Abstract
Itaconic acid, one of the most promising and flexible bio-based chemicals, is mainly produced by Aspergillus terreus. Previous studies to improve itaconic acid production in A. terreus through metabolic engineering were mainly focused on its biosynthesis pathway, while the itaconic acid-degrading pathway has largely been ignored. In this study, we used transcriptomic, proteomic, bioinformatic, and in vitro enzymatic analyses to identify three key enzymes, itaconyl-CoA transferase (IctA), itaconyl-CoA hydratase (IchA), and citramalyl-CoA lyase (CclA), that are involved in the catabolic pathway of itaconic acid in A. terreus. In the itaconic acid catabolic pathway in A. terreus, itaconic acid is first converted by IctA into itaconyl-CoA with succinyl-CoA as the CoA donor, and then itaconyl-CoA is hydrated into citramalyl-CoA by IchA. Finally, citramalyl-CoA is cleaved into acetyl-CoA and pyruvate by CclA. Moreover, IctA can also catalyze the reaction between citramalyl-CoA and succinate to generate succinyl-CoA and citramalate. These results, for the first time, identify the three key enzymes, IctA, IchA, and CclA, involved in the itaconic acid degrading pathway in itaconic acid producing A. terreus. The results will facilitate the improvement of itaconic acid production by metabolically engineering the catabolic pathway of itaconic acid in A. terreus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aspergillus species are well-known for the production of a wide range of bio-products, such as organic acids, pharmaceuticals, and enzymes (Lubertozzi and Keasling 2009). Itaconic acid (methylene succinic acid) is an unsaturated dicarboxylic organic acid that is mainly produced by Aspergillus terreus (Bonnarme et al. 1995). It can be widely used as a monomer or co-monomer in the manufacture of many high value-added polymers (Okabe et al. 2009; Tate 1981).
In an effect to improve the application potential of itaconic acid, a great deal of research related to itaconic acid biosynthesis (Bonnarme et al. 1995; Jaklitsch et al. 1991; Li et al. 2011) and its regulation mechanism (Huang et al. 2014b; Lin et al. 2004; Reddy and Singh 2002; Tevz et al. 2010) have been carried out to improve the production efficiency of itaconic acid. In addition to enhancing the biosynthesis pathway, disrupting or weakening the degradation pathway by metabolic engineering is also an effective strategy to promote the accumulation of target products. Unfortunately, there are only scarce studies on the itaconic acid degradation pathway in A. terreus. As early as in 1958, Arpai identified itaconicoxidase, which could oxidize itaconic acid into itatartaric acid in intracellular extracts of A. terreus (Arpai 1958). However, Jakubowska and Metodiewa did not find itaconicoxidase in the cytoplasm of A. terreus and thus neglected this potential degradation pathway of itaconic acid (Jakubowska and Metodiewa 1974). Since then, there have been no further studies on the catabolism of itaconic acid in A. terreus.
Almost simultaneously, the itaconic acid degradation phenomenon was also found in both mammals and some bacteria in the 1960s. These studies demonstrated that mammalian mitochondria (Wang et al. 1961) and bacteria, such as Micrococcus sp. (Cooper et al. 1965), Salmonella spp. (Martin et al. 1961), and Pseudomonas spp. (Cooper and Kornberg 1964; Martin et al. 1961), were able to metabolize itaconic acid, although some of them even did not possess the ability to produce itaconic acid. However, the degradation pathway of itaconic acid was not experimentally confirmed until the genes encoding the three involved enzymes were identified both in Yersinia pestis and Pseudomonas aeruginosa in 2014 (Sasikaran et al. 2014). The two species possessed similar pathways to degrade itaconic acid. In the pathway, itaconic acid is first thought to be transformed into itaconyl-CoA by itaconyl-CoA transferase (Ict) using succinyl-CoA as the CoA donor. Then, itaconyl-CoA is hydrated into (S)-citramalyl-CoA by itaconyl-CoA hydratase (Ich) subsequently. Finally, (S)-citramalyl-CoA is cleaved into acetyl-CoA and pyruvate by (S)-citramalyl-CoA lyase (Ccl) (Sasikaran et al. 2014). However, the sequence identities of the three enzymes between the two species were quite low (12 % for Ict, 12 % for Ich, and 28 % for Ccl). Consistent with the low identities, the substrate specificities between the enzymes from these two species varied considerably. For example, the first enzyme in Y. pestis, YpIct, could catalyze the conversion of itaconic acid into itaconyl-CoA with other CoA esters in addition to succinyl-CoA, while Ict from P. aeruginosa, PaIct, could only use succinyl-CoA as a specific CoA donor (Sasikaran et al. 2014). For pathogens, such as Y. pestis and P. aeruginosa, the itaconic acid catabolic pathway is required for their survival inside the eukaryotic host, which can produce itaconic acid as one of the antimicrobial agents to protect them against pathogenic infections (Michelucci et al. 2013).
