Abstract
Culture-independent methods have granted the possibility to study microbial diversity in great detail, but technical issues pose a threat to the accuracy of new findings. Biases introduced during DNA extraction can result in erroneous representations of the microbial community, particularly in samples with low microbial biomass. We evaluated the DNA extraction method, initial sample biomass, and reagent contamination on the assessment of the human gut microbiota. Fecal samples of 200 mg were subjected to 1:10 serial dilutions; total DNA was obtained using two commercial kits and the microbiota assessed by 16S ribosomal RNA (rRNA) gene sequencing. In addition, we sequenced multiple technical controls. The two kits were efficient in extracting DNA from samples with as low as 2 mg of feces. However, in instances of lower biomass, only one kit performed well. The number of reads from negative controls was negligible. Both DNA extraction kits allowed inferring microbial consortia with similar membership but different abundances. Furthermore, we found differences in the taxonomic profile of the microbial community. Unexpectedly, the effect of sample dilution was moderate and did not introduce severe bias into the microbial inference. Indeed, the microbiota inferred from fecal samples was distinguishable from that of negative controls. In most cases, samples as low as 2 mg did not result in a dissimilar representation of the microbial community compared with the undiluted sample. Our results indicate that the gut microbiota inference is not much affected by contamination with laboratory reagents but largely impacted by the protocol to extract DNA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Advancements in culture-independent methods have granted the possibility to study microbial diversity with unprecedented detail, revolutionizing our understanding of life on Earth (Brazelton et al. 2010) and transforming the comprehension we have of our own relation with the microbial world (The Human Microbiome Project Consortium 2012). The most thoroughly studied human-associated microbial community, the gut microbiome, has been shown to be critical for human health (de Vos and De Vos 2012; Pflughoeft and Versalovic 2012), and the number of studies looking into the association between the gut microbiome and a myriad of factors has greatly increased in recent years. However scrutinized, several technical issues remain underexplored in the processing of gut samples and pose a serious threat to the accuracy of the new findings (de la Cuesta-Zuluaga and Escobar 2016). Among them, the acquisition of genetic material has been shown to be a critical step, on which multiple biases can be introduced and that can result in an erroneous representation of the microbial community under scrutiny.
Differences in DNA extraction protocols are known to affect the studied microbial community, causing the overrepresentation or underrepresentation of given microbial groups (Ariefdjohan et al. 2010; Guo and Zhang 2013; Smith et al. 2011). For instance, some DNA extraction protocols require a bead-beating step that contributes to the disruption of the cell wall, whereas others rely on chemical lysis for this procedure (Ariefdjohan et al. 2010). The mechanical disruption of microbial cells has been shown to yield higher amounts of DNA of better quality (Feinstein et al. 2009; Smith et al. 2011; Wesolowska-Andersen et al. 2014) and was the preferred method of benchmark collaborative projects, such as MetaHIT and the human microbiome project (HMP) (Wesolowska-Andersen et al. 2014). However, performance differs between mechanical disruption-based protocols (Ariefdjohan et al. 2010; Feinstein et al. 2009; Guo and Zhang 2013; Smith et al. 2011; Wesolowska-Andersen et al. 2014) and even among lots of the same DNA extraction kit (Salter et al. 2014), making data comparison among studies a heartache challenge.
An additional issue that has been highly publicized but less explored in the analysis of fecal samples is the effect of reagent contamination and sample biomass. Both 16S ribosomal RNA (rRNA) gene and metagenomic analyses are non-specific and sensitive to contamination with foreign microbial DNA (Tanner et al. 1998). Sources of contamination include laboratory environment (Salter et al. 2014), DNA extraction reagents (Corless et al. 2000; Tanner et al. 1998), PCR reagents (Rand and Houck 1990; Shen et al. 2006), and even ultrapure water (Kulakov et al. 2002; Mcalister et al. 2002). Foreign DNA contamination has been downplayed when dealing with feces compared with other biological samples (e.g., blood, lung, meconium) because of their high microbial biomass. Samples with low biomass are more prone to being affected by contaminant genetic material, as they provide little template DNA to compete with that contained in laboratory reagents (Grahn et al. 2003; Salter et al. 2014). Furthermore, there is a critical point in which sample biomass becomes so low that the contaminating DNA dominates sequence libraries (Salter et al. 2014).
