Abstract
Biological removal of sulfide, nitrate, and phenol at loading rates of 600 g S/(m3 day), 900 g N/(m3 day), and 450 g C/(m3 day), respectively, from synthetic wastewaters was achieved in an expanded granular sludge bed (EGSB) reactor, whose rates are much higher than literature works and are considered feasible for handling high-strength petrochemical wastewaters without dilution. Effects of C/S ratio (2–2.5:1) on EGSB performance were noted insignificantly. The strains Bacillus sp., Thauera sp., and Pseudomonas sp. were the heterotrophic denitrifiers and the strains Thiobacillus sp., Azoarcus sp., and Sulfurovum sp. were the autotrophic denitrifiers in the EGSB granules. The EGSB reactor experienced biological breakdown at loadings higher than 1200 g S/(m3 day), 1800 g N/(m3 day), and 900 g C/(m3 day) by the following mechanism: high sulfide first inhibits heterotrophic denitrifies (Bacillus sp. and Pseudomonas sp.), thereby accumulating nitrite in the system; then, the accumulated nitrite inhibits autotrophic denitrifiers (Thiobacillus sp., Azoarcus sp., and Sulfurovum sp.) to complete breakdown of the system.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chemical and petrochemical industrial wastewaters such as spent caustic and sour waters frequently contain high levels of sulfide, ammonium, and aromatic compounds (Olmos et al. 2004; Wu et al. 2015; Yang et al. 2015). Sulfide is a toxic, odorous, and corrosive substance in acidic environments. Ammonium contributes mainly to eutrophication of water bodies, besides the risks associated with its toxicity and offensive odors. High concentration phenolic compounds are toxic, carcinogenic, mutagenic, and teratogenic to living forms.
Biological removal of sulfur, nitrogen, or phenolic compounds is a cost-effective and environmentally friendly process (Cadena and Peters 1988). Biological phenol removal has been reported under heterotrophic denitrifying condition (Thomas et al. 2002; Eiroa et al. 2005; Spence et al. 2001). Beristain-Cardoso et al. (2009a) proposed that when both autotrophic and heterotrophic denitrifiers were presented in the reactor with unlimited nitrate, simultaneous removal of nitrate, sulfide (S2− → SO4 2−), and phenol can occur. Beristain-Cardoso et al. (2009b) reported that at sulfide of 32 mg/L, oxidation of sulfide and phenol by the organo-lithoautotrophic microbial culture occur sequentially; with sulfide being rapidly oxidized to elemental sulfur and then to sulfate, following which phenol was oxidized to carbon dioxide; during this process nitrate was mostly convert to nitrogen gas (N2). The maximum loadings rates reported by Beristain-Cardoso et al. (2009a) using biofilm reactor were 37 g S/(m3 day) for sulfide, 168 g N/(m3 day) for nitrate, and 168 g C/(m3 day) for phenol.
Considering the very high concentrations of sulfur, nitrogen, and carbon pollutants presented in petrochemical wastewaters such as spent caustic and sour waters (Conner et al. 2000), biological treatment that can effectively handle these wastewaters are desired. An expanded granular sludge bed (EGSB) reactor allows wastewaters to pass through it at high upflow velocity, which is proposed to treat wastewaters of high loadings at acceptable removal rates. This study tested the performance of EGSB reactor at very high loadings (600 g S/(m3 day) for sulfide, 900 g N/(m3 day) for nitrate, and 450 g C/(m3 day) for phenol) and further specified the broke down loading, which was hardly achieved in a level real petrochemical wastewater. We identified the functional strains in granules responsible for the supreme capability noted for wastewater treatment.
Materials and methods
Experimental setup and startup
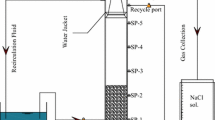
A modified EGSB reactor of that used by Liu et al. (2015a) was utilized, which is 60 mm in diameter and 120 cm in height. The working volume of EGSB reactor was 1.0 L, and the total volume was 2.25 L. Reactor internal temperature was maintained at 30 ± 1 °C by a temperature controller connecting to the reactor. Feed wastewater was introduced from the reactor bottom. A gas-washing device at the column top collected the H2S gas (if any) generated. A three-phase separator was also installed at reactor top to retain the biomass from washout. The recirculation ratio was fixed at 6.2:1 to mix the sludge and the wastewater in the reactor.