In our previous study, we found that the itaconic acid producing strain A. terreus LYT10 also possesses the ability to catabolize itaconic acid. When 40 g/L of exogenous itaconic acid was added into the medium at the beginning of fermentation (the condition was named as I40 for short), the biosynthesis of itaconic acid was significantly inhibited. The decrease in itaconic acid concentration showed that A. terreus LYT10 was able to catabolize itaconic acid (Fig. S1).
In the present study, we identified three key enzymes, IctA, IchA, and CclA, involved in itaconic acid catabolism in A. terreus by transcriptomic, proteomic, and bioinformatic analyses. Furthermore, the three key enzymes were heterologously expressed and purified, and the catabolic pathway was reconstructed and confirmed in vitro.
Materials and methods
Strains and growth conditions
A. terreus LYT10 was an industrial itaconic acid producing strain from Qingdao Langyatai (Group) Co., Ltd. (Qingdao, Shandong, China). The spores were cultured on potato dextrose agar (PDA) at 30 °C for 5 days. The 2.5 × 107 spores were inoculated into 55 ml itaconic acid production medium (IPM, pH 3.25) (Huang et al. 2014a) in a 500-ml shake flask, and the mycelium precultivation was carried out on a rotary shaker at 220 rpm and 37 °C. After cultivation for 20 h, 5 ml of the seed cultures were directly transferred into 50 ml of IPM (the normal condition) or IPM with extra 40 g/L of itaconic acid (the I40 condition, pH 3.25) in 500-ml shake flasks and cultivated on a rotary shaker at 220 rpm and 37 °C. The supernatants were collected for itaconic acid determination and the mycelia cultured in both conditions for 36 h were harvested for transcriptome and proteome analysis.
Escherichia coli DH5α was used for plasmid preparation and gene cloning, and E. coli BL21 (DE3) was used to express recombinant proteins. All E. coli cells containing plasmids were grown aerobically in Luria-Bertani (LB) medium (1 % peptone, 1 % NaCl, and 0.5 % yeast extract powder, pH 6.8–7.0) or on LB agar plates at 37 °C, supplemented with 50 μg/ml kanamycin (Solarbio, China).
Transcriptome and proteome analyses
Total RNA was extracted using Trizol reagents (Invitrogen, USA) according to the manufacturer’s protocol. The Beijing Genomics Institute (BGI) in Shenzhen, China performed digital gene expression profiling experiments based on RNA-Seq with an Illumina HiSeq 2000 System. Approximately 12 M clean reads of each sample were mapped to the reference genome of A. terreus NIH 2624 (http://www.cadre-genomes.org. uk/Aspergillus_terreus/Info/Index), allowing two mismatches per read. Gene expression was measured as the number of aligned reads to reference genes and was normalized to RPKM values (Reads Per Kb per Million reads). Genes with significant differential expression were identified with combined thresholds (a false discovery rate (FDR) ≤ 0.001 and fold change ≥2). The raw data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with the reference series number GSE73033.
Proteome identification based on Isobaric Tags for Relative and Absolute Quantitation (iTRAQ) was performed by BGI with a TripleTOF™ 5600 System (AB SCIEX, Concord, ON, Canada). Proteins identification was performed using a Mascot search engine 2.3.02 (Matrix Science, UK) against a reference database of A. terreus NIH2624. Protein quantitation required that a protein contained at least two unique peptides. The quantitative protein ratios were weighted and normalized by the median ratio in Mascot. We considered proteins with p values ≤0.05 and fold changes ≥1.2 as significant. Both the transcriptome and proteome analyses were performed once.
