid desaturase from arachidonic acid-producing mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur J Biochem 261(3):812–820
Santomartino R, Riego-Ruiz L, Bianchi MM (2017) Three, two, one yeast fatty acid desaturases: regulation and function. World J Microbiol Biotechnol 33(5):89. https://doi.org/10.1007/s11274-017-2257-y
Shanklin J, Guy JE, Mishra G, Lindqvist Y (2009) Desaturases: emerging models for understanding functional diversification of diiron-containing enzymes. J Biol Chem 284(28):18559–18563. https://doi.org/10.1074/jbc.R900009200
Shanklin J, Whittle E (2003) Evidence linking the Pseudomonas oleovorans alkane omega-hydroxylase, an integral membrane diiron enzyme, and the fatty acid desaturase family. FEBS Lett 545(2–3):188–192
Shanklin J, Whittle E, Fox BG (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33(43):12787–12794. https://doi.org/10.1021/bi00209a009
Shi H, Chen H, Gu Z, Song Y, Zhang H, Chen W, Chen YQ (2015) Molecular mechanism of substrate specificity for delta 6 desaturase from Mortierella alpina and Micromonas pusilla. J Lipid Res 56(12):2309–2321. https://doi.org/10.1194/jlr.M062158
Shimiziu S, Kawashima H, Shinmen Y, Akimoto K, Yamada H (1988) Production of eicosapentaenoic acid by Mortierella fungi. J Am Oil Chem Soc 65(9):1455–1459. https://doi.org/10.1007/BF02898307
Shinmen Y, Shimizu S, Akimoto K, Kawashima H, Yamada H (1989) Production of arachidonic acid by Mortierella fungi. Appl Microbiol Biotechnol 31(1):11–16. https://doi.org/10.1007/BF00252518
Strittmatter P, Rogers MJ, Spatz L (1972) The binding of cytochrome b5 to liver microsomes. J Biol Chem 247(22):7188–7194
Strittmatter P, Spatz L, Corcoran D, Rogers MJ, Setlow B, Redline R (1974) Purification and properties of rat liver microsomal stearyl coenzyme A desaturase. Proc Natl Acad Sci U S A 71(11):4565–4569. https://doi.org/10.1073/pnas.71.11.4565
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. https://doi.org/10.1093/molbev/mst197
Wang H, Klein MG, Zou H, Lane W, Snell G, Levin I, Li K, Sang BC (2015) Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat Struct Mol Biol 22(7):581–585. https://doi.org/10.1038/nsmb.3049
Wang L, Chen W, Feng Y, Ren Y, Gu Z, Chen H, Wang H, Thomas MJ, Zhang B, Berquin IM, Li Y, Wu J, Zhang H, Song Y, Liu X, Norris JS, Wang S, Du P, Shen J, Wang N, Yang Y, Wang W, Feng L, Ratledge C, Zhang H, Chen YQ (2011) Genome characterization of the oleaginous fungus Mortierella alpina. PLoS One 6(12):e28319. https://doi.org/10.1371/journal.pone.0028319
Wang M, Chen H, Gu Z, Zhang H, Chen W, Chen YQ (2013) ω3 fatty acid desaturases from microorganisms: structure, function, evolution, and biotechnological use. Appl Microbiol Biotechnol 97(24):10255–10262. https://doi.org/10.1007/s00253-013-5336-5
Weylandt KH, Chen YQ, Lim K, HM S, Cittadini A, Calviello G (2015) ω-3 PUFAs in the prevention and cure of inflammatory, degenerative, and neoplastic diseases 2014. Biomed Res Int 2015:695875. https://doi.org/10.1155/2015/695875
Whittle E, Cahoon EB, Subrahmanyam S, Shanklin J (2005) A multifunctional acyl-acyl carrier protein desaturase from Hedera helix L. (English ivy) can synthesize 16- and 18-carbon monoene and diene products. J Biol Chem 280(31):28169–28176. https://doi.org/10.1074/jbc.M504205200
Zhu G, Koszelak-Rosenblum M, Connelly SM, Dumont ME, Malkowski MG (2015) The crystal structure of an integral membrane fatty acid α-hydroxylase. J Biol Chem 290(50):29820–29833. https://doi.org/10.1074/jbc.M115.680124
Acknowledgements
We thank Lihong Shi, Jill Clodfelter, and Aaron Graff for their technical support. We thank Dr. Edward Pryor for his advice in the design of the expression constructs.
Funding
This research was supported by the National Natural Science Foundation of China (NSFC) (31722041, 21276108), the China Scholarship Council, the Fundamental Research Funds for the Central Universities (JUSRP51702A), the Crystallography and Computational Biophysics Shared Resource of the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (NCI CCSG P30CA012197) and Collaborative innovation center of food safety and quality control in Jiangsu Province.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 982 kb)
Rights and permissions
About this article
Cite this article
Wang, M., Chen, H., Ailati, A. et al. Substrate specificity and membrane topologies of the iron-containing ω3 and ω6 desaturases from Mortierella alpina . Appl Microbiol Biotechnol 102, 211–223 (2018). https://doi.org/10.1007/s00253-017-8585-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8585-x