Despite the importance recognized to the DNA extraction step, the impact of reagent contamination and sample biomass remains largely unaddressed in microbial ecology; to the best of our knowledge, no study has explored this on the inference of the human gut microbiota. Here, we assess the sequencing yield, composition, and diversity of the gut microbiota studied from human fecal samples, which were serially diluted and processed using two commercially available DNA extraction kits to determine the impact of DNA extraction, initial sample biomass, and reagent contamination on the evaluation of this microbial community using the 16S rRNA gene as marker.
Materials and methods
DNA extraction, serial dilutions, and 16S rRNA gene sequencing
Microbial DNA was obtained from fecal samples from five human donors (Online Resource 1). Samples were kept at − 80 °C since collection until DNA extraction. We assessed two commonly used commercial kits for fecal DNA extraction: PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA) and QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). We mostly followed the manufacturers’ instructions but introduced changes to improve DNA yield with mechanical lysis. For the MoBio kit, the bead-beating step (step 5) was reduced from 10 to 5 min (with 10-s rest intervals every minute) at 71 Hz using the original beads contained in the kit; for the Qiagen kit, a bead-beating step at 41 Hz for 8 s was introduced between steps 4 and 5 of the Isolation of DNA from Stool for Pathogen Detection protocol, using 1-mm sterile zirconium beads. In both cases, bead beating was performed in a Bead Blaster 24 (Benchmark Scientific, NJ). In both protocols, we started with 200 mg of homogenized feces (no dilution; D0). Then, we performed five 1:10 serial dilutions with autoclaved PBS, resulting in an initial biomass of 20 mg (dilution D1), 2 mg (dilution D2), 0.2 mg (dilution D3), 0.02 mg (dilution D4), and 0.002 mg (dilution D5). In all cases, DNA extraction was performed from a final volume of 200 μl, and DNA concentrations were obtained with a Synergy HT Microplate Reader (Bio-Tek Instruments, Winooski, VT).
Additionally, we obtained DNA sequences from nine negative controls to track potential sources of contamination. “Blank” DNA extractions were obtained with the two kits using autoclaved PBS as starting material (no feces)—blMBPBS and blQIAPBS; in the case of MoBio, it was possible to make an additional blank extraction from the solution originally contained in the PowerSoil Bead Tubes, adding no PBS (blMB). Blank extractions were expected to gather all sources of contamination introduced during DNA extraction protocols. We additionally obtained sequences from the MoBio C6 (buffC6) and Qiagen AE (buffAE) elution buffers, and autoclaved PBS (PBS). Sequences from buffC6 and buffAE served to distinguish the contamination introduced by these specific reagents from that introduced at other points during DNA extraction; since PBS was used to homogenize and dilute samples, as well as starting material in blMBPBS and blQIAPBS, we considered important to determine its microbial signature to identify instances of contamination by this reagent. We also obtained sequences from three additional negative controls (ultrapure water—H2O, and two “empty” wells—empA1 and empB1—with no sample but following the same sequencing treatment as samples); H2O informed about contamination introduced during PCR, library generation, and indexing, whereas empA1 and empB1 corresponded to pure sequencing noise. Finally, sequences from one positive control (mock community HM-782D, BEI Resources, Manassas, VA) served to get an estimate of the sequencing error rates.
