Abstract
Increased pentose phosphate pathway flux, relative to total substrate uptake flux, is shown to enhance succinic acid (SA) yields under continuous, non-growth conditions of Actinobacillus succinogenes biofilms. Separate fermentations of glucose and xylose were conducted in a custom, continuous biofilm reactor at four different dilution rates. Glucose-6-phosphate dehydrogenase assays were performed on cell extracts derived from in situ removal of biofilm at each steady state. The results of the assays were coupled to a kinetic model that revealed an increase in oxidative pentose phosphate pathway (OPPP) flux relative to total substrate flux with increasing SA titre, for both substrates. Furthermore, applying metabolite concentration data to metabolic flux models that include the OPPP revealed similar flux relationships to those observed in the experimental kinetic analysis. A relative increase in OPPP flux produces additional reduction power that enables increased flux through the reductive branch of the TCA cycle, leading to increased SA yields, reduced by-product formation and complete closure of the overall redox balance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Succinic acid is well established as a top potential value-added product derived from biomass (Bozell and Petersen 2010), and its microbial production continues to receive substantial attention. Successful bulk, microbial production of succinic acid that is cost-competitive with the traditional petrochemical route, will augment its current market and create opportunities for expansion into other traditionally petrochemical markets such as, for example, maleic anhydride, adipic acid and butanediol (Zeikus et al. 1999). Furthermore, succinic acid can serve as a precursor for the production of biopolymers such as polybutylene succinate and polyurethane (Bechthold et al. 2008), which provides additional incentive for microbial production from an environmentally friendly standpoint. Various microbial strains have been explored for succinic acid production (Song and Lee 2006; Beauprez et al. 2010) including Actinobacillus succinogenes, Anaerobiospirillum succiniciproducens, Basfia succiniciproducens (Scholten et al. 2009), Mannheimia succiniciproducens and modified strains of Escherichia coli. Of the prominent succinate producers, wild-type A. succinogenes is proving to be one of the most promising candidates as it has shown competitive performance on both a variety of monosaccharides (Liu et al. 2008; Maharaj et al. 2014; Yan et al. 2014; Bradfield and Nicol 2016) and scalable biorefinery streams (Bradfield et al. 2015; Salvachúa et al. 2016) and is able to tolerate high acid titres (Lin et al. 2008) while necessarily fixing CO2 during succinate production (Guettler et al. 1999).
Reduction in by-product formation is important in improving process economics as it decreases downstream separation costs and improves substrate utilization. It is desirable to achieve homosuccinate fermentation where succinate is the sole end product without associated by-product formation. However, despite the promising processing potential of wild-type A. succinogenes, it is unable to achieve homosuccinate fermentation. Homosuccinate fermentation on typical carbohydrate substrates (e.g. glucose and xylose) is possible in microbial hosts containing a complete TCA cycle or a glyoxylate shunt. These pathways, together with glycolysis, generate sufficient reduction power in a closed cycle to balance that which is required for succinate production without leading to by-product formation (Van Heerden and Nicol 2013a). A. succinogenes has an incomplete TCA cycle and lacks a glyoxylate shunt (McKinlay et al. 2010); therefore, by-product formation via fermentative pathways, particularly acetate and formate, is necessary to balance redox in succinate production.
However, it has been shown that succinic acid production occurs in preference to the two by-products at elevated succinic and total acid concentrations under continuous biofilm conditions, resulting in an increase in the succinic acid yield on substrate (Bradfield and Nicol 2014; Brink and Nicol 2014). The improvement in yield has been linked to a shift in metabolic flux distribution where the organism enters a non-growth or stationary phase (Brink and Nicol 2014). Furthermore, mass and redox balance closures based on excreted metabolites have been shown to be incomplete under these conditions. The unique metabolic behaviour and incomplete mass and redox balance closures have been observed on glucose (Bradfield and Nicol 2014), xylose (Bradfield and Nicol 2016) and biorefinery hydrolysate streams (Bradfield et al. 2015) and are therefore not exclusive to a particular carbohydrate substrate. In our previous studies, we hypothesized that the incomplete redox balance closures are due to increased carbon flux through the oxidative pentose phosphate pathway (OPPP), since no additional metabolites or sources of reduction power were detected. Increased OPPP flux generates reduction power as NADPH which can be converted to NADH by transhydrogenase in A. succinogenes (McKinlay et al. 2007) and used in the reductive branch of the TCA cycle. The OPPP has the potential to produce the same outcome as a complete TCA cycle or glyoxylate shunt, as it is able to generate sufficient reduction power for homosuccinate fermentation.
Metabolic flux analysis (MFA) or flux balancing (Gombert and Nielsen 2000) can be used to demonstrate that the OPPP is a plausible explanation for the observed metabolic behaviour. However, MFA alone cannot be used to confirm OPPP flux because CO2 is the only end product of the OPPP and CO2 production is not unique to the OPPP. Therefore, a more direct method is needed to detect OPPP flux. Nevertheless, with available metabolite measurements, MFA can be used to estimate OPPP fluxes thereby providing a theoretical basis for comparison to experimentally determined fluxes. In performing MFA, continuous operation is the most suitable mode (Stephanopoulos 1999) as it offers both process and metabolic steady-state conditions. These conditions allow for more accurate and simplified flux analysis compared to batch operation. In addition, continuous operation of biofilm reactors leads to enhanced productivities (Qureshi et al. 2005) making it the likely choice for industrial-scale succinic acid production by A. succinogenes. Therefore, gaining insight into the metabolism of the chosen microbial host under conditions similar to those of bulk-scale production is particularly advantageous as it aids process optimization and hints at potential targets for strain improvement.
In the current study, the hypothesis of OPPP flux serving as a source of additional reduction power in continuous glucose and xylose fermentations was explored at four different dilution rates. OPPP flux was estimated experimentally by means of glucose-6-phosphate dehydrogenase assays of biofilm extracts coupled to a kinetic model. In addition, MFA was used to determine the expected or theoretical flux relationships when the OPPP supplies the reduction power needed to satisfy the redox balance. The flux relationships determined from the kinetic model were then compared to the expected/theoretical flux relationships.