To investigate the performance of EGSB reactor, the concentration of all compounds was doubled for three times at the hydraulic retention time (HRT) of 8 h; at last, HRT was shortened from 8 to 5 h. The detailed operation schemes of EGSB reactor is described in Table 1. To investigate the effects of C/S ratio on EGSB performance, the influent concentrations for sulfide and nitrate were maintained at 100 mg S/L and 150 mg N/L, respectively, to make the S/N of 0.29:1. Then, three C/S ratios, 2:1, 2.25:1, and 2.5:1, were tested in the present EGSB reactor by maintaining the phenol concentration of 75, 84, and 93 mg C/L, respectively. The corresponding loadings of sulfide and nitrate were 300 g S/(m3 day) and 450 g N/(m3 day), respectively, while that of phenol was 225, 253, and 280 g C/(m3 day). Water samples were collected from the reactor, and then, NO3-N, NO2-N, S2−, S2O3 2−, SO4 2−, and phenol were measured.
The inoculated granules which acclimated from anaerobic methanogenic granules were acquired from a lab-scale denitrifying sulfide removal reactor, which treated sulfide, nitrate, and acetate wastewaters over a period of 1.5 months. The granule removal efficiency for the three contaminations was >95 %. The average diameter of granules was 2.25 mm. The shape of granules was near-round, and the surface has numerous microcolonies and channels. The suspended solids (SS) and volatile suspended solids (VSS) in seed granules were 42.1 and 28.5 g/l, respectively. Synthetic wastewater contained (/L): 50 mg S2−; 75 mg N NO3 −; 37.5 mg C as phenol; 375 mg NaHCO3; 25 mg NH4Cl; 4 mg P K2HPO4. The pH of synthetic wastewater was adjusted to 7.5. Other operational parameters of the EGSB reactor were available in Liu et al. (2015b).
Microbial population
Sludge samples were collected from the EGSB reactor and immediately stored at −80 °C. Total genomic DNA was extracted in duplicate from each sample using PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA) according to the manufacturer’s instructions. The extract was subsequently pooled, and the quality of the DNA extracted was examined by 1 % (w/v) agarose gel electrophoresis. The V3–V4 region of the extracted 16S rRNA gene was amplified by PCR (GeneAmp 9700 thermocycler, ABI, USA) in triplicate using bacterial primers 338F (5′-ACT CCT ACG GGA GGC AGC AG-3′) and 806R (5′-GGA CTA CHV GGG TWT CTA AT-3′), with the reverse primer containing a 6-bp barcode used to tag each sample. The PCR mix (in each 20 μL) contained 4 μL of 5× PCR buffer, 10 ng of template DNA, 0.2 μM of each primer, 0.25 mM of each dNTP, and 1 U FastPfu polymerase (TransGen, China). The PCR amplification was conducted at denaturation at 95 °C for 2 min, subsequent 30 cycles of temperature gradient at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and final extension at 72 °C for 5 min. The amplicos were electrophoresed on a 2 % (w/v) agarose gel and recovered using an AxyPrep DNA Gel Extraction Kit (AXYGEN, China).
The purified amplicon was quantified using a QuantiFluor-ST Fluorometer (Promega, USA), and then, a composite sequencing library was constructed by combining equimolar ratios of amplicons from all samples. The resulting library for paired end sequencing (2 × 250 bp) was analyzed on an Illumina Miseq platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Raw sequence data of this study have been deposited to the NCBI Sequence Read Archive with accession no. SRP067731.