All samples and controls were sent to the University of Michigan Medical School Host Microbiome Initiative (Ann Arbor, MI) for PCR amplification and sequencing of the 16S rRNA gene on the MiSeq platform. Several steps were followed in preparing samples for sequencing on this platform. Broadly, these included library generation and indexing, quality control, normalization and pooling, quantification, and sequencing. Specifically, the V4 hypervariable region of the 16S rRNA gene was amplified using the F515 (5′-CACGGTCGKCGGCGCCATT-3′) and R806 (5′-GGACTACHVGGGTWTCTAAT-3′) primers. We preferred to sequence this region (~ 250 bp) because its length allowed for fully overlapping forward and reverse reads, diminishing sequencing error rates. Multiplex PCR was performed with dual indices (Kozich et al. 2013) using 17 μl of Accuprime™ Pfx SuperMix (Thermo Fisher Scientific, Carlsbad, CA), 1 or 5 μl of template DNA (see Online Resource 2), and 2 μl of each paired set of index primers. PCR cycling condition consisted of an initial denaturation of 2 min at 95 °C; 30 cycles of 20 s at 95 °C, 15 s at 55 °C, and 5 min at 72 °C; and a final step at 72 °C for 10 min. All samples underwent electrophoresis on 96-well SYBR Safe E-gels (Invitrogen, Carlsbad, CA) to ensure amplification proceeded normally. Post-PCR products were quantified using the KAPA qPCR Library Quantification Kit (Kapa Biosystems, Wilmington, MA), cleaned up, and normalized with the SequalPrep Normalization Plate Kit (Invitrogen, Carlsbad, CA). Amplicons were finally pooled (5 μl per sample; average working concentration = 7.2 nM, range = 7.04–7.45 nM) into single wells. To assess the quality of the library, an Agilent Bioanalyzer Trace was performed. Finally, the pooled library was sequenced using the Illumina MiSeq sequencing platform with V2 chemistry. Detailed laboratory protocols are available at https://github.com/SchlossLab/MiSeq_WetLab_SOP/blob/master/MiSeq_WetLab_SOP_v4.md.
Bioinformatic processing of sequences
Raw 16S rRNA gene sequences were processed using Mothur v.1.38 (Schloss et al. 2009) following its Illumina MiSeq standard operating procedure (Kozich et al. 2013). Briefly, we first extracted sequences and quality scores from the paired fastq files and assembled the reads to form contigs. We eliminated reads containing bases with a quality score below 20, ambiguous bases, or shorter than 275 bp long. Next, we aligned the reads using the Silva reference alignment v.123 (Quast et al. 2013) and removed reads with a homopolymer run > 8 nucleotides and reads that did not overlap the region of the alignment spanning the V4 region. Then, we carried a preclustering step in which reads with an identity ≥ 99% (i.e., sequences differing in two nucleotides or less) were merged. Chimeric sequences were detected and discarded by UCHIME (Edgar et al. 2011). After that, we assigned the taxonomic classification to reads using the Greengenes 13_8_99 (DeSantis et al. 2006) and removed reads classified as mitochondria, eukaryota, or unknown. Next, we calculated the sequencing error rates using reads from the mock community (average error rate = 0.0083). Using the average neighbor algorithm, we generated operational taxonomic units (OTUs) at 97% sequence identity which were then taxonomically classified by consensus using Greengenes 13_8_99. A relaxed neighbor -joining tree with one representative sequence per OTU was finally obtained with Clearcut (Evans et al. 2006) after calculating uncorrected pairwise distances between aligned reads.
Statistical analyses
To quantify the overall impact of the origin of sample (donor), DNA extraction kit and dilution on the inferred gut microbiota diversity, membership (presence/absence), and abundance, we performed permutational multivariate analysis of variance (PERMANOVA) using the adonis function (analysis of variance using distance matrices) implemented in the Vegan package of R (Oksanen et al. 2015). PERMANOVAs were performed on three β-diversity distances—weighted and unweighted UniFrac obtained with the GUniFrac package of R (Chen et al. 2012) and Bray-Curtis with Vegan (Oksanen et al. 2015)—and three alpha-diversity metrics obtained with Mothur—number of observed OTUs, Shannon diversity index, and the inverse Simpson index. UniFrac analyses were performed on unrarefied read counts. Rarefication is a necessary step in ecological analyses to control for sampling unevenness; however, rarefication was undesirable in this case because negative controls and some highly diluted samples had too few 16S rRNA gene reads (< 100 reads/sample). Rarefying at very low depths made results extremely inconsistent. Alternatively, we performed Bray-Curtis analyses on relative abundances to partially control for unevenness in read counts. In addition, we performed analyses on samples with > 10,000 reads/sample to test the sensitivity of PERMANOVA to sampling unevenness. Principal correspondence analysis (PCoA) and taxonomic plots were obtained. Alpha-diversity analyses were performed on the subset of undiluted samples (D0) after rarefying the dataset to 13,000 reads/sample.
Availability of data and material
Raw 16S rRNA gene sequences were deposited at the NCBI’s Short Read Archive (SRA) under BioProject PRJNA385915. The Mothur logfile and R code built to obtain the results reported here are available in https://github.com/jsescobar/kitome.
