Abstract
We previously identified the aur1 gene cluster which produces the angucycline antibiotic auricin. Preliminary characterisation of auricin revealed that it is modified by a single aminodeoxysugar, d-forosamine. Here we characterise the d-forosamine-specific genes. The four close tandem genes, aur1TQSV, encoding enzymes involved in the initial steps of the deoxysugar biosynthesis, were located on a large operon with other core auricin biosynthetic genes. Deleting these genes resulted in the absence of auricin and the production of deglycosylated auricin intermediates. The two final d-forosamine biosynthetic genes, sa59, an NDP-hexose aminotransferase, and sa52, an NDP-aminohexose N-dimethyltransferase, are located in a region rather distant from the core auricin genes. A deletion analysis of these genes confirmed their role in d-forosamine biosynthesis. The Δsa59 mutant had a phenotype similar to that of the cluster deletion mutant, while the Δsa52 mutant produced an auricin with a demethylated d-forosamine. Although auricin contains a single deoxyhexose, two glycosyltransferase genes were found to participate in the attachment of d-forosamine to the auricin aglycon. An analysis of the expression of the d-forosamine biosynthesis genes revealed that the initial d-forosamine biosynthetic genes aur1TQSV are regulated together with the other auricin core genes by the aur1Ap promoter under the control of the auricin-specific activator Aur1P. The expression of the other d-forosamine genes, however, is governed by promoters differentially dependent upon the two SARP family auricin-specific activators Aur1PR3 and Aur1PR4. These promoters contain direct repeats similar to the SARP consensus sequence and are involved in the interaction with both regulators.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gram-positive Streptomyces bacteria are characterised by their ability to produce a wide range of bioactive secondary metabolites, including many known antibiotics, during their complex life cycle. The genes responsible for the biosynthesis of these metabolites are usually clustered, and these clusters contain their own pathway-specific transcriptional activators. The expression of these regulatory genes is controlled, in turn, by global regulators in a complex regulatory mechanism, which determines the onset of antibiotic production (Bibb 2005; Martin and Liras 2010).
A large number of these secondary metabolites are polyketides. These structurally diverse compounds are synthesised by polyketide synthases (PKSs) which catalyse repeated decarboxylative condensation cycles between acyl thioesters. Three types of PKSs (I, II and III) are known. The type II, aromatic PKSs are responsible for the biosynthesis of aromatic polyketides. A minimal type II PKS contains a ketoacyl synthase (α subunit), a chain length factor (β subunit) and an acyl carrier protein. Additional PKS subunits, including ketoreductases, aromatases and cyclises, modify the nascent chain to form a particular cyclic polyketide. Natural PKS pathways may also include other enzymes, such as oxygenases, methylases and glycosyltransferases, which tailor the polyketide backbone to create its final, bioactive form (Hertweck et al. 2007). Although a large repertoire of aromatic polyketides has been identified, they all belong to just a few common structural types. One of the dominant groups is the angucyclines, which have a broad range of biological activities, including antibiotic, antitumor, antiviral, antifungal, platelet aggregation and enzyme inhibitory effects. They are characterised by the angular shape of their tetracyclic aromatic ring (Kharel et al. 2012).
Sugars are frequent structural components of natural products which normally participate in interactions between the given compound and its cellular target. Their presence is often essential for the biological activity of many natural products. A great majority of these sugars are 6-deoxyhexoses, which are synthesised from nucleoside diphosphate (NDP)-activated hexoses via 4-keto-6-deoxy intermediates. Two common enzymatic steps leading to the biosynthesis of these intermediates are catalysed by NDP-hexose synthases and NDP-hexose-4,6-dehydratases. Further modifications of these key intermediates (by, for example, deoxygenation, transamination and methylation) lead to a variety of different 6-deoxyhexoses. These deoxysugars are then transferred to the corresponding aglycon by glycosyltransferases (GTs), which are generally sugar-, aglycon- and site-specific (Salas and Méndez 2005). Many angucyclines possess deoxysugars attached to their aglycon core, and in many cases (e.g. landomycin, urdamycin and jadomycin), they are essential for their biological activities (Kharel et al. 2012).
We previously identified a type II PKS gene cluster, aur1, which is responsible for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM3239 (Novakova et al. 2002). Recently, we found that the cluster is not located in the chromosome of S. aureofaciens CCM3239 but on the large linear plasmid pSA3239 (Novakova et al. 2013). Auricin is produced during a very narrow growth phase interval of several hours after entry into stationary phase, after which it is degraded into inactive metabolites due to its instability at the high pH values reached later in stationary phase. Preliminary structural analysis revealed that auricin is a unique angucycline polyketide conjugated with the aminodeoxyhexose d-forosamine (Kutas et al. 2013; Kormanec et al. 2014). No other angucyclines have yet been found which contain this amino deoxyhexose, though it has been found in the macrolides spinosyn, spiramycin and dunaimycin and in the naphtoquinone antibiotic forosamilylgriseucin A (referred to in Hong et al. 2008).