Chemical analysis
After filtration by 2.5-μm PTFE membranes, the concentrations of nitrate, nitrite, sulfate, and thiosulfate in the samples were measured by an ion chromatography (ICS-3000; Dionex, USA) equipped with IonOac AG4AAS4A-SC 4-mm analytical column with carbonate/bicarbonate eluent (1.8 mmol/dm3 Na2CO3/1.7 mmol/L NaHCO3 at 1 cm3/min) and sulfuric regeneration (H2SO4 25 mmol/L at 5 cm3/min). The sulfide concentration was determined using the methylene blue method (Truper and Schlegel 1964), including dissolved H2S, HS−, and S2−. Other sulfur compounds apart from sulfide, sulfate, and thiosulfate were generally unstable and are neglected in measurements. The production quantity of elemental sulfur (S0) was estimated by mass balance calculation (Chen et al. 2009). Phenol concentration was determined using a high-performance liquid chromatography (Pekin Elmer series 200UV) with a C-18 reverse-phase column (Sigma-Aldrich, USA), column size 300 × 3.90 mm (Beristain-Cardoso et al. 2009a). The pH of liquid samples was measured using a pHS-25 meter (Shanghai, China). The concentrations of suspended solids (SS) and volatile suspended solids (VSS) were measured according to Standard Methods (APHA 2005).
Results
Performance of EGSB reactor
The reactor was startup with HRT of 8 h for 21 days (Table 1). Removals of nitrate, sulfide, and phenol all reached 100 %, on days 4, 3, and 2, respectively (stage I in Fig. 1). No nitrite and S0 accumulation was observed in this stage. About 50 mg S/L of sulfate was noted in the effluent (Fig. 1b) which indicated all fed sulfide was converted to sulfate (Fig. 1c). Negligible quantity of p-OH benzoic acid was accumulated under denitrifying condition.
Performance of EGSB reactor for simultaneous removal of S2− (a), NO3 −-N (b), and phenol-C (c). Conditions for stages I–V present in Table 1
From the 22nd day on, the concentration of all compounds in influent were doubled and maintained for another 22 days. The removal efficiencies of sulfide, phenol, and nitrate were maintained at 100 % (stage II in Fig. 1), with sulfur end-product being sulfate. On day 45, the concentration of all compounds in influent were doubled again, reaching 200 mg/L S2−; 300 mg N/L for NO3 −; 150 mg C/L for phenol; 1500 mg/L NaHCO3; 100 mg/L NH4Cl; 16 mg P/L K2HPO4, equivalent to loading rates at 600 g S/(m3 day) for sulfide, 900 g N/(m3 day) for nitrate, and 450 g C/(m3 day) for phenol. Nitrate removal efficiency was dropped to 92.1 %, although the removal efficiencies of sulfide and phenol were remained perfect. All sulfides were still completely converted to sulfates. Little nitrite and p-OH benzoic acid was accumulated in the reactor (stage III in Fig. 1).
On day 60, influent sulfide, nitrate, and phenol was further increased to 400 mg S2−/L, 600 mg N/L, and 300 mg C/L, respectively, to make the loading rates of 1200 g S/(m3 day) for sulfide, 1800 g N/(m3 day) for nitrate, and 900 g C/(m3 day). The removal efficiencies for nitrate and phenol were reduced significantly to 25 and 30 %, respectively, whereas that of sulfide was decreased to 90 %. The effluent nitrite concentration became 20 mg N/L but still little p-OH benzoic acid accumulation was noted at this stage. About 100 mg S/L sulfate was in the effluent and S0 conversion efficiency was nearly 65 %. On day 75, the HRT of the EGSB reactor was reduced from 8 to 5 h to increase the loading rates further to 1920 g S/(m3 day) for sulfide, 2880 g N/(m3 day) for nitrate, and 1440 g C/(m3 day). The effluent nitrite accumulated was increased to 84 mg N/L. The removal efficiencies for nitrate and phenol were maintained 25 and 30 %, respectively, whereas that of sulfide reduced to 71 %. The EGSB reactor performance was seriously deteriorated at these loadings (stages IV and V in Fig. 1).
Effects of C/S ratio on EGSB performance
With C/S = 2:1, removal efficiencies of sulfide, phenol, and nitrate were all >96 %( Fig. 2). Little nitrite or p-OH benzoic acid was accumulated in the reactor while all sulfide was converted to sulfate. At C/S = 2.25:1, little sulfide was converted to S0 (Fig. 2b), but the removal efficiency of phenol was decreased to 91 %. Further increase the C/S ratio to 2.5:1, still little S0 accumulation was observed and most sulfides were converted to sulfate, but the removal efficiency of phenol was further decreased 82 %. These observations indicated that sulfate was the main end-product at the C/S ratio of 2:1–2.5:1.