Results
16S rRNA gene reads
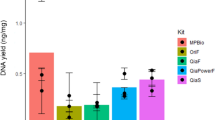
We first examined the number of 16S rRNA gene reads across DNA extraction kits and dilutions (Fig. 1, Online Resource 2). Note that the number of reads is not expected to be proportional to the initial sample biomass because libraries were normalized before sequencing. Instead, a high number of reads indicates that samples progressed appropriately in the laboratory workflow (DNA extraction, indexing, PCR, purification, normalization, and sequencing). In our experiments, the number of reads is of importance since it informs about the ability of each kit to extract sufficient microbial DNA in instances of high and low initial sample biomass.
As shown in Fig. 1a, the number of reads was high (> 10,000 reads/sample) in most samples extracted with MoBio and even in instances of high dilution, although this might depend upon the considered sample. In contrast, the Qiagen kit failed treating samples with very low biomass but was as efficient as the MoBio kit with samples with as low as 2 mg of human feces (Fig. 1b). This indicates that fecal microbial inference is jeopardized when using a starting biomass < 2 mg. Still, DNA from samples with as low as 2 mg of human feces (D2) was enough to get a high number of reads independent of the DNA extraction kit (mean ± SD: MoBio = 34,885 ± 12,201; Qiagen = 39,433 ± 20,393). All negative controls resulted in a very low number of reads independent of the DNA extraction kit (range 25–42 reads/sample) (Online Resource 2). This number is several orders of magnitude lower than the number of reads obtained in most samples, regardless of dilution. Any signal that may be introduced by laboratory reagents would be drowned by the sample’s microbial signal and is, thus, negligible.
DNA extraction affects the gut microbiota
As said above, the number of reads only informs about the ability of the kits to extract DNA in sufficient quantity and quality for the PCR and sequencing reactions to appropriately perform. Hence, it is not a good predictor of the precision of the microbial community inference. To appropriately evaluate the impact of the tested parameters on the gut microbiota membership and abundance, we performed PERMANOVA with β-diversity measures, either dependent or independent of the reconstruction of phylogenetic trees—UniFrac and Bray-Curtis, respectively. These analyses showed a pervasive and significant effect of the origin of the sample (i.e., donor) in the inferred microbiota; this factor explained the largest portion of the total variance in β-diversity (Table 1). Noteworthy, analyses were not affected by unevenness in the number of reads among samples, as separate PERMANOVA on a restricted dataset considering only samples with > 10,000 reads/sample output similar results (Online Resource 3). Differences in sex or BMI among donors might be at the origin of variation in the gut microbiota in these datasets (Online Resource 1). Large inter-subject variation is a common observation in studies of the human gut microbiota (Costello et al. 2009; The Human Microbiome Project Consortium 2012).
The two DNA extraction kits allowed inferring microbial consortia with similar membership but different abundances. This is illustrated by the smaller unweighted UniFrac distances observed between pairs of undiluted samples (D0) treated with either kit (Fig. 2a) but the larger distances observed with weighted UniFrac (Fig. 2b) or Bray-Curtis (Fig. 2c). A comparison of the taxonomic assignments of OTUs among undiluted samples (D0) showed that, at the phylum level, there were more OTUs from Firmicutes and Actinobacteria and less from Bacteroidetes with MoBio than Qiagen (Fig. 3a–b; Online Resource 4). At the class level, there were more OTUs classified as Clostridia (Firmicutes), Coriobacteriia (Actinobacteria), and class Actinobacteria (phylum Actinobacteria) with MoBio than Qiagen which, in turn, resulted in an increased number of OTUs belonging to Bacteroidia (Bacteroidetes), unclassified Firmicutes (Firmicutes), and Mollicutes (Tenericutes) (Fig. 3c, d; Online Resource 4). At the genus level, the pattern was variable among samples; among the most abundant genera, more reads from Blautia, Coprococcus, unclassified Lachnospiraceae, and Collinsella were obtained with MoBio, whereas Oscillospira and unclassified Ruminococcaceae were overrepresented with Qiagen (Online Resource 4).