Cloning, heterologous expression, and purification of recombinant IctA, IchA, and CclA
The three genes (Sequences Data S1) involved in itaconic acid degradation, ictA(GenBank: KU559320), ichA (GenBank: KU559321), and cclA (GenBank: KU559322), were amplified from the complementary DNA (cDNA) of A. terreus LYT10 and cloned into the Nco I and Hind III sites of the pET-28a(+) expression vector containing a C-terminal 6 × His-tag. The primers used for gene cloning are summarized in Table S1. The expression vectors pMC-1, pMC-2, and pMC-3 were confirmed by nucleotide sequencing and transformed into the E. coli BL21 (DE3) strain for protein expression.
The recombinant E. coli were cultured in LB liquid medium and induced by 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) when the OD600 of the culture reached ~0.8. After culturing for 20 h at 18 °C, the cells were harvested by centrifugation and resuspended in lysis buffer containing 20 mM imidazole, 500 mM NaCl, and 20 mM potassium phosphate buffer (pH 7.4). The cells were then broken by ultrasonication, and cell debris was removed by centrifugation. The extracts were loaded onto Nickel chelating resin (Qiagen, Germany) and then eluted with elution buffer containing 300 mM imidazole, 500 mM NaCl, and 20 mM potassium phosphate buffer (pH 7.4) to obtain the recombinant proteins with His-tags. The enzymes were desalted using HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer (100 mM HEPES, 100 mM KCl, and 10 % glycerinum, pH 7.0) by 30 kDa Microsep Advance Centrifugal Device (Pall Corporation, Dreieich, Germany).
The protein concentration was determined by the method of Bradford (Bradford 1976). SDS-PAGE was performed in 12 % polyacrylamide gels using Coomassie Blue R-250 staining.
In vitro reconstruction of the itaconic acid catabolic pathway
In vitro reconstruction using heterologously produced IctA, IchA, and CclA was performed in a reaction mixture (100 μL) containing 200 mM MOPS-KOH (pH 6.5), 1 mM itaconic acid, and 1 mM succinyl-CoA (Sigma, USA). Then, 10 μg of IctA, IchA, and CclA were consecutively added into the reaction mixture over an interval of 5 min. The reactions were performed at 37 °C. Reaction mixtures of each step were placed on ice, and the reactions were stopped by the addition of 3 μL of 25 % HCl. The reaction products were analyzed by HPLC (high-performance liquid chromatography) for organic acids identification and quantification, and LC-MS (liquid chromatography-mass spectrometry) for CoA esters identification.
Analysis of organic acids and CoA esters
The organic acids were determined by HPLC using an Aminex HPX-87H column (7.8 × 300 mm, Bio-Rad, USA) and detected at 210 nm. The column was operated at 35 °C with a mobile phase of 5 mM H2SO4 at a flow rate of 0.5 ml/min. Authentic itaconic acid (Qingdao Langyatai Group Co.,Ltd. China), succinate (Sigma, USA), pyruvate (Sigma, USA), and citramalate (Sigma, USA) were used as the standards to calculate the final concentration of organic acids, and the retention times were 9.4 min (pyruvate), 9.8 min (citramalate), 12.1 min (succinate), and 13.5 min (itaconic acid), respectively.
The CoA esters were identified by LC-MS using a BEH Amide Column (2.1 × 100 mm, 1.7 μm, Waters, USA). A gradient of 45 to 90 % acetonitrile in 10 mM ammonium formate buffer with a flow rate of 0.2 ml/min over 25 min was used. The M/Z values of the CoA esters were 766 (free CoA), 807 (acetyl-CoA), 866 (succinyl-CoA), 878 (itaconyl-CoA), and 896 (citramalyl-CoA), respectively.
Real-time quantitative PCR (qRT-PCR)
Total RNA was extracted using the RNAiso™ reagent (TaKaRa, Japan), and the first-strand cDNA was synthesized using a PrimeScript® RT Reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa, Japan) according to the manufacturer’s protocol.
The qRT-PCR amplifications were performed in a total volume of 20 μL containing 6 μL of H2O, 1 μL of each primer (5 μM), 10 μL of SYBR®Premix Ex Taq™ (Perfect Real Time) (TaKaRa, Japan), and 1 μL of template cDNA. The primers used for real-time quantitative PCR are summarized in Table S2. The amplifications were run on a LightCycler 480 instrument with software version 4.0 (Roche, Mannheim, Germany). The thermal cycling was performed as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s. All of the reactions were performed in triplicate. The transcript levels of the target genes were normalized against the level of the actin gene.