Microbial community shift at different loadings
The parameters related to the alpha diversity of microbial community for each sample at distance cutoff level of 0.03 are shown in Table 2. The phylogenetic classification of bacterial 16S rRNA sequence from the granular sludge samples at different loadings was illustrated based on phylum in Fig. 3 on genus in Fig. 4. The bacterial sequences affiliated with Chloroflexi, Proteobacteria, Firmicutes, Bacteroidetes, and Synergistetes were abundant in all samples, following Synergistetes > Firmicutes > Chloroflexi > Bacteroidetes > Proteobacteria. At 150 g S/(m3 day), 225 g N/(m3 day), and 112.5 g C/(m3 day) loading rate (stage I in Fig. 1), the abundance of Synergistetes decreased from 34.41 to 7.24 %. On the contrary, the abundances of Firmicutes, Chloroflexi, Bacteroidetes, and Proteobacteria were increased from 22.96, 17.27, 11.81, and 7.03 % to 30.25, 31.61, 16.54, and 8.34 %, respectively. When the loading rate was further increased to 600 g S/(m3 day), 900 g N/(m3 day), and 450 g C/(m3 day) (stage III), the abundance of Synergistetes was further decreased to 4.96 % and that of Chloroflexi further increased to 63.43 %. At stage V, the abundances of Bacteroidetes, Firmicutes, and Chloroflexi were decreased to 43.49, 1.36, and 3.52 %, respectively, while the abundances of Proteobacteria and Synergistetes were increased to 45.36 and 2.58 %, respectively.
Taxonomic classification of the bacterial communities at phylum level at sulfide loading rate of 150 g S/(m3 day) (day 0), 300 g S/(m3 day) (day 22), 600 g S/(m3 day) (day 60), 1200 g S/(m3 day) (day 75), and 1920 g S/(m3 day) (day 92). Phylum making up less than 0.5 % of total composition in the sample was classified as “others”
Taxonomic classification of the bacterial communities at genus level at sulfide loading rate of 150 g S/(m3 day) (day 0), 300 g S/(m3 day) (day 22), 600 g S/(m3 day) (day 60), 1200 g S/(m3 day) (day 75), and 1920 g S/(m3 d) (day 92). Phylum making up less than 0.5 % of total composition in the sample was classified as “others”
At genus level, a few amounts of sequences could not be classified (2.0, 2.0, 1.56, 16.7, and 5.40 % at the tested five loading rates) indicating the presence of many taxa unknown bacteria in the reactor (Fig. 3). Phylum sequences affiliated with Caldilineaceae_uncultured, vadinHA17_norank, Synergistaceae_uncultured, Anaerolineaceae_uncultured, Clostridium, Bacillus, Thiobacillus, Thauera, Azoarcus, Sulfurovum, and Pseudomonas, SB-1_norank were the main genera detected. The genera Bacillus, Thauera, and Pseudomonas were reported commonly to hetetrophic denitrifiers (Verbaendert et al. 2011; Liu et al. 2013; Rezaee et al. 2008). The genus Bacillus accounted only 0.03 % in the inoculated sludge and increased to 0.33 % after setup. At 600 g S/(m3 day), 900 g N/(m3 day), 450 g C/(m3 day) loading rates (stage III), the abundance of Bacillus was further increased to 6.61 %. In stages IV and V, the abundance of Bacillus was decreased to 0.16 and to 0.18 %, respectively. A similar pattern was noted for the genus Pseudomonas, which was 0.12 % in the inoculated sludge and increased to 2.67 % after setup; further increase in loading rates decreased its abundance 0.11– 0.27 %. On the contrary, the abundance of Thauera in the inoculated granular sludge was 0.35 % and was increased to 0.92 % after setup. In stage IV and stage V, the abundance of Thauera was increased to 2.91 and to 1.49 %, respectively.
The OTUs most closely related to Thiobacillus, Azoarcus, and Sulfurovum, the common autotrophic denitrifier in DSR sludge (Wang et al. 2005; Huang et al. 2015; Lee et al. 2013) were 0, 0.04, and 0.03 %, respectively, in the granular sludge. They increased with loading rates and peaked in abundance at 8.44, 3.63, and 3.24 %, respectively, in stage IV. Further increasing the influent loading rates to 1920 g S (m3 day) 2880 g N (m3 day), and 1440 g C (m3 day) (stage V), the abundance of Thiobacillus, Azoarcus, and Sulfurovum adversely decreased to <1 %.