Principal correspondence analyses (PCoA) on unweighted UniFrac (a), weighted UniFrac (b), and Bray-Curtis (c) distances. Colors and shapes represent the origin of sample (MoBio (circles); Qiagen (triangles); negative controls (crosses)). In panels b and c, the point size is proportional to the number of sequenced reads. Undiluted samples (D0) (filled points); diluted samples (D1–D5) (open points)
In terms of alpha-diversity, the analysis of the subset of undiluted samples (D0) indicated that most of the microbes of these samples were captured in our sequencing (Online Resource 5). In addition, the two DNA extraction kits allowed inferring communities with similar diversity, suggesting that their coverage of the gut microbiota was similar (number of observed OTUs: MoBio = 243, Qiagen = 285, p = 0.10; Shannon diversity index: MoBio = 3.52, Qiagen = 3.70, p = 0.10; inverse Simpson index: MoBio = 19.05, Qiagen = 19.70, p = 0.69).
Dilution of human feces has a moderate impact on the gut microbiota inference
The composition of the microbial community inferred from fecal samples, independently of dilution, was significantly different to that of negative controls, with the exception of dilution D5 treated with Qiagen (Online Resource 6). Regarding fecal samples only, dilution, in general, had a statistically significant effect on microbial inference, explaining 6–14% of the observed variance, depending on the β-diversity measure evaluated (Table 1; Online Resource 3). Unexpectedly, however, this effect was moderate and did not introduce severe bias into the microbial inference. The effect was minor in terms of presence/absence of OTUs, as demonstrated by the unweighted UniFrac analysis, especially in samples treated with MoBio (Fig. 2a). Analyses that take OTU abundance into account (weighted UniFrac and Bray-Curtis) confirmed that fecal samples treated with MoBio had more similar composition than samples treated with Qiagen, especially in instances of high dilution (Fig. 2b, c). In most cases, samples with as low as 2 mg (D2) or 0.2 mg (D3) of feces resulted in a reduced introduction of bias into the representation of the gut microbiota. Taxonomic analyses confirmed these results, indicating overrepresentation of Firmicutes (Clostridia class) and underrepresentation of Actinobacteria (Coriobacteriia class) with dilution in MoBio and a more irregular pattern in Qiagen (Fig. 3). At the genus level, distortion of the community was idiosyncratic of each sample (Online Resource 7).
Discussion
Previous studies have explored the impact of DNA extraction on the inferred microbial communities (Ariefdjohan et al. 2010; Guo and Zhang 2013; Smith et al. 2011). However, the impacts of reagent contamination and sample biomass have been poorly addressed; yet, it has been demonstrated that when microbial biomass is low, the signal coming from samples is overwhelmed by the signal emerging from laboratory reagents (note that reagents are not DNA-free, and autoclave does not make DNA disappear). The extent of this contamination can be very large; the contaminating species are characteristic to each laboratory, reagent, and reagent’s lot (Salter et al. 2014), reason why it is highly recommended to include several negative controls in experiments. In the case of human feces serving to infer processes occurring in the gut microbiome, there are no reports determining the threshold at which biomass is so low that the inferred microbial community may be artifactual. Low fecal biomass might be a considerable issue in forensics, when dealing with ancient specimens or in some animal models. In this report, we demonstrate that contamination introduced by laboratory reagents is probably limited within the wide range of tested biomasses and that the variance introduced by the protocol employed to extract DNA is a bigger source of bias in human gut microbiome studies.
We show that the two tested kits were efficient in extracting DNA from samples with 100 times lower biomass than suggested by the manufacturers’ protocols. However, the gut microbiota inferred with the two kits was different, especially when considering the abundance of microbial groups. In general, the dissimilar nature of the two DNA extraction protocols could explain the differences in microbial community profiles (Guo and Zhang 2013). It has been repeatedly reported that protocols incorporating bead-containing lysing matrix and vigorous shaking extract larger amounts of DNA and produce a more comprehensive microbial profile than protocols based on chemical lysis alone (Ariefdjohan et al. 2010; Guo and Zhang 2013; Smith et al. 2011). In our experiments, the protocols for DNA extraction incorporated both chemical and mechanical cell lysis, although each followed specific procedures and used proprietary reagents. Pointing out exactly which procedures/components of the kits might be responsible of the differences we found seems too speculative without specific tests. Furthermore, the fact that differences have even been shown to exist among kit lots of the same brand (Salter et al. 2014) suggests that sources of variance are not easy to control for and raise the question whether data from different studies should be compared at all (Schloss 2008; Wesolowska-Andersen et al. 2014). The fundamental puzzle with kit and lot effects is that, in real samples, there is no means to know the actual microbial community composition, so that there is no way to evaluate the kit/lot accuracy. To be conservative, in a given study, total microbial DNA should be extracted with the same kit, ideally the same kit lot, and caution must be taken when comparing data from different studies.