Materials and methods
Organism and fermentation medium
A. succinogenes 130Z (DSM 22257; ATCC 55618) was acquired from the German Collection of Microorganisms and Cell Cultures (DSMZ). Cultures were maintained in 66 % v/v glycerol solutions at −40 °C. Inoculum was incubated at 37 °C and 150 rpm for 16–24 h in 30-mL sealed vials containing 15 mL sterilized tryptone soy broth at 30 g L−1. Prior to inoculation, the inoculum was analysed by HPLC to ensure culture purity and consistent metabolite distributions.
The fermentation medium comprised three parts: (1) a nutrient and salts mixture, (2) a carbohydrate solution and (3) a phosphate buffer. The nutrient and salts mixture consisted of (in g L−1): 6.0 yeast extract, 10.0 clarified corn steep liquor (Sigma-Aldrich, USA) (Bradfield and Nicol 2014), 0.5 NaCl, 0.2 MgCl2·6H2O, 0.2 CaCl2·2H2O and 0.5 mL L−1 Antifoam SE-15 (Sigma-Aldrich, Germany). The phosphate buffer comprised 1.6 g L−1 KH2PO4 and 0.8 g L−1 K2HPO4. Xylose and glucose solutions were prepared at 60 g L−1. All chemicals were obtained from Merck KgaA unless stated otherwise.
Continuous fermentations
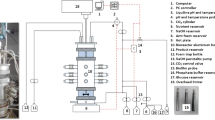
Independent fermentations on glucose and xylose were performed at dilution rates between 0.02 and 0.3 h−1 in a custom, externally recycled bioreactor, similar to that used in Bradfield and Nicol (2016). The range of dilution rates allowed for four distinct steady states (i.e. SA titres), which were common to both fermentations (i.e. substrates). Reactor volume was maintained at 360 mL by means of an overflow tube connected to an exit pump. pH was measured by a Ceragel CPS71D glass electrode (Endress + Hauser, Germany) connected to a Liquiline CM442 unit (Endress + Hauser, Germany) and controlled at 6.80 ± 0.01 by the addition of 10 N NaOH. Temperature was measured by the pH electrode and controlled at 37.0 ± 0.1 °C by means of a custom PID feedback controller coupled to a hotplate. A 10 % v/v solution of Antifoam SE-15 (Sigma-Aldrich, Germany) was dosed onto the liquid headspace to suppress foam as needed. The average flow rates of NaOH and antifoam were determined in real time over a 4-h period and used to adjust feed concentrations due to dilution of the feed (Van Heerden and Nicol 2013b). Furthermore, the time profile of the average NaOH flow rate was used in estimating steady-state conditions. CO2 gas (Afrox, South Africa) was fed directly into the recycle line at 0.1 vvm to ensure CO2 saturation of the fermentation broth—essential for succinic acid production. To ensure anaerobic conditions, CO2 venting from the reactor was recycled to the feed medium reservoir to create a CO2 headspace which also assisted in CO2 saturation of the feed and prevented air from entering the system. The reactor and liquid reservoirs (excluding NaOH) were autoclaved together at 121 °C for 60 min, with the three parts of the feed medium kept separate to prevent unwanted interactions.
Steady-state conditions were based on constant metabolite concentrations over time and a flat time-averaged NaOH dosing profile, as described in Bradfield and Nicol (2016). The glucose fermentation had a total time of 960 h with an estimated medium consumption of 35 L, and the xylose fermentation lasted 550 h and consumed approximately 25 L of medium. The extended fermentation times and large volumes of medium required, highlight the difficulties of performing continuous fermentations of appreciable volume and at reasonable dilution rates at the lab scale. This is especially true of fermentations involving biofilm as it is first necessary to form a stable biofilm, achieve steady state, then regenerate the biofilm after sampling. For each substrate, steady-state conditions at an intermediate succinic acid concentration (∼30 g L−1) were performed in duplicate to show repeatability of fermentation performance. The duplicate sets occurred at the beginning and end of the fermentation. The duplicate for glucose was excluded in the assay analyses because it was taken 2 weeks after the final steady state, and there appeared to be a substantial amount of dead biomass in the fermenter, despite good reproducibility of fermentation performance. As a result, the scrubbed dry cell weight (DCW) was 212 % greater than the initial value but at an equivalent reactor productivity. The repeat point for the xylose fermentation was taken within 4 days after the final steady state, and no signs of extensive dead biomass were observed.
To increase cell density and facilitate biofilm growth in the reactor, four wooden sticks wrapped in mutton cloth and fixed to a top distributor plate were inserted into the reactor. The stick structure served as a support surface for biofilm attachment, growth and stability. The number of sticks was limited to four to prevent biofilm overgrowth, blockage of the reactor and obstruction of mass transfer. As a result, less biofilm was present in the fermenter compared to previous fermentations using a similar fermentation setup (Bradfield and Nicol 2014; Maharaj et al. 2014; Bradfield and Nicol 2016).
Analytical methods
Fermentation broth was sampled via the outlet line onto a bed of ice to suppress metabolic activity of the cells once outside the fermenter. The concentration of organic acids, ethanol and carbohydrate substrates present in the feed medium and fermentation broth was measured by means of high-performance liquid chromatography (HPLC). The system comprised an Agilent 1260 Infinity HPLC (Agilent Technologies, USA), equipped with a 300 × 7.8 mm Aminex HPX-87H ion-exchange column (Bio-Rad Laboratories, USA) and an RI detector. Two methods were used for HPLC analysis to overcome co-elution of xylose and pyruvate (Bradfield and Nicol 2016). In the first method, a 5-mM solution of H2SO4 at a flow rate of 0.6 mL min−1 served as the mobile phase with column and RID temperatures of 60.0 and 55.0 °C, respectively. In the second method, a 20-mM solution of H2SO4 was used with all other parameters remaining the same as the first method.
