Abstract
The fungal cyclic dipeptide prenyltransferase FtmPT1 from Aspergillus fumigatus catalyzes a regular C2-prenylation of brevianamide F (cyclo-l-Trp-l-Pro) and is involved in the biosynthesis of a number of biologically active natural products including tryprostatins, spirotryprostatins, verruculogen, and fumitremorgins. FtmPT1, like other members of the dimethylallyltryptophan synthase superfamily, was shown to have high substrate promiscuity for tryptophan-containing cyclic dipeptides and a few other aromatic substrates. A previous study demonstrated the acceptance of 1-naphthol by FtmPT1, but with very low product yield. In this study, we report the significantly increased acceptance of 1-naphthol and other hydroxynaphthalenes by FtmPT1_G115A and six FtmPT1_Y205X single mutants as well as FtmPT1_G115A_Y205C. These results provided an example for creation of biocatalysts with improved catalytic activity by site-directed mutagenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prenyltransferases are ubiquitous in nature and catalyze the transfer reactions of prenyl moieties from different prenyl donors to various aliphatic or aromatic acceptors, including small-molecule natural products, peptides, proteins, and t-RNAs. Therefore, they play important roles in both primary and secondary metabolism (Dumelin et al. 2012; Heide 2009; Winkelblech et al. 2015). According to their amino acid sequences, structures, biochemical properties, and functions, prenyltransferases are classified into different subgroups (Bonitz et al. 2011; Heide 2009; Li 2009; Winkelblech et al. 2015). One of the most investigated subgroups is the dimethylallyltryptophan synthase (DMATS) superfamily from fungi, which share sequence homology with the dimethylallyltryptophan synthase involved in the biosynthesis of ergot alkaloids in Claviceps sp. (Tsai et al. 1995). Until now, more than 40 such enzymes have been identified and characterized biochemically (Winkelblech et al. 2015). The majority of the DMATS superfamily is involved in the biosynthesis of prenylated indole alkaloids and takes indole derivatives including tryptophan and tryptophan-containing cyclic dipeptides as substrates (Winkelblech et al. 2015). They contribute greatly to the structure diversity of a large number of natural products (Heide 2009; Li 2010). In comparison to their non-prenylated precursors, prenylated derivatives often show clearly distinct biological and pharmacological activities and are important sources for drug discovery and development (Botta et al. 2005; Heide 2009; Li 2010; Wollinsky et al. 2012a; Yazaki et al. 2009), which make prenyltransferases including DMATSs valuable biocatalysts in the structural modification of small molecules (Fan et al. 2015a).
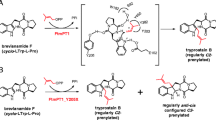
DMATSs usually demonstrate high substrate promiscuity toward aromatic substrates and catalyze mostly regiospecific Friedel–Crafts alkylations on the aromatic ring system (Fan et al. 2015a; Winkelblech et al. 2015). For example, FtmPT1 from the ascomycetous fungus Aspergillus fumigatus catalyzes a regular C2 - prenylation on the indole ring of brevianamide F (cyclo-l-Trp-l-Pro) in the presence of DMAPP and is involved in the biosynthesis of several biologically active natural products including tryprostatins and fumitremorgins (Fig. 1a) (Grundmann and Li 2005; Li 2011). FtmPT1 also accepted a large number of tryptophan-containing cyclic dipeptides and other aromatic compounds like 1-naphthol for Friedel–Crafts alkylations (Chen et al. 2012; Grundmann and Li 2005; Wollinsky et al. 2012b; Yu et al. 2011). However, the product yield of 1-naphthol with FtmPT1 was significantly lower than those with other members of the DMATS superfamily such as FgaPT2, 7-DMATS, CdpNPT, and AnaPT (Yu et al. 2011). Therefore, prenylation of hydroxynaphthalenes by FtmPT1 was not further investigated in that study.
In the last years, structures of several DMATSs including FgaPT2 (Metzger et al. 2009), FtmPT1 (Jost et al. 2010), CdpNPT (Schuller et al. 2012), and AnaPT (Yu et al. 2013) were determined and used as the basis for understanding the catalytic mechanism and for creation of new biocatalysts. Comparing these structures revealed that these enzymes share similar folds consisting of five repeating “αββα” (ABBA) barrel folds. Their active sites are located in the center of the barrel. The amino acid residues in the DMAPP-binding sites seem to be fairly conserved, while the binding sites of the aromatic substrates differ from each other. Based on the structures of FgaPT2 and by molecular modeling-guided site-directed mutagenesis, we created several derivatives with distinct or strongly increased catalytic activity (Fan et al. 2015b; Fan and Li 2016). Recently, Mori et al. (2016) reported the alteration of the preference for prenyl chain length as well as regioselectivities and stereoselectivities of TleC and MpnD by structure-based enzyme engineering. These results demonstrated a convenient route for new enzyme design by mutagenesis.