Importantly, we demonstrate that highly diluted samples exhibit a microbial community profile distinguishable, in most cases, from that of laboratory reagents. Highly diluted samples in our study had as few as 20 μg (D4) or 2 μg (D5) of feces. Although samples with such low biomass are unsuitable for robust microbial inference, and unrealistic in most study designs, their microbial signature is still distinguishable from that of laboratory reagents, especially looking at the presence/absence of data. This limited effect of dilution in the inferred gut microbial community is unexpected since previous reports demonstrated that samples with low microbial biomass basically amplify contaminating DNA (Salter et al. 2014). In our case, samples with as low as 2 mg of feces allowed obtaining both high numbers of 16S rRNA gene reads and a representation of the gut microbiota similar to that obtained with samples with 100 times more biomass.
Our results are particularly relevant now that multiple calls for the standardization of procedures have been made in the microbial ecology community (Alivisatos et al. 2015; Blaser et al. 2016), as they provide insights on the steps researchers must focus on when dealing with DNA extraction from fecal samples. As general recommendations, in both 16S rRNA gene and metagenomic experiments, the inclusion of as many technical controls as possible is highly encouraged to control for potential sources of noise in the final dataset; this includes laboratory and reagent contaminants and sequencing errors. Furthermore, researchers should avoid mixing different DNA extraction kits within a study and must include the kit’s lot as an additional metadatum for each sample. The question of how to address comparisons between studies and meta-analyses remains open; however, the fact that measurements between samples extracted with different kits can be statistically significant strengthens the idea that comparisons between studies that employed different extraction methods should be discouraged (Henderson et al. 2013).
References
Alivisatos AP, Blaser MJ, Brodie EL, Chun M, Dangl JL, Donohue TJ, Dorrestein PC, Gilbert JA, Green JL, Jansson JK, Knight R, Maxon ME, McFall-Ngai MJ, Miller JF, Pollard KS, Ruby EG, Taha SA (2015) A unified initiative to harness Earth’s microbiomes. Science 350:507–508. https://doi.org/10.1126/science.aac8480
Ariefdjohan MW, Savaiano DA, Nakatsu CH (2010) Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens. Nutr J 9:23. https://doi.org/10.1186/1475-2891-9-23
Blaser MJ, Cardon ZG, Cho MK, Dangl JL, Donohue TJ, Green JL, Knight R, Maxon ME, Northen TR, Pollard KS, Brodie EL (2016) Toward a predictive understanding of Earth’s microbiomes to address 21st century challenges. Am Soc Microbiol 7:1–16. https://doi.org/10.1128/mBio.00714-16
Brazelton WJ, Ludwig K a, Sogin ML, Andreishcheva EN, Kelley DS, Shen CC, Edwards RL, Baross J a (2010) Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci U S A 107:1612–1617. https://doi.org/10.1073/pnas.0905369107
Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD, Collman RG, Bushman FD, Li H (2012) Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28:2106–2113. https://doi.org/10.1093/bioinformatics/bts342
Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ (2000) Contamination and sensitivity issues with a real-time universal 16s rRNA PCR. J Clin Microbiol 38:1747–1752
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R (2009) Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. https://doi.org/10.1126/science.1177486
de la Cuesta-Zuluaga J, Escobar JS (2016) Considerations for optimizing microbiome analysis using a marker gene. Front Nutr 3:26. https://doi.org/10.3389/fnut.2016.00026
de Vos WM, De Vos Eaj (2012) Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr, Rev 70:S45–S56. https://doi.org/10.1111/j.1753-4887.2012.00505.x
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Evans J, Sheneman L, Foster J (2006) Relaxed neighbor joining: a fast distance-based phylogenetic tree construction method. J Mol Evol 62:785–792. https://doi.org/10.1007/s00239-005-0176-2
Feinstein LM, Woo JS, Blackwood CB (2009) Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl Environ Microbiol 75:5428–5433. https://doi.org/10.1128/AEM.00120-09
Grahn N, Olofsson M, Ellnebo-Svedlund K, Monstein HJ, Jonasson J (2003) Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol Lett 219:87–91. https://doi.org/10.1016/S0378-1097(02)01190-4