DCW was determined by centrifuging a known volume of fermentation broth, washing the resulting cell pellets with distilled water, then centrifuging again and discarding the supernatant. The remaining pellets were dried at 80 °C until constant mass and the concentration of cell mass (or biomass) was determined. To minimize the effect of biofilm fluctuations on suspended DCWs, samples were taken for the duration of at least one volume turnover. DCWs were performed on the fermentation broth after biofilm scrubbing (see below) to determine the biomass content present in the biofilm sample and the enzyme assays. Attempts to separate cells and extracellular polymeric substance from the biofilm showed poor repeatability compared to a previous study (Brink and Nicol 2014). Therefore, the DCW of the scrubbed biofilm was used as a measure of the total biomass content of each sample and was assumed to be an accurate representation of the overall biomass content in the reactor.
Preparation of cell extracts
In situ removal of biofilm (i.e. scrubbing) was achieved by varying the direction and increasing the speed of the recycle pump to create turbulence, thereby shearing off portions of the attached biofilm. In addition, a magnetic stirrer bar located below the biofilm support structure was activated to increase turbulence during biofilm scrubbing. Biofilm-rich fermentation broth was then extracted from the fermenter via a suction line that ran from the head of the reactor to the lower portion of the support structure, positioned to maximize contact with the wooden sticks and extract biofilm in preference to fermentation broth. The fermenter extract therefore contained a mixture of biofilm and suspended cells, where the biofilm comprised 74 to 98 % of the total DCW from the glucose fermentation and 79 to 90 % of the DCW from the xylose fermentation (as determined by the difference in DCWs before and after scrubbing, i.e. total DCW minus suspended DCW).
Samples of scrubbed biofilm (40 mL) were centrifuged at 4000×g for 25 min at room temperature. The resulting cell pellets were combined and resuspended in Tris–HCl (pH 7.4) to a volume of 10 mL. From the 10-mL suspension, three 2-mL samples were centrifuged at 10,000×g for 10 min at 4 °C. The resulting cell pellets were then combined and resuspended in Tris–HCL (pH 7.4) to a final volume of 6 mL. The suspension was then disrupted on ice by sonication (Branson 250 probe sonicator, USA) using ten pulsed cycles of 15 s with a 1-min resting period. The sonicator was set to power level 4 and 40 % duty cycle. Cell extracts were then centrifuged at 16,000×g for 30 min at 4 °C. The supernatant, containing soluble cell extracts, was then used for enzyme assays.
Enzyme assays
The presence and activity of glucose-6-phosphate dehydrogenase (G6PDH) was determined spectrophotometrically by observing the increase in absorbance at 340 nm which corresponds to NADPH production (Noltmann et al. 1961). The assay solution consisted of reaction cocktail, cell extracts and assay substrate. The reaction cocktail (pH 7.4) consisted of 100 mM glycylglycine buffer (Sigma-Aldrich, Germany), 3.2 mM NADP+ (Melford, UK) and 18 mM MgCl2 (Sigma-Aldrich, Germany). Since phosphoglucose isomerase (PGI) activity interferes with G6PDH activity by reacting with glucose-6-phosphate (G6P), a kinetic model that included both enzymatic reactions was developed. Five different combinations of G6P (Sigma-Aldrich, Germany) and fructose-6-phosphate (F6P) (Sigma-Aldrich, Germany) starting concentrations were used in the assay system (Table 1) to fit kinetic parameters to the model.
Enzyme assays were performed in 24-well plates (Cellstar, Greiner Bio-one, Germany) at 37 °C using a SpectraMax Paradigm microplate reader (Molecular Devices, USA). In each assay, reagent cocktail was added to the well first followed by 100 μL of cell extracts; the substrate was then added to a final volume of 1.5 mL corresponding to a path length of 1 cm. The volume of the reagent cocktail was dependent on the volume of the substrate added. Assays were performed in succession at each substrate concentration condition. No change in absorbance was observed when adding cell extracts to the reaction cocktail without substrate, which implies that any observed activity was a result of enzyme specificity for the added substrate. Absorbance readings were taken every 15 s over a 10-min interval at each condition, and an extinction coefficient of 6.22 mM−1 cm−1 was used to relate absorbance readings to NADPH concentration.
Mass and redox balances
Mass and redox balances were performed as described in Bradfield and Nicol (2016) and summarized here for convenience. To determine mass balance closures, the stoichiometric amount of substrate required to produce the measured metabolite concentrations based on elemental balances was compared to the actual amount of substrate consumed. Elemental balances were performed on C, H, O and N between the substrates and products, since for each element appearing in the substrates, an equal amount must appear in the products (i.e. succinic, acetic, formic and pyruvic acids, biomass and H2O). CO2 can serve as both a substrate and a metabolite and its net production or consumption will be reflected in the mass balance solution. Elemental balances can be solved using linear algebra techniques and metabolite concentrations. Once the substrate consumption rate is determined, it is divided by the actual (measured) substrate consumption rate and expressed as a percentage that represents the mass balance closure. The standard formula of biomass, CH1.8O0.5N0.2 (Villadsen et al. 2011), was used in the elemental balance to account for cell growth from DCW measurements. A generalized approach to elemental balancing is presented in Villadsen et al. (2011).
Redox balances (as NADH balances) were performed by using overall metabolic pathways for the formation of excreted metabolites and comparing the net production and consumption of NADH. In the formation of acetic, formic and pyruvic acids, a net production of NADH occurs (oxidative pathways) while for succinic acid, a net consumption of NADH occurs (reductive pathway). NADPH is viewed as NADH in the redox balance, since they can be interconverted by transhydrogenase in A succinogenes (McKinlay et al. 2007), and both co-factors carry the same redox currency and contribute equally to the redox balance. Biomass was excluded from the redox balance due to the low DCWs and the uncertainty in the associated amount of NADH.