As for FtmPT1, several amino acid residues including Gly115 and Tyr205 were proposed to be involved in the binding of brevianamide F (Jost et al. 2010). Based on the structure, a modifiable reaction chamber was identified and a mutant FtmPT1_G115T was obtained, which still accepted brevianamide F as substrate, but catalyzed mainly a reversely syn-cis C3 prenylation instead of the regularly C2- prenylation (Jost et al. 2010). In another study, several Tyr205 mutants showed comparable consumption of brevianamide F as the non-mutated FtmPT1, but with clearly increased activity for the formation of C3-prenylated derivatives (Zhou et al. 2016) (Fig. 1b). Due to the low product yields, the prenylation of hydroxynaphthalenes by FtmPT1 was not studied in detail in our previous study (Yu et al. 2011). After availability of Gly115 and Tyr205 mutants, their activities toward hydroxynaphthalenes were further investigated.
Materials and methods
Chemicals
DMAPP was synthesized according to the method described for geranyl diphosphate reported previously (Woodside et al. 1988). Naphthalene derivatives were purchased from Fluka, TCI, Acros Organics, Aldrich, and Alfa Aesar.
Bacterial strains, plasmids, and culture conditions
Escherichia coli XL1-Blue MRF’ (Stratagene, Heidelberg, Germany) was used for cloning and expression experiments. pAG12 containing ftmPT1 in pQE70 was constructed previously (Grundmann and Li 2005) and used as expression vector for FtmPT1 and as DNA template for site-directed mutagenesis experiments. E. coli cells harboring plasmids were grown in liquid Lysogeny Broth (LB) medium and on solid LB medium with 1.5% (w/v) agar at 37 °C. Fifty micrograms of carbenicillin per milliliter was used for selection of recombinant E. coli strains.
Site-directed mutagenesis
Constructs for expression of FtmPT1 Tyr205 mutants were reported elsewhere (Zhou et al. 2016). The expression constructs pST2 and pST9–11 were described previously (Jost et al. 2010). For creation of additional Gly115 mutants, site-directed mutagenesis was carried out as described previously (Zhou et al. 2016). Degenerated primers at bps 343–345 were used to generate mutants of FtmPT1 listed in Table 1. The Expand Long Template PCR system (Roche Diagnostics, Mannheim, Germany) was used for creation of the plasmids pKZ1–pKZ17, pKZ22, and pKZ24. pKZ3 and pKZ11 were used as templates for construction of plasmids for expression of the double mutants G115T_Y205C, G115T_Y205M, G115A_Y205C, and G115T_Y205M.
Overproduction and purification of the recombinant proteins
Protein overproduction and purification of FtmPT1 and its mutants were carried out as described previously (Grundmann and Li 2005; Zhou et al. 2016) and analyzed on SDS-PAGE. Protein concentration was estimated by measurement of the absorption at 280 nm on a NanoDrop 2000c UV–Vis spectrophotometer and by comparison of their intensities with those of protein markers on SDS-PAGE (Fig. S1).
Enzyme assays with purified recombinant proteins
All enzyme assays contained 50 mM Tris–HCl (pH 7.5), 5 mM CaCl2, 0.15–5% (v/v) glycerol, 5% (v/v) DMSO, and 2 mM DMAPP and were incubated at 37 °C. To determine the enzyme activity of FtmPT1 and its mutants toward hydroxynaphthalenes, 1-naphthol (1a), 2-naphtol (2a), 1,6-dihydroxynaphthalene (3a), 1,7-dihydroxynaphthalene (4a), 2,3-dihydroxynaphthalene (5a), 2,6-dihydroxynaphthalene (6a), 2,7-dihydroxynaphthalene (7a), 1-hydroxy-5-aminonaphthalene (8a), and 1-amino-7-hydroxynaphrhalene (9a) at a concentration of 1 mM were incubated with 0.1 μg μL−1 of purified recombinant protein in a total volume of 100 μL. After incubation for 2 h, the reactions were terminated by addition of 100 μL methanol. Proteins were removed by centrifugation at 13,000 rpm for 20 min, and the supernatants were analyzed on HPLC. In order to determine the kinetic parameters of the prenyl transfer reactions, aromatic substrates at concentrations of up to 2 mM were used. Varied protein amounts and incubation times were used for assays as shown in Table 3.
For isolation of the enzyme products, reactions were carried out in large scales (15 mL) containing 1 mM aromatic substrate and 2–4 mg recombinant protein. After incubation for 16 h, the reaction mixtures were extracted with 15 mL EtOAc for four times. The extracts were concentrated on a rotating vacuum evaporator at 35 °C and then dissolved in 1 mL methanol for injection into HPLC.