Results
Transcriptomic and proteomic analyses on the candidate enzymes involved in itaconic acid degradation in A. terreus LYT10
In order to explore if there is a similar itaconic acid degradation pathway in A. terreus just like that in P. aeruginosa and Y. pestis, the published genome of A. terreus NIH 2624 was used to search for the homologs of the three key enzymes involved in the degradation pathway in A. terreus. In result, seven hypothetical CoA transferases belonging to CoA transferase family III, two hypothetical CoA hydratases, and one hypothetical citramalyl-CoA lyase were preliminarily selected (Table S3). As the degradation phenomenon of itaconic acid in A. terreus LYT10 was much more significant under the I40 condition compared to the normal condition, we assumed that the expression of the degrading enzymes would also be higher under the I40 condition. It is found that only one hypothetical itaconyl-CoA transferase (IctA, Gene ID: ATET_06299), one itaconyl-CoA hydratase (IchA, Gene ID: ATET_03709), and the citramalyl-CoA lyase (CclA, Gene ID: ATET_03186) in the selected enzymes were notably up-regulated at both transcription and protein levels under the I40 condition according to the transcriptomic and proteomic analyses of A. terreus LYT10 (Table 1). The real-time quantitative PCR results also confirmed this variation trend at the transcript level under the I40 condition (Table 1). These results indicated the potential roles of IctA, IchA, and CclA in itaconic acid catabolism in A. terreus.
Bioinformatic analysis on the candidate enzymes involved in itaconic acid degradation in A. terreus LYT10
To further confirm the potential enzymes for itaconic acid degradation, we also searched the homologs of itaconyl-CoA transferase, itaconyl-CoA hydratase and citramalyl-CoA lyase from P. aeruginosa, and Y. pestis in the genome of A. terreus NIH 2624 by BLASTP (Basic Local Alignment Search Tool for protein) (Fig. S2). Coincidentally, the three homologs with the highest identity were exactly the enzymes selected using the transcriptomic and proteomic data above.
Like PaIct and YpIct, IctA was annotated as a CoA transferase family III protein and possessed 66 and 12 % identity with PaIct and YpIct, respectively (Table S3). IchA was a hypothetical enoyl-CoA hydratase (having 30 and 32 % identity with PaIch and YpIch, respectively), which can catalyze the syn-addition of a water molecule across the double bond of unsaturated CoA-esters (Willadsen and Eggerer 1975). CclA (having 31 and 31 % identity with PaCcl and YpCcl, respectively) was actually annotated as a citrate lyase (CitE)-like protein. The CitE-like proteins had been demonstrated to carry out the citramalyl-CoA lyase activity to cleave citramalyl-CoA into acetyl-CoA and pyruvate in Clostridium tetanomorphum and Homo sapiens (Dimroth et al. 1977; Sasikaran et al. 2014), indicating the potential roles of CclA in catalyzing citramalyl-CoA.
Although it is difficult to propose exact enzymatic functions of these proteins only by the above bioinformatic sequence analysis, the high identities of the conserved domains compared with those of P. aeruginosa and Y. pestis indicated that those enzymes possessed possible core catalytic functions for itaconic acid degradation in A. terreus.
In vitro reconstruction of the itaconic acid degradation pathway of A. terreus
To assess the functions of IctA, IchA, and CclA in itaconic acid catabolism, the three candidate genes ictA, ichA, and cclA were heterologously expressed in E. coli BL21 (Fig. S3). The catalytic activities of the recombinant proteins were measured in vitro.
IctA is the first enzyme in the pathway and showed activity toward itaconic acid, with succinyl-CoA as the CoA donor to generate itaconyl-CoA. The reaction proceeded almost immediately with the addition of IctA. As the reaction progressed, the product itaconyl-CoA was transformed into its isomer after 5 min, which might be mesaconyl-CoA or citraconyl-CoA (Fig. S4). To understand the substrate selectivity of IctA more comprehensively, some other CoA-esters were also tested as CoA donors besides succinyl-CoA. However, IctA could not transfer the CoA group from CoA, acetyl-CoA, and propionyl-CoA or butyryl-CoA (Fig. S5), which is similar with the substrate selectivity of PaIct in P. aeroginoasa.