Flux models
Metabolic flux models, including the OPPP, were developed for glucose (Fig. 1a) and xylose (Fig. 1b) based on the established metabolic pathways and the genome of A. succinogenes (McKinlay et al. 2007; McKinlay and Vieille 2008; McKinlay et al. 2010). A matrix-based description of the metabolic network, as reviewed in Gombert and Nielsen (2000), was used to perform metabolic flux analysis of each network. The method entails defining a unique flux (v) for each pathway in the network, on a carbon-mole (C-mole) basis, then using nodal, NADH and stoichiometric balances together with known metabolite production rates to define the system. For the system to be fully defined, the number of equations and specifications must equal the number of fluxes. Since the system is linear, it can be solved by linear algebra techniques using Eq. 1 (Varma and Palsson 1994). The complete matrix system for each flux model is provided below each model in Fig. 1:
Simplified metabolic networks based on McKinlay et al. (2005), McKinlay et al. (2007), McKinlay et al. (2010) and McKinlay and Vieille (2008) used for calculating oxidative pentose phosphate pathway flux from experimental metabolite measurements for a glucose and b xylose. Each flux is denoted by v and represents the rate of carbon moles through a particular branch of the network. Nodes are given by ringed capital letters. The related matrix system (Eq. 1) for each network is given below each diagram. In the matrices, capital letters correspond to the ringed letters in the diagram and denote nodal balances, NADH denotes the redox balance, PPP denotes a stoichiometric specification for the pentose phosphate pathway and S denotes a metabolite specification. Specifications were given as per the measured metabolite rates (r). Not all intermediates are shown and biomass formation is excluded. AA acetic acid, FA formic acid, G3P glyceraldehyde-3-phosphate, G6P glucose-6-phosphate, Glc glucose, PEP phosphoenolpyruvate, Pyr pyruvate, Pyr ex extracellular pyruvate, Ru5P ribulose-5-phosphate, SA succinic acid, Xu5P xylulose-5-phosphate, Xyl xylose
Enzyme kinetics
Parameters of the Michaelis-Menten model, K m and Vʹ max, for the G6PDH reaction were determined by fitting the model to NADPH concentration profiles generated from the enzyme assays. Vʹ max represents the specific reaction rate and is normalized to biomass concentration in the assay (C x ) for each steady state. Fits were performed on the entire dataset (all five steady states) for each substrate using non-linear, least-squares optimization in Matlab. Only data from the first 5 min of each assay were included in each fit to ensure that the NADP+ concentration was sufficiently high and did not influence the rate of the reaction. This allows for a simplified kinetic model where the rate dependency on NADP+ is zero order and therefore can be excluded from the kinetic expression. Glucose datasets were used initially to solve for K m , which was then fixed in determining the Vʹ max values for the xylose datasets to maintain consistency. The mean absolute percentage error between the model and the experimental data for an entire dataset was used to indicate the quality of the fit.
Results
Continuous glucose and xylose fermentations
Similar to previous continuous biofilm studies on A. succinogenes (Bradfield and Nicol 2014; Maharaj et al. 2014; Brink and Nicol 2014; Bradfield and Nicol 2016) in the current study, the succinic acid yield on substrate (Fig. 2a) and the ratio of succinic acid to acetic acid (YAASA; Fig. 2b) increased with increasing succinic acid titre (CSA). Furthermore, the formic acid-to-acetic acid ratio (YAAFA) decreased with increasing CSA (Fig. 2b), indicating increased pyruvate or formate dehydrogenase activity (Bradfield and Nicol 2014). In addition, YAASA values exceeded the limits defined by the active metabolism where full pyruvate formate-lyase activity allows for a maximum YAASA value of 1.97 g g−1, while full pyruvate/formate dehydrogenase activity allows for a maximum YAASA value of 3.93 g g−1 (Bradfield and Nicol 2014).
Similar trends were observed compared to previous studies in a the yield of succinic acid on glucose (YGlcSA; circles) and xylose (YXylSA; diamonds) and b the metabolite ratios (YAASA = succinic acid/acetic acid with circles for glucose and diamonds for xylose; YAAFA = formic acid/acetic acid with squares for glucose and triangles for xylose)
It has been suggested that these trends are linked to a titre-dependent shift in metabolism from a growth to a non-growth or stationary mode (Bradfield and Nicol 2014; Brink and Nicol 2014), with associated incomplete closures of mass and redox balances. In the current study, average mass balance closures (Fig. 3a) were 91 and 84 % for glucose and xylose, respectively, and decreased with increasing CSA, which is in agreement with previous observations. In addition, redox balance closures (Fig. 3b) showed greater deviations at higher CSA values. Moreover, the shift from growth to non-growth metabolism at increasing titres is exemplified by Fig. 4 where dry cell weights (DCWs) of the fermentation broth decreased with increasing CSA. Samples for determination of suspended DCWs were collected over an extended period to account for biomass fluctuations due to biofilm dynamics. In each fermentation, the duplicate point near a CSA of 30 g L−1 showed good repeatability and conformed to the trend of the other steady-state points. Pyruvic acid concentrations were between 0.80 and 3.21 g L−1 for the xylose fermentation and between 0.34 and 0.99 g L−1 for the glucose fermentation. SA yields and mass balance closures in the xylose fermentation were distinctly lower than those for glucose, in accordance with previous observations (Bradfield and Nicol 2016).
a Mass balance closures (glucose circles, xylose diamonds) and b a redox (as NADH) parity plot showed similar trends to previous studies. Production and consumption of NADH (glucose circles, xylose diamonds) are based on overall pathways and metabolite measurements on a molar basis. In b, the black arrow indicates the direction of increasing succinic acid concentration. The blue diagonal line gives equimolar consumption
The parity plot, which reflects incomplete closure of the redox balance (Fig. 3b), reveals that more NADH is being consumed in succinic acid production than is being generated by the central metabolic network of A. succinogenes based on the concentrations of excreted metabolites (Fig. 1; excluding the pentose phosphate pathway). Furthermore, the deviation increases with increasing CSA. Biomass is an unlikely source of the additional reduction power since the deviation in the redox balance closure increases as the suspended DCW decreases with increasing CSA (Fig. 4). Furthermore, biomass (as suspended DCW) only accounts for 0.35 % of total glucose consumption when redox balance deviations are at their highest. In addition, A. succinogenes has an incomplete TCA cycle and lacks Entner-Doudoroff and glyoxylate pathways (McKinlay et al. 2010), and no additional metabolites or sources of reduction power were detected. These observations are in agreement with previous studies and, together with the limited central metabolic network of Actinobacillus succinogenes, led to the hypothesis that the oxidative pentose phosphate pathway (OPPP) is a plausible source of the unaccounted reduction power (Bradfield and Nicol 2014; Bradfield et al. 2015; Bradfield and Nicol 2016).