HPLC analysis and isolation of the enzyme products
The enzyme reaction mixtures were analyzed on an HPLC (Agilent series 1200, Böblingen, Germany) by using a Multospher 120 RP-18 column (250 × 4 mm, 5 μm, CS-Chromatographie Service, Langerwehe, Germany) at a flow rate of 1 mL min−1. Water and methanol, both containing 0.5% trifluoroacetic acid, were used as solvents. For analysis of the enzyme products, a linear gradient of 30–100% (v/v) methanol in 20 min was used. The column was then washed with 100% (v/v) methanol for 5 min and equilibrated with 30% (v/v) methanol for 5 min. For analysis of the enzyme products as shown in Figs. S2 and S4, water and acetonitrile, both containing 0.5% trifluoroacetic acid, were used as solvents. A linear gradient of 40–100% (v/v) acetonitrile in 15 min was used for analysis. The column was then washed with 100% acetonitrile for 5 min and equilibrated with 40% acetonitrile in water for another 5 min. Detection was carried out with a photodiode array detector. For isolation of the enzyme products, the same HPLC equipment with a Multospher 120 RP-18 column (250 × 10 mm, 5 μm, CS-Chromatographie Service, Langerwehe, Germany) was used. A linear gradient of 30–100% (v/v) of methanol in water in 40–60 min was carried out with a flow rate at 2.5 mL min−1. The column was then washed with 100% (v/v) methanol for 10 min and equilibrated with 30% (v/v) methanol for 10 min.
NMR and mass spectrometric analyses
For structure elucidation, the isolated enzyme products were subjected to 1H NMR and LC-MS analyses. High-resolution electrospray ionization mass spectrometry (HR-ESI-MS) data were obtained on a Bruker microTOF-Q III mass spectrometer. The spectrometer was equipped with an Agilent 1260 HPLC system. 1H NMR spectra were recorded at room temperature with an ECX-500 spectrometer (JEOL, Tokyo, Japan) equipped with a broadband probe with z-gradient. All spectra were processed with MestReNov. 5.2.2 (Mestrelab Research, Santiago de Compostella, Spain). Chemical shifts were referenced to the solvent signal at 3.30 ppm for MeOH. For NMR spectra, please see Figs. S13, S14, S15, S16, S17, S18, S19, and S20 in Electronic Supplementary Material.
-
Compound 1b: HR-ESI-MS: m/z 213.1279 ([M+H]+, calculated for C15H17O: 213.1274). 1H NMR (500 Hz, CD3OD) δ H 8.21 (ddd, 1H, J = 8.3, 1.4, and 0.6 Hz), 7.90 (br d, 1H, J = 8.3 Hz), 7.45 (td, 1H, J = 7.5 and 1.4 Hz), 7.40 (td, 1H, J = 7.5 and 1.3 Hz), 7.09 (d, 1H, J = 7.7 Hz), 6.71 (d, 1H, J = 7.7 Hz), 5.33 (tsept, 1H, J = 7.0 and 1.4 Hz), 3.65 (d, 2H, J = 7.0 Hz), 1.80 (d, 3H, J = 1.0 Hz), 1.74 (d, 3H, J = 1.2 Hz). These data correspond well to those published previously (Yu et al. 2011).
-
Compound 3b: HR-ESI-MS: m/z 229.1213 ([M+H]+, calculated for C15H17O2: 229.1223). 1H NMR (500 Hz, CD3OD) δ H 8.07 (d, 1H, J = 9.0 Hz), 7.17 (d, 1H, J = 2.3 Hz), 6.98 (m, 2H), 6.51 (d, 1H, J = 7.6 Hz), 5.33 (tsept, 1H, J = 6.8 and 1.4 Hz), 3.54 (d, 2H, J = 6.8 Hz), 1.79 (d, 3H, J = 1.0 Hz), 1.74 (d, 3H, J = 1.2 Hz). These data correspond well to those published previously (Yu et al. 2011).
-
Compound 4b: HR-ESI-MS: m/z 229.1209 ([M+H]+, calculated for C15H17O2: 229.1223). 1H NMR (500 Hz, CD3OD) δ H 7.77 (d, 1H, J = 9.1 Hz), 7.49 (d, 1H, J = 2.6 Hz), 7.04 (dd, 1H, J = 9.1 and 2.6 Hz), 6.87 (d, 1H, J = 7.6 Hz), 6.64 (d, 1H, d, J = 7.6 Hz), 5.31 (br t, 1H, J = 7.6 Hz), 3.59 (d, 2H, J = 7.0 Hz), 1.79 (s, 3H), 1.73 (s, 3H). These data correspond well to those published previously (Yu et al. 2011).