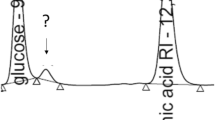
When the second enzyme IchA was added into the reaction system, the itaconyl-CoA was further converted into citramalyl-CoA (Fig. 1a). Moreover, the addition of the second enzyme facilitated the reaction rate of the first step, and the concentration of itaconic acid decreased dramatically (Fig. 1b). Interestingly, an unexpected product citramalate was also detected in the reaction system (Fig. 1b). There might be two ways to generate citramalate: either through cleavage of citramalyl-CoA because of the instability of CoA-ester or through the transfer of the CoA-group from citramalyl-CoA to other organic acids, such as itaconic acid or succinate, by IctA.
In vitro analysis of the metabolites during the degradation of itaconic acid. The reaction mixture (100 μL) contained 200 mM MOPS buffer (pH 6.5), 1 mM itaconic acid, and 1 mM succinyl-CoA. Then, 10 μg of IctA, IchA, and CclA were consecutively added into the reaction mixture over an interval of 5 min. a LC-MS analysis of the CoA-esters in the reaction: (1) The control reaction without adding of any enzymes; (2) the first step of the degrading reaction catalyzed by 10 μg of IctA for 5 min; (3) the second step reaction with 10 μg of IchA added into the system of the first step for another 5 min; (4) the third step reaction with 10 μg of CclA added into the system of the second step. b HPLC analysis of the organic acids in the reaction. The retention times were 9.5 min (pyruvate), 9.9 min (citramalate), 12.2 min (succinate), and 13.4 min (itaconic acid). c The time course of organic acid concentrations during the reaction. IctA itaconyl-CoA transferase, IchA itaconyl-CoA hydratase, CclA citramalyl-CoA lysase, Suc-CoA succinyl-CoA, Ita-CoA itaconyl-CoA, Cit-CoA citramalyl-CoA, Ace-CoA acetyl-CoA
The last candidate enzyme, CclA, is supposed to catalyze the formation of acetyl-CoA and pyruvate from citramalyl-CoA. Agreeing well with the theoretical value, approximately 1 mM pyruvate, was generated, while 1 mM itaconic acid and 1 mM succinyl-CoA were consumed completely (Fig. 1c). However, only 0.4 mM succinate was detected, much lower than the theoretical value (1 mM). As citramalate was generated concomitantly, this observation could be explained by assuming that IctA was capable of transferring the CoA-group from citramalyl-CoA to succinate. Thus, the appearance of citramalate and rapid consumption of itaconic acid in the second step could also be explained.
Based on the step by step experiment, the pathway from itaconic acid and succinyl-CoA to acetyl-CoA and pyruvate was verified by testing metabolites in each step of the reactions, except that the process from citramalyl-CoA to citramalate was still uncertain.
To further explore the destinations of citramalyl-CoA in the pathway, degrading reactions with various proportions of succinyl-CoA and itaconic acid were performed. Equimolar mixture of IctA, IchA, and CclA was used to catalyze succinyl-CoA and itaconic acid with mole ratios of 1:0.5, 1:1, 1:2, or 1:3. If citramalyl-CoA could not be used as CoA donor by IctA, the excessive amounts of itaconic acid would remain in the system and the final maximal concentration of citramalate would not be over 1 mM. But in practice, itaconic acid was consumed almost completely in all reactions (Fig. 2), implying that there were other CoA donors besides succinyl-CoA involved in catalyzing itaconic acid in the reaction system. In addition, the generation of citramalate increased proportionately along with the increasing percent of itaconic acid, and it was over 1.8 mM in the reaction with 1 mM succinyl-CoA and 3 mM itaconic acid (Fig. 2). These results proved that citramalyl-CoA can be used as CoA donor in itaconic acid catabolization. Moreover, the amounts of succinate remained around 0.5 mM in all systems, which is much lower than the theoretical value (1 mM) (Fig. 2). This indicated that the transfer of CoA-group from citramalyl-CoA to itaconic acid was performed through succinate (Fig. 3). Surprisingly, there were very small amounts of pyruvate detected in all reactions, and it even decreased gradually along with the increasing percent of itaconic acid (Fig. 2). This is quite different with the results of the step by step reaction, in which pyruvate was generated as the main product (Fig. 1c).