Glycolysis, together with a complete TCA cycle or a glyoxylate shunt, can generate sufficient reduction power to achieve homosuccinate fermentation (Van Heerden and Nicol 2013a). The OPPP is also capable of generating the required reduction power for homosuccinate fermentation by carbon cycling between glycolysis and the OPPP (Villadsen et al. 2011). Therefore, there are three routes coupled to glycolysis by which homosuccinate fermentation can be attained with a theoretical maximum yield of succinic acid on glucose of 1.12 g g−1 (Bradfield and Nicol 2014). With regard to the OPPP, homosuccinate fermentation can be achieved if 85.7 % of total glucose (on a molar basis) is channelled through the OPPP (Fig. 5) and back to glycolysis after a minor loss of carbon from the pathway as CO2. However, there is a net consumption of CO2 (decrease in the CO2 yield on glucose) in succinic acid production which increases with increasing succinic acid yield, or increasing OPPP-to-substrate uptake flux (Fig. 5), defined here as α for the flux models (Eq. 2):
Therefore, carbon is not lost from the system as CO2 even though CO2 is produced concomitant with NADPH production.
The yield of CO2 (solid line), succinic acid (dashed line) and acetic acid (dotted line) on glucose as a function of increasing values of α (Eq. 2). The ratio of fluxes (α) corresponds to Fig. 1a where a similar matrix system was used. The model assumes exclusive pyruvate dehydrogenase activity (i.e. no pyruvate formate-lyase activity) and no pyruvate excretion, for simplification, and to define the system for maximum CO2 production. The theoretical maximum yield (homosuccinate fermentation) occurs when 85.7 % of total carbon (molar basis) splits to the OPPP from glycolysis. This can be calculated by specifying zero acetic acid formation and solving for α. A decrease in CO2 yield implies that CO2 consumption increased with an increase in OPPP flux
Glucose-6-phosphate dehydrogenase and enzyme kinetics
In glucose metabolism, glucose-6-phosphate (G6P) serves as the branch point from glycolysis to the OPPP (Fig. 1a), while for xylose, G6P forms part of the recycle loop that channels carbon from glycolysis back to the pentose phosphate pathway via the OPPP (Fig. 1b). In both cases, glucose-6-phosphate dehydrogenase (G6PDH) is involved in catalysing the first reaction of the OPPP (G6P → 6-phosphoglucono-δ-lactone (6PGL)); consequently, the activity of this enzyme provides an indication of the flux through the OPPP. In addition, the reaction converts NADP+ to NADPH, allowing the activity of the enzyme to be determined spectrophotometrically (Noltmann et al. 1961).
G6P is the required substrate for G6PDH assays; however, since cell extracts contain a number of soluble proteins, phosphoglucose isomerase (PGI) is likely to be present and react with G6P. Therefore, the assay system included F6P and G6P at various concentrations (Table 1) in order to develop a kinetic model in vitro that would account for the PGI reaction and allow for a more accurate determination of the rate of the G6PDH reaction (Fig. 6). An additional concern is that 6PGL can be further converted to 6-phosphogluconate (6PG) by lactonase (LAC), which in turn is converted to ribulose-5-phosphate (Ru5P) by 6-phosphogluconate dehydrogenase (6PGDH) with associated formation of NADPH. However, since the G6PDH reaction is conventionally considered the rate-limiting step in the OPPP (Stanton 2012), it was assumed that the LAC and 6PGDH reactions were not limiting and relatively instantaneous. Consequently, the rate of NADPH formation was considered to be twice that of the G6PDH reaction to simplify fitting of the kinetic model. In addition, the concentration of NADP+ was maintained in excess to ensure a zero-order rate dependency on NADP+, thereby further simplifying the kinetic model.
Reaction scheme used for developing the kinetic model from enzyme assay data. Reactions could not proceed beyond G6P and F6P in glycolysis as the required co-factors (ADP and ATP) were excluded from the assays. The rates of the LAC and 6PGDH reactions were assumed to be instantaneous relative to the G6PDH reaction since this is considered the rate-limiting step of the OPPP. 6PG 6-phosphogluconolactone, 6PGL 6-phosphoglucono-δ-lactone, F6P fructose-6-phosphate, G6P glucose-6-phosphate, Ru5P ribulose-5-phosphate, 6PGDH 6-phosphogluconate dehydrogenase, G6PDH glucose-6-phosphate dehydrogenase, LAC lactonase, PGI phosphoglucose isomerase
Michaelis-Menten kinetics was used to model both the G6PDH and the reversible PGI reactions. The hypothesis of OPPP activity implies that the rate of the G6PDH reaction should vary across dilution rates (i.e. different acid titres), and therefore, the enzyme concentration and activity will be different in each assay. Furthermore, the biomass concentration in each assay (C x ) is different and, in combination with the assumed differences in enzyme concentration, implies that the maximum specific reaction rate (Vʹ max) is unique for each steady-state condition. Therefore, separate fits are required for Vʹ max at each steady state. Kinetic parameters for the PGI reaction were obtained from Dyson and Noltmann (1968), where maximum rates for the forward and reverse directions were related by the Haldane relationship and remained variable in the model.