-
Compound 7b: HR-ESI-MS: m/z 229.1203 ([M+H]+, calculated for C15H17O2: 229.1223). 1H NMR (500 Hz, CD3OD) δ H 7.56 (d, 1H, J = 8.8 Hz), 7.44 (d, 1H, J = 8.8 Hz), 7.12 (d, 1H, J = 2.4 Hz), 6.87 (d, 1H, J = 8.8 Hz), 6.83 (dd, 1H, d, J = 8.8 and 2.4 Hz), 5.17 (tsept, 1H, J = 6.6 and 1.4 Hz), 3.63 (d, 2H, J = 6.6 Hz), 1.88 (d, 3H, J = 0.9 Hz), 1.68 (d, 3H, J = 1.3 Hz). These data correspond well to those published previously (Yu et al. 2011).
-
Compound 7c: HR-ESI-MS: m/z 229.1225 ([M+H]+, calculated for C15H17O2: 229.1223). 1H NMR (500 Hz, CD3OD) δ H 7.59 (dd, 2H, J = 8.9 and 1.1 Hz), 7.00 (m, 2H), 6.88 (dd, 2H, J = 8.9 and 2.5 Hz), 5.52 (br t, 1H, J = 6.6 Hz), 4.61 (d, 1H, J = 6.6 Hz), 1.81 (br s, 3H), 1.79 (br s, 3H). These data correspond well to those published previously (Yu et al. 2011).
-
Compound 7d: HR-ESI-MS: m/z 297.1844 ([M+H]+, calculated for C20H25O2: 297.1849). 1H NMR (500 Hz, CD3OD) δ H 7.60 (d, 1H, J = 8.9 Hz), 7.47 (d, 1H, J = 8.7 Hz), 7.13 (d, 1H, J = 2.4 Hz), 6.92 (d, 1H, J = 8.7 Hz), 6.88 (dd, 1H, d, J = 8.9 and 2.4 Hz), 5.52 (tsept, 1H, J = 6.6 and 1.3 Hz), 5.17 (tsept, 1H, J = 6.7 and 1.4 Hz), 4.62 (d, 2H, J = 6.5 Hz), 3.69 (d, 2H, J = 6.7 Hz), 1.90 (br s, 3H), 1.81 (br s, 3H), 1.79 (br s, 3H), 1.70 (d, 3H, J = 1.2 Hz). These data correspond well to those published previously (Yu et al. 2011).
-
Compound 7e: HR-ESI-MS: m/z 365.2467 ([M+H]+, calculated for C25H33O2: 365.2475). 1H NMR (500 Hz, CD3OD) δ H 7.39 (d, 1H, J = 8.7 Hz), 7.38 (s, 1H), 7.08 (s, 1H), 6.89 (d, 1H, J = 8.7 Hz), 5.54 (br t, 1H, J = 6.5 Hz), 5.35 (br t, 1H, J = 7.4 Hz), 5.17 (br t, 1H, J = 6.7 Hz), 4.62 (d, 2H, J = 6.5 Hz), 3.68 (d, 2H, J = 7.0 Hz), 3.36 (d, 2H, J = 7.5 Hz), 1.91 (s, 3H), 1.82 (s, 3H), 1.79 (s, 3H), 1.75 (s, 3H), 1.73 (s, 3H), 1.70 (s, 3H).
-
Compound 9b: HR-ESI-MS: m/z 228.1384 ([M+H]+, calculated for C15H18NO: 228.1383). 1H NMR (500 Hz, CD3OD) δ H 7.81 (d, 1H, J = 9.1 Hz), 7.23 (d, 1H, J = 2.5 Hz), 7.05 (dd, 1H, J = 9.1 and 2.5 Hz), 6.89 (d, 1H, J = 7.5 Hz), 6.72 (d, 1H, d, J = 7.5 Hz), 5.31 (tsept, 1H, J = 7.0 and 1.5 Hz), 3.60 (d, 2H, J = 7.2 Hz), 1.79 (s, 3H), 1.73 (d, 3H, J = 1.0 Hz).
Results
Acceptance of 1-naphthol by single mutants of Gly115
Four FtmPT1 mutants G115A, G115T, G115I, and G115L were created in a previous study for proof of the importance of Gly115 and showed different influence of these enzymes on the prenyl transfer reaction for brevianamide F (Jost et al. 2010). G115A did not change the prenylation position, while G115T catalyzed a reverse C3 instead of a regular C2 prenylation at the indole ring of brevianamide F (Jost et al. 2010). Incubation of these mutants (10 μg protein) with 1-naphthol (1a) at 37 °C for 2 h and analysis of their reaction mixtures on HPLC also revealed different behaviors (Fig. S2). Under this condition, FtmPT1_G115A showed significantly higher activity toward 1a with a product yield of 13.5 ± 2.5% than FtmPT1 with 5.5 ± 0.1%. G115T showed a conversion yield of 2.1 ± 0.2%. No product formation was detected in the incubation mixtures of 1a with mutants G115I or G115L under the tested conditions.