Generation of organic acids from various proportions of succinyl-CoA and itaconic acid. The reaction mixture (100 μL) contained 200 mM MOPS buffer (pH 6.5), 1 mM succinyl-CoA, and 0.5 mM, 1 mM, 2 mM, or 3 mM itaconic acid. 1 μM of IctA, IchA, and CclA were added into the reaction mixtures simultaneously. The reactions were performed at 37 °C for 10 min
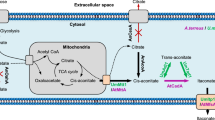
Finally, the itaconic acid catabolic pathway in A. terreus LYT10 can be proposed here (Fig. 3). Itaconic acid is first converted by IctA into itaconyl-CoA with succinyl-CoA as the CoA donor. Then, itaconyl-CoA is hydrated into citramalyl-CoA by IchA. Finally, citramalyl-CoA is cleaved into acetyl-CoA and pyruvate by CclA. In addition, IctA can also catalyze the reaction between citramalyl-CoA and succinate to generate succinyl-CoA and citramalate, which effectively accelerates the degradation of itaconic acid.
Discussion
Itaconic acid is not only an important biosynthetic chemical, but also plays important physiological roles in many organisms. In mammalian macrophages, itaconic acid is secreted as a part of an antibacterial response of mammalian macrophages during inflammation (Michelucci et al. 2013; Strelko et al. 2011) and acts as a potent inhibitor of isocitrate lyase, a key enzyme of the glyoxylate cycle in many pathogens, which is essential for the survival of many pathogens inside the eukaryotic host (Hillier and Charnetzky 1981; Honer Zu Bentrup et al. 1999; Williams et al. 1971). On the other hand, to survive in the macrophages, many pathogens possess the ability to metabolize itaconic acid as a result of convergent evolution (Sasikaran et al. 2014). In A. terreus, the most excellent producer of itaconic acid so far, there must also be important physiological functions for the existence of itaconic acid. One of the physiological roles might be providing appropriate low pH environment, as the optimal pH for growth of most A. terreus strains ranged from pH 3.0–3.5 (Rychtera and Wase 1981).
In this study, we observed the degradation phenomenon of itaconic acid in itaconic acid-producing strain A. terreus LYT10. However, the physiological roles of degrading itaconic acid for A. terreus were still unknown. Moreover, although A. terreus LYT10 could catabolize itaconic acid, it cannot grow in the medium using itaconic acid (with concentrations from 5 to 40 g/L) as the sole carbon source. This made the physiological roles of itaconic acid degradation more complex.
To confirm the key enzymes involved in catabolizing itaconic acid in A. terreus, we used the transcriptomic, proteomic, and bioinformatic analyses. All results indicated that three enzymes IctA, IchA, and CclA function in itaconic acid degradation. Although the identity between their protein sequences with those identified in P. aeruginosa and Y. pestis were quite low, which might be due to the far relationship between eukaryotes and prokaryotes. Compared with enzymes from Y. pestis, the sequence identities of IctA, IchA, and CclA with their homologs in P. aeruginosa were much higher, indicating the similar characterizations of the enzymes between A. terreus and P. aeruginosa. The in vitro reconstruction experiment also proved that the degrading enzymes showed similar substrate specificities to those of P. aeruginosa. For example, the IctA could use succinyl-CoA as CoA-donor, but could not transfer the CoA group from CoA, acetyl-CoA, propionyl-CoA, or butyryl-CoA (Fig. S5), which was similar to PaIct but contrary to YpIct.
Even the pathway consisting of IctA, IchA, and CclA was proved to catabolize itaconic acid in vitro in this study, we do not exclude the possibility that there are other enzymes or pathways involved in degrading itaconic acid in A. terreus. For example, itaconic acid could also be activated by succinyl-CoA synthetase, an important enzyme in TCA cycle, while the itaconyl-CoA was likely hydrated by the methylglutaconyl-CoA hydratase in liver mitochondria (Adler et al. 1957; Sasikaran et al. 2014; Wang et al. 1961). Hence, there might be similar reactions existing in A. terreus, which needs to be confirmed or excluded by in vitro and in vivo experiments in future.