G6PDH activity was observed across all steady-state conditions in both glucose (Vʹ max between 8.17 × 10−8 and 2.82 × 10−7 molG6P gcells −1 s−1) and xylose (Vʹ max between 8.86 × 10−8 and 1.47 × 10−7 molG6P gcells −1 s−1) fermentations, indicating the presence of OPPP flux. Moreover, the presence of G6PDH activity in the xylose fermentations strongly supports the hypothesis, since G6P only features in the OPPP in xylose metabolism, whereas in glucose metabolism, G6P forms part of glycolysis. Therefore, the presence of G6PDH activity under non-growth conditions (as reflected by DCW (Fig. 4)) indicates that a definite OPPP flux occurred in the xylose-grown cells. Fits of the assay data to the kinetic model gave a mean absolute percentage error of 4.9 and 4.5 % across all glucose and xylose datasets, respectively (Fig. 7a). Also, the fitted K m value of 0.26 mM for G6P was similar to that reported for G6PDH from other bacteria (0.11 mM, Lee and Levy 1992; 0.17 mM, Scopes et al. 1985).
a The complete set of modelled and experimental (assay) NADPH concentrations (glucose circles, xylose diamonds), including a 5 % deviation region (+5 % dashed line, −5 % dotted line, equimolar solid line). The model was fitted to assay readings taken every 15 s over a comparison period of 300 s. b Comparison of the rate of the G6PDH reaction from glucose-grown biofilm when using only G6P or only F6P as substrates in the assay for high (blue circles) and low (green circles) concentration levels of each (equal rates solids line). For each concentration level, the points of the duplicate dilution rate overlap precisely; therefore, only four points are visible for each concentration level, but five points are actually present. The independence of the G6PDH reaction rate on starting substrate implies that the PGI reaction is relatively instantaneous and does not influence the rate of the G6PDH reaction
Furthermore, it was found that the rate of the PGI reaction was substantially greater than that of the G6PDH reaction since the fitted Vʹ max values of the G6PDH reaction were essentially the same for G6P-only and F6P-only starting concentrations, at each of the two concentration levels (Fig. 7b). This is further confirmed by maximum rates of up to 2500 times greater for the forward PGI reaction compared to the G6PDH reaction and indicates that the conversion of F6P to G6P is relatively instantaneous. Therefore, the PGI reaction did not have a considerable effect on the rate of the G6PDH reaction, but rapid equilibrium meant that a portion of G6P was constantly being lost to F6P, thus lowering the effective G6P concentration available for the G6PDH reaction.
Flux analysis at the glucose-6-phosphate node
The maximum specific rates (Vʹ max) for the G6PDH reaction determined from the assays and the kinetic model give an experimental indication of OPPP flux. In addition to the experimental rates, fluxes were determined from metabolic network models for each substrate (Fig. 1) and the measured metabolite concentrations. The models include the OPPP in order to provide a theoretical reference against which the experimental data can be compared as part of testing the hypothesis. Since overall substrate uptake flux regulates the magnitude of individual pathway fluxes, it is necessary to normalize individual fluxes to substrate uptake flux. To this end, α (Eq. 2) represents the ratio of OPPP flux to substrate flux (or substrate uptake rate) and gives the theoretical split of carbon into the OPPP at the G6P node on a molar rate basis.
In order to compare the experimentally determined rates to α, it is necessary to use the same basis. The ideal experimental equivalent is to use the biomass-based rate of G6P production divided by the biomass-based rate of substrate uptake (rʹ s ; [molsubs gbiomass −1 s−1]); however, these rates are not explicitly known. Nevertheless, the biomass-based G6P rate can be approximated by the experimentally determined specific reaction rate (i.e. Vʹ max), although the true rate is likely to be lower since intracellular metabolite concentrations in the central metabolism (including PPP) of E. coli have been shown to be near the K m value (Bennett et al. 2009). Also, since the total biomass content in the fermenter (C f x ) cannot be determined for each steady state, rʹ s is unknown. However, the volumetric rate of substrate consumption is known (r s ; [molsubs Lreactor −1 s−1]), which enables the definition of γ (Eq. 3) as an experimental approximation of α:
Given the uncertainty in both the intracellular metabolite concentrations and C f x , a direct quantitative comparison between α and γ is not possible. However, if one assumes that C f x is constant and that intracellular intermediate concentrations in the central metabolism are similar across all steady states, a trend comparison of the experimental and theoretical rate relationships is possible. In Fig. 8, the two ratios are compared with the underlying assumption that scalar differences between the two y-axes are based on unknowns that remained constant across all steady states.
Experimental (enzyme kinetic model, circles) and modelled (flux model, diamonds) ratios of OPPP flux-to-total substrate uptake flux versus succinic acid concentration for a glucose and b xylose. The experimental flux relationships are represented by γ (Eq. 3), while those of the flux model are given by α (Eq. 2), corresponding to the metabolic networks of Fig. 1
The trends in OPPP flux relative to total substrate uptake flux determined from the enzyme kinetic model (γ) compare well with the trends from the flux models (α) (Fig. 8). For glucose (Fig. 8a), the shape and relative magnitudes of α and γ are similar across all CSA values. For xylose (Fig. 8b), the shapes of the trends are similar but relative magnitudes are different, yet both show an increasing trend with increasing CSA. Importantly, since the experimentally observed flux behaviour on both substrates matches the increasing trend of the flux models, based on the inclusion of the OPPP in the models, it implies that the experimental trends do indeed reflect OPPP flux with the same characteristics as the models. Also, since the trends of the experimental fluxes match those of the models for two independent fermentations using different substrates, the hypothesis of increasing OPPP activity with increasing CSA is strongly supported. Therefore, evidence is provided of OPPP flux leading to the production of additional reduction power in biofilms of A. succinogenes.