The unusual pattern of auricin production likely arises from its strict, but complex regulatory mechanism, involving both feed-forward and feed-back control by auricin intermediates via several transcriptional regulators (Kutas et al. 2013; Kormanec et al. 2014). Four of these have already been characterised in detail. Aur1P, the key auricin-specific regulator from the atypical response regulator family, specifically activates expression of the core auricin biosynthetic gene cluster (aur1A-aur1U) (Fig. 1) from the aur1Ap promoter (Novakova et al. 2005). Aur1R, which belongs to the TetR family of transcriptional repressors, has been shown to repress the expression of the aur1P gene by directly binding the aur1Pp promoter (Novakova et al. 2010). The other two auricin-specific regulators, Aur1PR3 and Aur1PR4, belong to the SARP family of transcriptional activators. They are most likely involved in the regulation of putative auricin tailoring biosynthetic genes. Interestingly, they are under the differential control of Aur1R and Aur1P (Novakova et al. 2011b; Rehakova et al. 2013). In addition, at the global level, auricin production is regulated by the SagA-SagR γ-butyrolactone autoregulator-receptor system, which directly controls the expression of the auricin-specific regulatory genes aur1P and aur1R (Mingyar et al. 2015).
Genetic organisation of the auricin aur1 cluster. Thick arrows denote the direction of expression and the size of the genes. Dark-grey arrows correspond to the auricin genes, hatched arrows to the proposed d-forosamine biosynthetic and attachment genes, light-grey arrows to the putative PKS and accessory genes, black arrows to the regulatory genes and white arrows to genes of other functions. For details regarding individual genes and their products, see Table 2. Bent arrows indicate the positions and directions of those promoters which have been identified
The aim of the present study was to characterise the role of those genes within the auricin cluster which appear to encode deoxysugar biosynthetic enzymes and glycosyltransferases for the biosynthesis of d-forosamine and its attachment to the auricin aglycon. For this purpose, the genes were inactivated and auricin production was investigated in the resulting mutants. A gene expression analysis by S1 nuclease mapping was also used to characterise the expression of the genes and their dependence upon auricin-specific regulators. The results show that both SARP family regulators Aur1PR3 and Aur1PR4 directly regulate the expression of the d-forosamine biosynthetic and attachment genes.
Materials and methods
Bacterial strains, plasmids and culture conditions
The S. aureofaciens CCM3239 wild-type strain was from the Czechoslovak Collection of Microorganisms, Brno, Czech Republic. The S. aureofaciens Δaur1PR3 and Δaur1PR4 mutant strains were described in Novakova et al. (2011b) and Rehakova et al. (2013). Escherichia coli BW25113/pIJ790 was used as a host for PCR-targeted gene disruption using the apramycin resistance plasmid pIJ773 or the spectinomycin resistance plasmid pIJ778, and E. coli ET12567/pUZ8002 was used as a non-methylating host (Gust et al. 2003). E. coli BL21(DE3) pLysS and the plasmid pET28a, used for aur1PR3 and aur1PR4 overexpression, were obtained from Novagen. The growth of S. aureofaciens CCM3239 strains was carried out in rich Bennet medium (Horinouchi et al. 1983) as described in Novakova et al. (2010). For RNA isolation from liquid-grown cultures, 5 × 108 cfu of S. aureofaciens CCM3239 spores were inoculated in 50 ml Bennet medium in 250 ml Erlenmeyer flasks and grown on an orbital shaker at 270 rpm and 28 °C to different growth phases. E. coli growth and transformation conditions followed Ausubel et al. (1995).