Discussions
The above experimental findings suggest that the simultaneous biological removal of phenol, sulfide, and nitrate could be achieved in an EGSB reactor at 600 g S/(m3 day) for sulfide, 900 g N/(m3 day) for nitrate, and 450 g C/(m3 day) for phenol with >92 % efficiencies, much higher than those reported in Beristain-Cardoso et al. (2009a). Chen et al. (2008) investigated that simultaneous biological removal of sulfur, nitrogen, and carbon could be achieved in one EGSB reactor, and the loading rates could achieved at 3.0 kg S/(m3 day), 1.45 kg N/(m3 day), and 2.77 kg C/(m3 day). However, the carbon removed in the study of Chen et al. (2008) was in the form of acetate which was of little toxicity and easy biodegradation. Furthermore, acetate was scarcely present in chemical and petrochemical industrial wastewaters. Based on end-product distributions noted, most sulfides were converted to sulfate, nitrate to nitrogen gas, and phenol to carbon dioxide. This observation is welcome since the tested EGSB reactor has the potential to be applied for handling high-strength petrochemical wastewaters such as spent caustic and sour waters without dilution. Tests at C/S ratio of 2:1–2.5:1 demonstrated acceptable performances for simultaneous sulfide, nitrate, and phenol removal, although higher C/S ratio was noted unfavorable for phenol removal by the reactor. At even higher loading rates, the EGSB reactor was gradually broken down in performance (stage IV and stage V in Fig. 1).
The biomass of granules in this study was much higher than the biofilm in Beristain-Cardoso et al. (2009a). The bacteria in the granules could maintain high activities and resist high concentration of toxic contaminates. Furthermore, the EGSB reactor allows wastewater to pass through at high upflow velocity. The removal rates in this study are much higher than the literature (Beristain-Cardoso et al. 2009a).
The genus Bacillus, Thauera, and Pseudomonas, identified as the main heterotrophic denitrifiers in the reactor and the Thiobacillus, Azoarcus, and Sulfurovum, were the main autotrophic denitrifiers; in stage IV, nitrite was noted to accumulate with sulfide being preferable removed than phenol. That is, autotrophic denitrification was still active in this stage but the corresponding heterotrophic counterpart was inhibited to certain extents. Correspondingly, the genus Bacillus and Pseudomonas were diminished in abundance but that of Thauera was enriched for maintaining limited activity of hetrotrophic denitrification in the reactor. The Thiobacillus, Azoarcus, and Sulfurovum were predominant over heterotrophic denitrifiers in this stage IV. In stage V, nitrite was accumulated and the abundance of all autotrophic denitrifiers was significantly decreased. Restated, autotrophic denitrifiers were likely inhibited by the accumulated nitrite.
Hence, the reactor breakdown of the tested EGSB reactor is a sequential process by high sulfide concentration first inhibits heterotrophic denitrifiers to effectively remove nitrite as a metabolite by autotrophic denitrification, while the accumulated nitrite further inhibits the autotrophic denitrifiers to complete breakdown of the process.