Guo F, Zhang T (2013) Biases during DNA extraction of activated sludge samples revealed by high throughput sequencing. Appl Microbiol Biotechnol 97:4607–4616. https://doi.org/10.1007/s00253-012-4244-4
Henderson G, Cox F, Kittelmann S, Miri VH, Zethof M, Noel SJ, Waghorn GC, Janssen PH (2013) Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS One 8:e74787. https://doi.org/10.1371/journal.pone.0074787
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Kulakov LA, McAlister MB, Ogden KL, Larkin MJ, O’Hanlon JF (2002) Analysis of bacteria contaminating ultrapure water in industrial systems. Appl Environ Microbiol 68:1548–1555. https://doi.org/10.1128/AEM.68.4.1548-1555.2002
Mcalister M, Kulakov L, O’hanlon J, Larkin M, Ogden K (2002) Survival and nutritional requirements of three bacteria isolated from ultrapure water. J Ind Microbiol Biotechnol 29:75–82. https://doi.org/10.1038/sj.jim.7000273
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2015) Vegan: community ecology package. R Packag version:2.3–2.1. https://doi.org/10.4135/9781412971874.n145
Pflughoeft KJ, Versalovic J (2012) Human microbiome in health and disease. Annu Rev Pathol 7:99–122. https://doi.org/10.1146/annurev-pathol-011811-132421
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41. https://doi.org/10.1093/nar/gks1219
Rand KH, Houck H (1990) Taq polymerase contains bacterial DNA of unknown origin. Mol Cell Probes 4:445–450. https://doi.org/10.1016/0890-8508(90)90003-I
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. https://doi.org/10.1186/s12915-014-0087-z
Schloss PD (2008) Evaluating different approaches that test whether microbial communities have the same structure. Isme J 2:265–275. https://doi.org/10.1038/Ismej.2008.5
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Ryan A, Oakley BB, Parks DH, Courtney J, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF, Lesniewski RA, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Shen H, Rogelj S, Kieft TL (2006) Sensitive, real-time PCR detects low-levels of contamination by Legionella pneumophila in commercial reagents. Mol Cell Probes 20:147–153. https://doi.org/10.1016/j.colsurfa.2005.08.010
Smith B, Li N, Andersen AS, Slotved HC, Krogfelt KA (2011) Optimising bacterial DNA extraction from faecal samples: comparison of three methods. Open Microbiol J 5:14–17. https://doi.org/10.2174/1874285801105010014
Tanner MA, Goebel BM, Dojka MA, Pace NR (1998) Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol 64:3110–3113
The Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. https://doi.org/10.1038/nature11234
Wesolowska-Andersen A, Bahl MI, Carvalho V, Kristiansen K, Sicheritz-Pontén T, Gupta R, Licht TR (2014) Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2:19. https://doi.org/10.1186/2049-2618-2-19
Acknowledgements
We thank participants who agreed to donate samples, the APOLO Scientific Computing Center at EAFIT University for hosting part of the bioinformatic resources employed in analyses, and the University of Michigan Medical School Host Microbiome Initiative for sequencing support.
Funding
This work was funded by Grupo Empresarial Nutresa. The funder has not had any role in designing or conducting the study; in the analysis or interpretation of the data; in the writing, review, or approval of the manuscript; and in the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was conducted in accordance with the principles of the Declaration of Helsinki and had minimal risk according to the Colombian Ministry of Health (Resolution 008430 of 1993). This study was conducted with approval from the Bioethics Committee of SIU—University of Antioquia (approbation act 14-24-588 dated May 28, 2014).
Informed consent
The participants were assured of anonymity and confidentiality. Written informed consent was obtained from them before beginning the study.
Electronic supplementary material
ESM 1
(PDF 493 kb).
Rights and permissions
About this article
Cite this article
Velásquez-Mejía, E.P., de la Cuesta-Zuluaga, J. & Escobar, J.S. Impact of DNA extraction, sample dilution, and reagent contamination on 16S rRNA gene sequencing of human feces. Appl Microbiol Biotechnol 102, 403–411 (2018). https://doi.org/10.1007/s00253-017-8583-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8583-z