Our previous study showed that saturation mutagenesis could be a powerful tool for creation of enzyme derivatives (Fan and Li 2016). Following this experience, different oligonucleotides with wobbles at base pairs of 343 to 345 of ftmPT1 were used as degenerated primer pairs for mutagenesis (Table 1). After identification of 11 G115 mutants, the remaining mutants were obtained with more specific primers (Table 1). Constructs for additional 8 mutants were obtained in this way (Table 1). Satisfied purity and amount were achieved for nine mutants (Fig. S1). Very low protein yield and purity were obtained for G115Q, G115P, and G115K. HPLC analysis of the incubation mixtures of 1a with these mutants revealed that G115D, G115S, G115N, and G115F showed activities, but lower than that of the non-mutated FtmPT1. No product formation was detected in the incubation mixtures of 1a with G115H, G115Y, G115Q, G115W, G115K, G115P, G115V or G115R under the tested conditions (Fig. S2).
Acceptance of 1-naphthol by single mutants of Tyr205
In the presence of DMAPP (2 mM), 1-naphthol (1a) was incubated with 10 μg of recombinant FtmPT1 or its 19 mutants obtained from the previous study (Zhou et al. 2016) in 100 μL enzyme assays at 37 °C for 2 h. HPLC analysis of the incubation mixtures revealed that 1a was accepted by the tested enzymes with clearly different activities (Figs. 2 and S3). Under this condition, 1a was converted by FtmPT1 with a product yield of 5.5 ± 0.1%. Six mutants including Y205L, Y205M, Y205F, Y205I, Y205C, and Y205S showed higher activity toward 1a than FtmPT1, with product yields of 28.8 ± 1.1, 25.0 ± 2.7, 21.4 ± 1.2, 23.3 ± 1.3, 18.5 ± 3.5, and 11.7 ± 1.8%, respectively. The activities of the first five mutants are also significantly higher than that of G115A. These six Tyr205 mutants also accepted brevianamide F as well, but with activities not higher than the non-mutated FtmPT1 (Zhou et al. 2016). Six mutants Y205N, Y205V, Y205K, Y205P, Y205R, and Y205E accepted 1a with product yields between 0.6 and 6.0%. No product peak was observed from the reaction mixtures of 1a with Y205A, Y205T, Y205D, Y205G, or Y205W. One product peak each with the same retention time was detected for the reaction mixtures of FtmPT1 and its mutants, indicating the presence of the same product(s).
Acceptance of 1-naphthol by selected double mutants on Gly115 and Tyr205
The importance of Gly115 and Tyr205 for prenylation of brevianamide F has been demonstrated in previous studies. That is, G115T redirected the prenylation position and several Tyr205 mutants increased the product yields of C3-prenylated derivatives (Jost et al. 2010; Zhou et al. 2016). As mentioned above, G115A, Y205L, Y205M, Y205F, Y205I, Y205C, and Y205S accepted 1a much better than the wild type of FtmPT1. We were curious to prove the effect of mutations on both residues. Therefore, double mutants G115A_Y205C, G115A_Y205M, G115T_Y205C, and G115T_Y205M were created by site-directed mutagenesis (Table 1). HPLC analysis of the incubation mixtures of 1a with the purified proteins under the same conditions as for the single mutants revealed that FtmPT1_G115A_Y205C showed higher activities toward 1a than the wild type of FtmPT1, with a product yield of 19.7 ± 0.2%. This activity is slightly higher than that of G115A of 13.5 ± 2.5% and comparable with that of Y205C of 18.5 ± 3.5%. Product yields of 4.2 ± 0.5 and 5.3 ± 0.5% were observed for G115T_Y205C and G115T_Y205M, respectively, which are lower than those of Y205C and Y205M. No product peak was detected in the incubation mixture of 1a with G115A_Y205M.