References
APHA (2005) Standard Methods for the Examination of Water and Wastewater, 21st edn. American Public Health Association, Washington
Beristain-Cardoso R, Texier AC, Alpuche-Solis A, Gomez J, Razo-Flores E (2009a) Phenol and sulfide oxidation in a denitrifying biofilm reactor and its microbial community analysis. Process Biochem 44(1):23–28
Beristain-Cardoso R, Texier AC, Sierra-Alvarea R, Razo-Flore EJ, Field A, Gomez J (2009b) Effect of initial sulfide concentration on sulfide and phenol oxidation under denitrifying conditions. Chemosphere 74:200–205
Cadena F, Peters RW (1988) Evaluation of chemical oxidizer for hydrogen sulfide control. J Water Pollut Control Fed 60(7):1259–1263
Chen C, Ren NQ, Wang AJ, Yu ZG, Lee DJ (2008) Simultaneous biological removal of sulfur, nitrogen and carbon using EGSB reactor. Appl Microbiol Biotechnol 78:1057–1063
Chen C, Wang A, Ren N, Lee DJ, Lai JY (2009) High-rate denitrifying sulfide removal process in expanded granular sludge bed reactor. Bioresour Technol 100(7):2316–2319
Conner JA, Beitle RR, Duncan K, Kolhatkar R, Sublette KL (2000) Biotreatment of refinery spent-sulfidic caustic using an enrichment culture immobilized in a novel support matrix. Appl Biochem and Biotechnol Part A Enzym Eng Biotechnol 84–86:707–719
Eiroa M, Vilar A, Amor L, Kennes C, Veiga MC (2005) Biodegradation and effect of formaldehyde and phenol on the denitrification process. Water Res 39(2–3):449–455
Huang C, Li Z, Chen F, Liu Q, Zhao Y, Zhou J, Wang A (2015) Microbial community structure and function in response to the shift of sulfide/nitrate loading ratio during the denitrifying sulfide removal process. Bioresour Technol 197(2):227–234
Lee DJ, Wong BT, Adav SS (2013) Azoarcus taiwanensis sp. nov., a denitrifying species isolated from a hot spring. Appl Microbiol Biotechnol 98(3):1–7
Liu B, Mao Y, Bergaust L, Bakken LR, Frostegard A (2013) Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ Microbiol 15(10):2816–2828
Liu C, Zhao C, Wang A, Guo Y, Lee DJ (2015a) Denitrifying sulfide removal process on high-salinity wastewaters. Appl Microbiol Biotechnol 99(15):6463–6469
Liu C, Zhao D, Yan L, Wang A, Gu Y, Lee DJ (2015b) Elemental sulfur formation and nitrogen removal from wastewaters by autotrophic denitrifiers and anammox bacteria. Bioresour Technol 191(5):332–336
Olmos A, Olguıin P, Fajardo C, Razo-Flores E, Monroy O (2004) Physicochemical characterization of spent caustic from the OXIMER process and source waters from Mexican oil refineries. Energ Fuel 18:302–304
Rezaee A, Godini H, Dehestaniathar S, Yazdanbakhsh A, Mosavi G, Kazemnejad A (2008) Biological denitrification by Pseudomonas stutzeri immobilized on microbial cellulose. World J Microb Biot 24(11):2397–2402
Spence J, Bottrell H, Higgo W, Harrison I, Fallick AE (2001) Denitrification and phenol degradation in a contaminated aquifer. J Contam Hydrol 53(3):305–318
Thomas S, Sarfaraz S, Mishra LC, Iyengar L (2002) Degradation of phenol and henolic compounds by a defined denitrifying bacterial culture. World J Microbiol Biotechnol 8(1):57–63
Truper HG, Schlegel HG (1964) Sulfur metabolism of Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Anton Leeuw Int 30(1):225–238
Verbaendert I, Boon N, Vos PD, Heylen K (2011) Denitrification is a common feature among members of the genus Bacillus. Syst Appl Microbiol 34(5):385–391
Wang A, Du D, Ren N, van Groenestijn JW (2005) An innovative process of simultaneous desulfurization and denitrification by Thiobacillus denitrification. J Environ Sci Health A 40(5):1939–1949
Wu C, Zhou Y, Wang P, Guo S (2015) Improving hydrolysis acidification by limited aeration in the pretreatment of petrochemical wastewater. Bioresour Technol 194(2):256–262
Yang Q, Xiong P, Ding P, Chu L, Wang J (2015) Treatment of petrochemical wastewater by microaerobic hydrolysis and anoxic/oxic processes and analysis of bacterial diversity. Bioresour Technol 196(5):169–175
Acknowledgments
The research was supported by the National Natural Science Foundation of China under Grant No. 21307160 and the Natural Science Foundation of Shandong Province under Grant No. ZR2013EEQ030.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Liu, C., Han, K., Lee, DJ. et al. Simultaneous biological removal of phenol, sulfide, and nitrate using expanded granular sludge bed reactor. Appl Microbiol Biotechnol 100, 4211–4217 (2016). https://doi.org/10.1007/s00253-016-7293-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7293-2