Discussion
Overall, the results presented above provide valuable insight into the metabolism of wild-type A. succinogenes and point to the benefits of a naturally upregulated OPPP in microbial succinic acid production. The benefits of upregulating OPPP for increased succinic acid production have been demonstrated with genetically modified strains of A. succinogenes (Guettler et al. 2015) where G6PDH and malate dehydrogenase were overexpressed, yet this behaviour has not previously been demonstrated in wild-type A. succinogenes. Guettler et al. (2015) report a yield of 0.89 g g−1 and a maximum succinic acid productivity of 2.19 g L−1 h−1 (100.6 g L−1 succinic acid) with a modified strain overexpressing G6PDH (FZ53/p830.60). In the current study, a yield of 0.74 g g−1 and a productivity of 0.80 g L−1 h−1 (35.2 g L−1 succinic acid) were achieved at a dilution rate of 0.022 h−1, while at a dilution rate of 0.289 h−1, a productivity of 3.2 g L−1 h−1 (11.0 g L−1 succinic acid) and a yield of 0.65 g g−1 were achieved. Therefore, the recombinant strain outperforms the wild strain in terms of yield and titre but the wild strain is able to achieve greater productivities. Furthermore, the wild-type organism does not require genetic modifications and has the ability to form a stable biofilm. However, it is not ideal to directly compare batch and continuous fermentations and it would be beneficial to see continuous fermentations performed using the genetically modified strains and to determine whether these strains also have the ability to form biofilm.
To date, no studies have been performed on metabolic flux distributions in non-growing A. succinogenes cells. 13C-metabolic flux analysis of growing cells of A. succinogenes from batch fermentations gave γ (or α) values of up to 6.9 % (McKinlay and Vieille 2008) and 5 % (McKinlay et al. 2007) on a defined medium. The flux models in the current study give α values between 5.8 and 21.2 % for glucose and between 6.2 and 14.0 % for xylose, where values increase with increasing CSA. Therefore, at low CSA conditions, where growth is minimal yet apparent (Fig. 4), the flux ratios compare well with the previous studies; however, as the cells enter a stationary phase due to growth inhibition at high CSA, flux through the OPPP increases as indicated experimentally. A similar result was observed in Rühl et al. (2012), where OPPP flux increased by 83 % in non-growing cells compared to growing cells of wild-type Bacillus subtilis. Under non-growing conditions, γ values in B. subtilis cells of up to 64 % were observed, suggesting that the values predicted by the models in the current study are reasonable.
Although OPPP activity can lead to closure of the redox balance, the mass balance remains unchanged since the only by-product of the OPPP is CO2 and there is a net consumption of CO2 within the system across all ratios of OPPP-to-substrate uptake flux (Fig. 5). In our previous work, we speculated that if OPPP flux was greater than the amount required to close the redox balance, additional reduced co-factors would be produced, which cannot be consumed in the central metabolism. To ensure an overall redox balance of the system, the reduced co-factors must necessarily be oxidized external to the cell, possibly by an electron accepter (oxidizing agent) in the feed such as, for example, yeast extract (Schindler et al. 2014). In such a scenario, CO2 consumption would be lower and there may be a net production of CO2 under sufficiently high OPPP fluxes, thereby accounting for the incomplete mass balance. Including an NADH sink in the flux models (Fig. 1) to account for this effect showed that the trends in OPPP flux versus substrate uptake flux is essentially the same as those in Fig. 8, but the magnitudes are substantially higher. Therefore, our speculation regarding the incomplete mass balance closures may be correct but does not dovetail quantitatively with the current results and more investigation is required.
References
Beauprez JJ, De Mey M, Soetaert WK (2010) Microbial succinic acid production: natural versus metabolic engineered producers. Process Biochem 45:1103–1114. doi:10.1016/j.procbio.2010.03.035
Bechthold I, Bretz K, Kabasci S, Kopitzky R, Springer A (2008) Succinic acid: a new platform chemical for biobased polymers from renewable resources. Chem Eng Technol 31:647–654. doi:10.1002/ceat.200800063
Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5:593–599. doi:10.1038/nchembio.186
Bozell JJ, Petersen GR (2010) Technology development for the production of biobased products from biorefinery carbohydrates—the US Department of Energy’s “top 10” revisited. Green Chem 12:539. doi:10.1039/b922014c
Bradfield MFA, Nicol W (2014) Continuous succinic acid production by Actinobacillus succinogenes in a biofilm reactor: steady-state metabolic flux variation. Biochem Eng J 85:1–7. doi:10.1016/j.bej.2014.01.009
Bradfield MFA, Nicol W (2016) Continuous succinic acid production from xylose by Actinobacillus succinogenes. Bioprocess Biosyst Eng 39:233–244. doi:10.1007/s00449-015-1507-3
Bradfield MFA, Mohagheghi A, Salvachúa D, Smith H, Black BA, Dowe N, Beckham GT, Nicol W (2015) Continuous succinic acid production by Actinobacillus succinogenes on xylose-enriched hydrolysate. Biotechnol Biofuels 8:181. doi:10.1186/s13068-015-0363-3
Brink HG, Nicol W (2014) Succinic acid production with Actinobacillus succinogenes: rate and yield analysis of chemostat and biofilm cultures. Microb Cell Factories 13:111. doi:10.1186/s12934-014-0111-6
Dyson JED, Noltmann EA (1968) The effect of pH and temperature on the kinetic parameters of phosphoglucose isomerase. J Biol Chem 243:1401–1414
Gombert AK, Nielsen J (2000) Mathematical modelling of metabolism. Curr Opin Biotechnol 11:180–186. doi:10.1016/S0958-1669(00)00079-3
Guettler M, Rumler D, Jain M (1999) Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int J Syst Bacteriol 49:207–216
Guettler M, Hanchar R, Kleff S, Jadhav S (2015) Recombinant microorganisms for producing organic acids. EP2904104A1.