Subcellular localization of the metabolic enzymes is an important factor for efficient catalysis. For biosynthesis of itaconic acid, the reactions accrued in different cell compartments in A. terreus. The formation of citrate and aconitate catalyzed by aconitase took place in the mitochondria, while the glycolysis and the final formation of itaconic acid catalyzed by cis-aconitate decarboxylase (CAD) progressed in the cytosol (Jaklitsch et al. 1991). There was no report yet about the effect of changing the targeting position of the biosynthetic enzymes for itaconic acid production in A. terreus. However, it did work in A. niger that heterologous expression of both aconitase and CAD in the mitochondria doubled the productivity of itaconic acid compared to strains expressing the enzymes in the cytosol (Blumhoff et al. 2013). It indicated that the proper targeting position of enzymes is important for the production of itaconic acid. For itaconic acid degradation, all the three enzymes were predicted to be localized in the mitochondria using the subCELlular LOcalization predictor CELLO v.2.5 (Yu et al. 2006). The mitochondrial localization of the three proteins implies that succinyl-CoA needed for the degradation of itaconic acid could be continuously generated via the α-ketoglutarate dehydrogenase. However, as the last step of itaconic acid biosynthesis took place in the cytosol, the catabolism of itaconic acid in vivo through the pathway identified in this paper might be restricted to some extent.
The in vivo experiments by blocking the three key enzymes involved in degrading pathway or targeting them to proper compartment would be efficient methods to further explore the mechanism and physiological roles of this pathway and to enhance the production of itaconic acid. Although the most significant progress in improving itaconic acid production is made by classical process optimization techniques until now (Hevekerl et al. 2014), the construction of mutant strains by genetic engineering is also promising for improving the itaconic acid production. For example, overexpression of CAD in A. terreus LYT10 enhanced the itaconic acid production level by nearly 10 % in shake flasks, and the production stage was shortened by around 4 h at the pilot scale (35 m3 fermentor) (Huang et al. 2014b). We do believe that the combination of classical process optimization and genetic engineering will make greater progress in improving itaconic acid production.
In this study, we identified a proposed catabolic pathway of itaconic acid in A. terreus that can catalyze the transformation of itaconic acid into pyruvate and acetyl-CoA in vitro. This is the first report to identify an itaconic acid degradation pathway in itaconic acid-producing A. terreus. The results provide a new perspective for further genetic engineering of A. terreus to improve itaconic acid production in the future.
References
Adler J, Wang SF, Lardy HA (1957) The metabolism of itaconic acid by liver mitochondria. J Biol Chem 229(2):865–879
Arpai J (1958) Itaconicoxidase: an enzyme from an ultra-violet-induced mutant of Aspergillus terreus. Nature 182(4636):661–662. doi:10.1038/182661a0
Blumhoff ML, Steiger MG, Mattanovich D, Sauer M (2013) Targeting enzymes to the right compartment: metabolic engineering for itaconic acid production by Aspergillus niger. Metab Eng 19:26–32. doi:10.1016/j.ymben.2013.05.003
Bonnarme P, Gillet B, Sepulchre AM, Role C, Beloeil JC, Ducrocq C (1995) Itaconate biosynthesis in Aspergillus terreus. J Bacteriol 177(12):3573–3578
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi:10.1016/0003-2697(76)90527-3
Cooper RA, Itiaba K, Kornberg HL (1965) The utilization of aconate and itaconate by Micrococcus sp. Biochem J 94:25–31. doi:10.1042/bj0940025
Cooper RA, Kornberg HL (1964) The utilization of itaconate by Pseudomonas sp. Biochem J 91(1):82–91. doi:10.1042/bj0910082
Dimroth P, Buckel W, Loyal R, Eggerer H (1977) Isolation and function of the subunits of citramalate lyase and formation of hybrids with the subunits of citrate lyase. Eur J Biochem 80(2):469–477. doi:10.1111/j.1432-1033.1977.tb11902.x
Hevekerl A, Kuenz A, Vorlop KD (2014) Influence of the pH on the itaconic acid production with Aspergillus terreus. Appl Microbiol Biotechnol 98:10005–10012. doi:10.1007/s00253-014-6047-2
Hillier S, Charnetzky WT (1981) Glyoxylate bypass enzymes in Yersinia species and multiple forms of isocitrate lyase in Yersinia pestis. J Bacteriol 145(1):452–458