Different behaviors of seven mutants of FtmPT1 toward hydroxynaphthalenes
As mentioned above, seven mutants Y205L, Y205M, Y205F, Y205C, Y205I, Y205S, and G115A_Y205C showed high activities toward to 1a. Therefore, FtmPT1 and the seven mutants were afterwards assayed with nine hydroxynaphthalenes for 2 h at 37 °C with 10 μg proteins. These included 1a, 2-naphtol (2a), 1,6-dihydroxynaphthalene (3a), 1,7-dihydroxynaphthalene (4a), 2,3-dihydroxynaphthalene (5a), 2,6-dihydroxynaphthalene (6a), 2,7-dihydroxynaphthalene (7a), 1-hydroxy-5-aminonaphthalene (8a), and 1-amino-7-hydroxynaphthalene (9a). HPLC analysis of the incubation mixtures showed that the product yields of 1a converted by FtmPT1 and its mutants were reproducible (Fig. S3). With 2a as substrate, the conversion yield of 0.3 ± 0.1% was observed with FtmPT1, while the tested mutants clearly accepted it with the highest product yield of 2.4 ± 0.3% by G115A_Y205C (Table 2, Fig. S5). 3a and 4a were consumed by FtmPT1 with product yields of 2.1 ± 0.3 and 5.5 ± 0.7%, respectively, while the seven mutants demonstrated clearly higher catalytic activities than FtmPT1, with the highest product yield of 10.4 ± 0.1% for 3a with Y205L and 13.2 ± 2.0% for 4a with Y205L, respectively (Table 2 and Figs. S6 and S7). This means a fivefold increase of activity for 3a and a threefold increase for 4a. 2,3-Dihydroxynaphthalene (5a) and 2,6-dihydroxynaphthalene (6a) were poor substrates for both FtmPT1 and its mutants with product yields of less than 7% (Table 2 and Figs. S8 and S9). Toward 7a, the seven mutants demonstrated low activities with product yields in the range of 3.8–8.6% (Table 2 and Fig. S10). 8a was accepted by FtmPT1 and the seven mutants with product yields of less than 1% (Table 2 and Fig. S11). 9a was a good substrate for both FtmPT1 and its mutants. The best product yield of approximately 20% was achieved with FtmPT1_Y205F (Table 2 and Fig. S12). Taking the results mentioned above together, some of the selected mutants Y205L, Y205M, Y205F, Y205C, Y205I, Y205S, and G115A_Y205C were found to demonstrate clearly higher catalytic activities toward 1a, 2a, 3a, 4a, 5a, and 9a than the non-mutated FtmPT1 (Table 2).
Structure elucidation of the detected enzyme products
To obtain enough enzyme products for structure elucidation, the best mutants were used for large-scale reactions (15 ml), i.e., FtmPT1_Y205M for 1a, 3a, and 7a; FtmPT1_Y205L for 4a; and FtmPT1_Y205F for 9a. The enzyme products 1b, 3b, 4b, and 9b were isolated from the reaction mixtures of 1a, 3a, 4a, and 9a, respectively. 7b, 7c, 7d, and 7e were obtained as products of 7a. The isolated products were then subjected to MS and NMR analyses (Figs. S13 S14, S15, S16, S17, S18, S19, and S20 in Electronic Supplementary Material). The obtained spectroscopic data of 1b, 3b, and 4b corresponded very well to those of the enzyme products of 1a, 3a, and 4a with other DMATS enzymes published previously (Yu et al. 2011) and confirmed the para-prenylation of 1-hydroxynaphhalenes as given in Fig. 3.
By comparison with the data in the literature (Yu et al. 2011), 7b and 7c were identified as C- and O-monoprenylated, respectively, while 7d is a C,O-diprenylated derivative (Fig. 3). MS data indicated a triprenylation in 7e. In the 1H NMR spectrum of 7e, signals at 5.54 (br t, 1H, 6.5 Hz), 5.35 (br t, 1H, 7.4 Hz), and 5.17 ppm (br t, 1H, 6.7 Hz) indicated the presence of three regularly attached prenyl moieties. One of them is attached to an oxygen atom due to the down-field shift of H-1 to 5.54 ppm. Considering the chemical shifts and coupling pattern of the four aromatic protons, the prenylation position is determined as O-2, C-3, and C-8 (Fig. 3). These results demonstrated that the products of the FtmPT1_Y205 mutants for a given substrate are very similar or identical to those obtained with other prenyltransferases of the DMATS superfamily (Yu et al. 2011). Detection of signals for five aromatic protons in the NMR spectrum of 9b indicated the attachment of the prenyl moiety to a C atom. Inspection of the NMR revealed the presence of two coupling systems. Three protons are coupled with coupling constants of 9.1 and 2.5 Hz and the other two with a coupling constant of 7.5 Hz. This proved that the prenylation had been taken place at the ortho-position of the amino group.
Comparing the results obtained for the pairs with different hydroxylation positions, i.e., 1a and 2a, 3a, and 6a as well as 4a and 7a, it seems that mutation of Tyr205 has a stronger positive influence on the acceptance of 1- than 2-hydroxynaphthalene derivatives. 1a, 4a, and 9a were good substrates for FtmPT1 and its mutants.
Kinetic parameters of the prenyl transfer reactions with three of the identified best Tyr205 mutants
To compare the catalytic activity of FtmPT1 and its mutated proteins toward hydroxynaphthalenes, kinetic parameters were determined for 1a and 3a with FtmPT1 and Y205M, 4a and 7a with FtmPT1 and Y205L, and 9a with FtmPT1 and Y205F by Eadie–Hofstee, Hanes–Woolf, and Lineweaver–Burk plots (Figs. S21, S22, S23, S24, S25, S26, S27, S28, S29, and S30 in Electronic Supplementary Material) as described under the “Materials and methods” section and summarized in Table 3. The K M values of the mutated proteins toward 1a, 3a, 4a, 7a, and 9a are found in the range of 0.19–0.52 mM, significantly lower than those with FtmPT1 in the range of 0.51–1.24 mM, indicating the increased affinity of the mutated proteins to these substrates. The turnover numbers of the reactions with the mutated proteins are about two to eight folds of those with FtmPT1. The kinetic parameters of 7a are comparable with FtmPT1 and Y205L. These results correspond well to the observed product yields (Table 2).