Lee WT, Levy HR (1992) Lysine-21 of Leuconostoc mesenteroides glucose 6-phosphate dehydrogenase participates in substrate binding through charge-charge interaction. Protein Sci 1:329–334
Lin SKC, Du C, Koutinas A, Wang R, Webb C (2008) Substrate and product inhibition kinetics in succinic acid production by Actinobacillus succinogenes. Biochem Eng J 41:128–135. doi:10.1016/j.bej.2008.03.013
Liu Y-P, Zheng P, Sun Z-H, Ni Y, Dong J-J, Zhu L-L (2008) Economical succinic acid production from cane molasses by Actinobacillus succinogenes. Bioresour Technol 99:1736–1742. doi:10.1016/j.biortech.2007.03.044
Maharaj K, Bradfield MFA, Nicol W (2014) Succinic acid-producing biofilms of Actinobacillus succinogenes: reproducibility, stability and productivity. Appl Microbiol Biotechnol. doi:10.1007/s00253-014-5779-3
McKinlay JB, Vieille C (2008) 13C-metabolic flux analysis of Actinobacillus succinogenes fermentative metabolism at different NaHCO3 and H2 concentrations. Metab Eng 10:55–68. doi:10.1016/j.ymben.2007.08.004
McKinlay JB, Zeikus JG, Vieille C (2005) Insights into Actinobacillus succinogenes fermentative metabolism in a chemically defined growth medium. Appl Environ Microbiol 71:6651–6656. doi:10.1128/AEM.71.11.6651-6656.2005
McKinlay JB, Shachar-Hill Y, Zeikus JG, Vieille C (2007) Determining Actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metab Eng 9:177–192. doi:10.1016/j.ymben.2006.10.006
McKinlay JB, Laivenieks M, Schindler BD, McKinlay AA, Siddaramappa S, Challacombe JF, Lowry SR, Clum A, Lapidus AL, Burkhart KB, Harkins V, Vieille C (2010) A genomic perspective on the potential of Actinobacillus succinogenes for industrial succinate production. BMC Genomics 11:680. doi:10.1186/1471-2164-11-680
Noltmann EA, Gubler CJ, Kuby SA (1961) Glucose 6-phosphate dehydrogenase (Zwischenferment). J Biol Chem 236:1225–1230
Qureshi N, Annous BA, Ezeji TC, Karcher P, Maddox IS (2005) Biofilm reactors for industrial bioconversion processes: employing potential of enhanced reaction rates. Microb Cell Factories 4:24. doi:10.1186/1475-2859-4-24
Rühl M, Le Coq D, Aymerich S, Sauer U (2012) 13C-flux analysis reveals NADPH-balancing transhydrogenation cycles in stationary phase of nitrogen-starving Bacillus subtilis. J Biol Chem 287:27959–27970. doi:10.1074/jbc.M112.366492
Salvachúa D, Mohagheghi A, Smith H, Bradfield MFA, Nicol W, Black BA, Biddy MJ, Dowe N, Beckham GT (2016) Succinic acid production on xylose-enriched biorefinery streams by Actinobacillus succinogenes in batch fermentation. Biotechnol Biofuels 9:28. doi:10.1186/s13068-016-0425-1
Schindler BD, Joshi RV, Vieille C (2014) Respiratory glycerol metabolism of Actinobacillus succinogenes 130Z for succinate production. J Ind Microbiol Biotechnol 41:1339–1352. doi:10.1007/s10295-014-1480-x
Scholten E, Renz T, Thomas J (2009) Continuous cultivation approach for fermentative succinic acid production from crude glycerol by Basfia succiniciproducens DD1. Biotechnol Lett 31:1947–1951. doi:10.1007/s10529-009-0104-4
Scopes RK, Testolin V, Stoter A, Griffiths-Smith K, Algar EM (1985) Simultaneous purification and characterization of glucokinase, fructokinase and glucose-6-phosphate dehydrogenase from Zymomonas mobilis. Biochem J 228:627–634. doi:10.1042/bj2280627
Song H, Lee SY (2006) Production of succinic acid by bacterial fermentation. Enzym Microb Technol 39:352–361. doi:10.1016/j.enzmictec.2005.11.043
Stanton RC (2012) Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64:362–369. doi:10.1002/iub.1017
Stephanopoulos G (1999) Metabolic fluxes and metabolic engineering. Metab Eng 1:1–11. doi:10.1006/mben.1998.0101
Van Heerden C, Nicol W (2013a) Continuous and batch cultures of Escherichia coli KJ134 for succinic acid fermentation: metabolic flux distributions and production characteristics. Microb Cell Factories 12:80. doi:10.1186/1475-2859-12-80
Van Heerden C, Nicol W (2013b) Continuous succinic acid fermentation by Actinobacillus succinogenes. Biochem Eng J 73:5–11. doi:10.1016/j.bej.2013.01.015
Varma A, Palsson BO (1994) Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technology 12:994–998. doi:10.1038/nbt1094-994
Villadsen J, Nielsen J, Lidén G (2011) Bioreaction engineering principles, Third edn. Springer US, Boston, MA
Yan Q, Zheng P, Tao S-T, Dong J-J (2014) Fermentation process for continuous production of succinic acid in a fibrous bed bioreactor. Biochem Eng J 91:92–98. doi:10.1016/j.bej.2014.08.002
Zeikus JG, Jain MK, Elankovan P (1999) Biotechnology of succinic acid production and markets for derived industrial products. Appl Microbiol Biotechnol 51:545–552. doi:10.1007/s002530051431
Acknowledgments
The authors wish to thank Professor Lyn-Marie Birkholtz and Mrs. Sandra van Wyngaard from the Department of Biochemistry at the University of Pretoria for the lab usage and assistance with the enzyme assay methodology. The financial assistance of the National Research Foundation (NRF) towards this research is hereby acknowledged. Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the NRF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Bradfield, M.F.A., Nicol, W. The pentose phosphate pathway leads to enhanced succinic acid flux in biofilms of wild-type Actinobacillus succinogenes . Appl Microbiol Biotechnol 100, 9641–9652 (2016). https://doi.org/10.1007/s00253-016-7763-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7763-6