Honer Zu Bentrup K, Miczak A, Swenson DL, Russell DG (1999) Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J Bacteriol 181(23):7161–7167
Huang X, Lu X, Li JJ (2014a) Cloning, characterization and application of a glyceraldehyde-3-phosphate dehydrogenase promoter from Aspergillus terreus. J Ind Microbiol 41(3):585–592. doi:10.1007/s10295-013-1385-0
Huang X, Lu X, Li Y, Li X, Li J (2014b) Improving itaconic acid production through genetic engineering of an industrial Aspergillus terreus strain. Microb Cell Factories 13:119. doi:10.1186/s12934-014-0119-y
Jaklitsch WM, Kubicek CP, Scrutton MC (1991) The subcellular organization of itaconate biosynthesis in Aspergillus terreus. J Gen Microbiol 137(3):533–539. doi:10.1099/00221287-137-3-533
Jakubowska J, Metodiewa D (1974) Studies on the metabolic pathway for itatartaric acid formation by Aspergillus terreus. II. Use of (−) citramalate, citraconate and itaconate by cell-free extracts. Acta Microbiol Pol B 6(2):51–61
Li A, van Luijk N, ter Beek M, Caspers M, Punt P, van der Werf M (2011) A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus. Fungal Genet Biol 48(6):602–611. doi:10.1016/j.fgb.2011.01.013
Lin YH, Li YF, Huang MC, Tsai YC (2004) Intracellular expression of Vitreoscilla hemoglobin in Aspergillus terreus to alleviate the effect of a short break in aeration during culture. Biotechnol Lett 26(13):1067–1072. doi:10.1023/B:BILE.0000032964.15178.7c
Lubertozzi D, Keasling JD (2009) Developing Aspergillus as a host for heterologous expression. Biotechnol Adv 27(1):53–75. doi:10.1016/j.biotechadv.2008.09.001
Martin WR, Frigan F, Bergman EH (1961) Noninductive metabolism of itaconic acid by Pseudomonas and Salmonella species. J Bacteriol 82:905–908
Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, Buttini M, Linster CL, Medina E, Balling R, Hiller K (2013) Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci U S A 110(19):7820–7825. doi:10.1073/pnas.1218599110
Okabe M, Lies D, Kanamasa S, Park EY (2009) Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol 84(4):597–606. doi:10.1007/s00253-009-2132-3
Reddy CS, Singh RP (2002) Enhanced production of itaconic acid from corn starch and market refuse fruits by genetically manipulated Aspergillus terreus SKR10. Bioresour Technol 85(1):69–71. doi:10.1016/S0960-8524(02)00075-5
Rychtera M, Wase DAJ (1981) The growth of Aspergillus terreus and the production of itaconic acid in batch and continuous cultures. The influence of pH. J Chem Technol Biotechnol 31(8):509–521. doi:10.1002/jctb.503310168
Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA (2014) Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10(5):371–377. doi:10.1038/nchembio.1482
Strelko CL, Lu W, Dufort FJ, Seyfried TN, Chiles TC, Rabinowitz JD, Roberts MF (2011) Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc 133(41):16386–16389. doi:10.1021/ja2070889
Tate H (1981) Assessing tumour markers. Br J Cancer 44(5):643–651
Tevz G, Bencina M, Legisa M (2010) Enhancing itaconic acid production by Aspergillus terreus. Appl Microbiol Biotechnol 87(5):1657–1664. doi:10.1007/s00253-010-2642-z
Wang SF, Adler J, Lardy HA (1961) The pathway of itaconate metabolism by liver mitochondria. J Biol Chem 236:26–30
Willadsen P, Eggerer H (1975) Substrate stereochemistry of the enoyl-CoA hydratase reaction. Eur J Biochem 54(1):247–252. doi:10.1111/j.1432-1033.1975.tb04134.x
Williams JO, Roche TE, McFadden BA (1971) Mechanism of action of isocitrate lyase from Pseudomonas indigofera. Biochemistry 10(8):1384–1390. doi:10.1021/bi00784a017
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64(3):643–651. doi:10.1002/prot.21018
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (2015AA021003), the National Natural Sciences Foundation of China (31500042 and 31400080), the Key Research Program of the Chinese Academy of Sciences (KSZD-EW-Z-016-2), and the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (KFJ-EW-STS077-RW10).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals.
Electronic Supplementary Material
ESM 1
(PDF 819 kb)
Rights and permissions
About this article
Cite this article
Chen, M., Huang, X., Zhong, C. et al. Identification of an itaconic acid degrading pathway in itaconic acid producing Aspergillus terreus . Appl Microbiol Biotechnol 100, 7541–7548 (2016). https://doi.org/10.1007/s00253-016-7554-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7554-0