Discussion
Prenyltransferases of the DMATS superfamily are capable of prenylation of various substrates, including indole, tyrosine, xanthone, flavonoid, and naphthalene derivatives. However, they show in many cases low activity toward their non-natural substrates. Several studies in the last years have demonstrated that protein engineering can also be considered as a powerful tool for the creation of prenyltransferases with altered features (Fan et al. 2015a; Winkelblech et al. 2015).
In a previous study, mutation of Gly115 to Thr reduced the formation of a C2-prenylated product tryprostatin B from its natural substrate brevianamide F. Instead, a reversely C3-prenylated derivative was identified as the main product. In contrast to G115T, G115A did not change the prenylation position (Jost et al. 2010). In another study, all 19 Tyr205 mutants were created by saturation mutagenesis (Zhou et al. 2016). Seven mutants, Y205L, Y205M, Y205F, Y205C, Y205I, Y205S, and Y205N, showed comparable or slightly lower activity than the non-mutated FtmPT1 toward its natural substrate brevianamide F. However, different products were identified in the reaction mixtures. Like FtmPT1, Y205M and Y205F converted brevianamide F mainly to a regularly C2-prenylated derivative, while Y205L, Y205C, Y205I, Y205S, and Y205N catalyzed both regular C2 and C3 prenylations of brevianamide F. The ratios of C3- and C2-prenylated derivatives varied between 1:1.5 and 2.3:1 (Zhou et al. 2016).
In this study, additional Gly115 mutants were created by saturation mutagenesis. HPLC analysis of the incubation mixtures of 1-naphthol with the obtained mutants revealed that G115A had a higher enzyme activity than the non-mutated FtmPT1 (Table 2 and Fig. S2). HPLC analysis of the reaction mixtures of 1a with Tyr205 mutants demonstrated that six of these mutants Y205L, Y205M, Y205F, Y205C, Y205I, and Y205S were found to have clearly higher catalytic activities. Then, we mutated the two positions and created four double mutants. HPLC analysis showed that G115A_Y205C had higher activity toward 1a than the wild type, but lower than several Tyr205 mutants. Other three tested double mutants showed no or lower activity than FtmPT1. This indicated that combination of both positions is not the best way for increasing the enzyme activity toward 1a. Afterwards, seven mutants, Y205L, Y205M, Y205F, Y205C, Y205I, Y205S, and G115A_Y205C, were assayed with nine hydroxynaphthalenes 1a–9a. Our results demonstrated that several of these mutants showed higher activity toward 1a, 2a, 3a, 4a, 5a, and 9a than the non-mutated FtmPT1 (Table 2). Isolation and structure elucidation revealed that for a given substrate, the products are very similar or identical to those obtained with other prenyltransferases of the DMATS superfamily (Yu et al. 2011). Since 3a–7a are dihydroxylated isomers, their acceptance by different mutants could be of significant importance for understanding the interactions between the hydroxynaphthalenes and enzymes as well as for further optimization of the enzyme activity toward those substrates.
Prenylated naphthalenes are not as common as prenylated flavonoids or indole derivatives in nature. Therefore, only a few hydroxylnaphthalene prenyltransferases have been reported (Haug-Schifferdecker et al. 2010; Kumano et al. 2008), which share no amino acid sequence homology with the members of the DMATS superfamily. Acceptance of hydroxynaphthalenes by DMATS enzymes and increasing their catalytic activity by mutagenesis expand the possibility for production of prenylated hydroxynaphthalenes by chemoenzymatic synthesis or synthetic biology. This could play an important role in the drug discovery and development process.
References
Bonitz T, Alva V, Saleh O, Lupas AN, Heide L (2011) Evolutionary relationships of microbial aromatic prenyltransferases. PLoS One 6:e27336
Botta B, Vitali A, Menendez P, Misiti D, Delle MG (2005) Prenylated flavonoids: pharmacology and biotechnology. Curr Med Chem 12:717–739
Chen J, Morita H, Wakimoto T, Mori T, Noguchi H, Abe I (2012) Prenylation of a nonaromatic carbon of indolylbutenone by a fungal indole prenyltransferase. Org Lett 14:3080–3083
Dumelin CE, Chen Y, Leconte AM, Chen YG, Liu DR (2012) Discovery and biological characterization of geranylated RNA in bacteria. Nat Chem Biol 8:913–919
Fan A, Li S-M (2016) Saturation mutagenesis on Arg244 of the tryptophan C4-prenyltransferase FgaPT2 leads to enhanced catalytic ability and different preferences for tryptophan-containing cyclic dipeptides. Appl Microbiol Biotechnol 100:5389–5399
Fan A, Winkelblech J, Li S-M (2015a) Impacts and perspectives of prenyltransferases of the DMATS superfamily for use in biotechnology. Appl Microbiol Biotechnol 99:7399–7415
Fan A, Zocher G, Stec E, Stehle T, Li S-M (2015b) Site-directed mutagenesis switching a dimethylallyl tryptophan synthase to a specific tyrosine C3-prenylating enzyme. J Biol Chem 290:1364–1373
Grundmann A, Li S-M (2005) Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology 151:2199–2207
Haug-Schifferdecker E, Arican D, Brueckner R, Heide L (2010) A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene dimethylallyltransferase reaction. J Biol Chem 285:16487–16494
Heide L (2009) Prenyl transfer to aromatic substrates: genetics and enzymology. Curr Opin Chem Biol 13:171–179
Jost M, Zocher G, Tarcz S, Matuschek M, Xie X, Li S-M, Stehle T (2010) Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity. J Am Chem Soc 132:17849–17858
Kumano T, Richard SB, Noel JP, Nishiyama M, Kuzuyama T (2008) Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorg Med Chem 16:8117–8126
Li S-M (2009) Applications of dimethylallyltryptophan synthases and other indole prenyltransferases for structural modification of natural products. Appl Microbiol Biotechnol 84:631–639
Li S-M (2010) Prenylated indole derivatives from fungi: structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat Prod Rep 27:57–78
Li S-M (2011) Genome mining and biosynthesis of fumitremorgin-type alkaloids in ascomycetes. J Antibiot 64:45–49
Metzger U, Schall C, Zocher G, Unsöld I, Stec E, Li S-M, Heide L, Stehle T (2009) The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci U S A 106:14309–14314
Mori T, Zhang L, Awakawa T, Hoshino S, Okada M, Morita H, Abe I (2016) Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases. Nat Commun 7:10849
Schuller JM, Zocher G, Liebhold M, Xie X, Stahl M, Li S-M, Stehle T (2012) Structure and catalytic mechanism of a cyclic dipeptide prenyltransferase with broad substrate promiscuity. J Mol Biol 422:87–99
Tsai HF, Wang H, Gebler JC, Poulter CD, Schardl CL (1995) The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem Biophys Res Commun 216:119–125
Winkelblech J, Fan A, Li S-M (2015) Prenyltransferases as key enzymes in primary and secondary metabolism. Appl Microbiol Biotechnol 99:7379–7397
Wollinsky B, Ludwig L, Hamacher A, Yu X, Kassack MU, Li S-M (2012a) Prenylation at the indole ring leads to a significant increase of cytotoxicity of tryptophan-containing cyclic dipeptides. Bioorg Med Chem Lett 22:3866–3869
Wollinsky B, Ludwig L, Xie X, Li S-M (2012b) Breaking the regioselectivity of indole prenyltransferases: identification of regular C3-prenylated hexahydropyrrolo[2,3-b]indoles as side products of the regular C2-prenyltransferase FtmPT1. Org Biomol Chem 10:9262–9270
Woodside AB, Huang Z, Poulter CD (1988) Trisammonium geranyl diphosphate. Org Synth 66:211–215
Yazaki K, Sasaki K, Tsurumaru Y (2009) Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70:1739–1745
Yu X, Xie X, Li S-M (2011) Substrate promiscuity of secondary metabolite enzymes: prenylation of hydroxynaphthalenes by fungal indole prenyltransferases. Appl Microbiol Biotechnol 92:737–748
Yu X, Zocher G, Xie X, Liebhold M, Schütz S, Stehle T, Li S-M (2013) Catalytic mechanism of stereospecific formation of cis-configured prenylated pyrroloindoline diketopiperazines by indole prenyltransferases. Chem Biol 20:1492–1501
Zhou K, Zhao W, Liu X-Q, Li S-M (2016) Saturation mutagenesis on Tyr205 of the cyclic dipeptide C2-prenyltransferase FtmPT1 results in mutants with strongly increased C3-prenylating activity. Appl Microbiol Biotechnol 100:9943–9953
Acknowledgements
We thank Lena Ludwig for synthesis of DMAPP, Rixa Kraut and Stefan Newel for taking MS and NMR spectra, respectively.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
The Bruker microTOF-Q III mass spectrometer was funded by the Deutsche Forschungsgemeinschaft (INST 160/620-1 to S.-M. L.). Kang Zhou is a recipient of a scholarship from the China Scholarship Council (201308440282).
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 1025 kb)
Rights and permissions
About this article
Cite this article
Zhao, W., Fan, A., Tarcz, S. et al. Mutation on Gly115 and Tyr205 of the cyclic dipeptide C2-prenyltransferase FtmPT1 increases its catalytic activity toward hydroxynaphthalenes. Appl Microbiol Biotechnol 101, 1989–1998 (2017). https://doi.org/10.1007/s00253-016-7966-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7966-x