Disruption of the S aureofaciens CCM3239 genes
The PCR-targeted REDIRECT procedure (Gust et al. 2003) was used to delete the entire coding region of the aur1TQSV, sa46, sa52, sa53 and sa59 genes in S. aureofaciens CCM3239. For the aur1TQSV region, the apramycin resistance cassette from the template plasmid pIJ773 was PCR amplified using primers Aur1TdDir and Aur1VdRev (Table 1). For the sa46, sa52, sa53 and sa59 genes, the same plasmid with primers Sa46dDir and Sa46dRev, Sa52dDir and Sa52dRev, Sa53dDir and Sa53dRev and Sa59dDir and Sa59dRev, respectively (Table 1) was used. The resulting PCR products were used to electroporate E. coli BW25113/pIJ790 containing cosmid pCosSA31 (Novakova et al. 2013) for sa46, sa52, sa53 and sa59 deletion, or cosmid pCosSA19 (Rehakova et al. 2013) for aur1TQSV deletion. The correct replacement of all genes in the resulting cosmids pCosSA19-aur1TQSV::AmR, pCosSA31-sa46::AmR, pCosSA31-sa52::AmR, pCosSA31-sa53::AmR and pCosSA31-sa59::AmR was verified by restriction mapping. The recombinant cosmids were transformed into the non-methylating E. coli ET12567/pUZ8002 strain and introduced into S. aureofaciens CCM3239 by conjugation; mutants resistant to apramycin and sensitive to kanamycin were selected. Four apramycin-resistant and kanamycin-sensitive colonies from each conjugation were selected, and the correct double-cross event was confirmed by Southern blot hybridization. All four clones from each disruption had similar phenotypes. One representative strain from each disruption, S. aureofaciens Δaur1TQSV, S. aureofaciens Δsa46, S. aureofaciens Δsa52, S. aureofaciens Δsa53 and S. aureofaciens Δsa59, was chosen for further study (Fig. S1).
The same strategy was used to prepare the S. aureofaciens Δsa46Δsa53 double mutant strain. The spectinomycin resistance cassette from the template plasmid pIJ778 was PCR amplified using the primers Sa46dDir and Sa46dRev (Table 1). The resulting PCR product was used to electroporate E. coli BW25113/pIJ790 containing cosmid pCosSA31. The correct replacement of the sa46 gene in the resulting cosmid pCosSA31-sa46::SpR was verified by restriction mapping. The recombinant cosmid was transformed into the non-methylating E. coli ET12567/pUZ8002 strain and introduced into S. aureofaciens Δsa53 by conjugation, and mutants resistant to spectinomycin and sensitive to kanamycin were selected. Four spectimycin-resistant and kanamycin-sensitive colonies were selected, and the correct double-cross event was confirmed by Southern blot hybridization. All four clones from each disruption had similar phenotypes. One representative strain, S. aureofaciens Δsa46 Δsa53, was chosen for further studies (Fig. S1).
RNA isolation and S1 nuclease mapping
Isolation of total RNA from S. aureofaciens CCM3239 and high-resolution S1 nuclease mapping were performed according to Kormanec (2001). Forty-microgram samples of RNA were hybridised to approximately 0.015 pmol of a suitable DNA probe labelled at one 5′ end with [γ-32P] ATP (producing approximately 106 dpm pmol−1 of probe) and treated with 120 U of S1 nuclease. The S1 probes were prepared by PCR amplification using chromosomal DNA from S. aureofaciens CCM3239 as a template and the following primers (Table 1): probe 1 (for the sa14p promoter) with the 5′ end-labelled primer Sa14S1Rev from the sa14 coding region and the unlabelled primer Sa14S1Dir from the sa14 upstream region, probe 2 (for the sa46p promoter) with the 5′ end-labelled primer Sa46S1Rev from the sa46 coding region and the unlabelled primer Sa46S1Dir from the sa46 upstream region, probe 3 (for the sa52p promoter) with the 5′ end-labelled primer Sa52S1Rev from the sa52 coding region and the unlabelled primer Sa52S1Dir from the sa52 upstream region and probe 4 (for the sa53p promoter) with the 5′ end-labelled primer Sa52S1Dir and the unlabelled primer Sa52S1Rev. The control hrdBp2 promoter probe was described in Kormanec and Farkasovsky (1993). The primers were labelled at the 5′ end with [γ-32P] ATP (ICN, 4500 Ci mmol−1) and T4 polynucleotide kinase as described in Ausubel et al. (1995). The protected DNA fragments were analysed on DNA sequencing gels together with G + A and T + C sequencing ladders derived from the end-labelled fragments (Maxam and Gilbert 1980).
Overexpression of the aur1PR3 and aur1PR4 genes in E. coli and protein purification
The aur1PR3 and aur1PR4 genes were amplified by PCR using S. aureofaciens CCM3239 chromosomal DNA as a template and the primer pairs Ar1PR3Nde–Aur1PR3Hind and Aur1PR4Nde–Aur1PR4Hind, respectively (Table 1). These introduced an NdeI site overlapping the translation initiation codon and a HindIII site downstream of the stop codon of both genes. The resulting PCR DNA fragments were digested with NdeI and HindIII and cloned into a pET28a vector cut with the same enzymes, resulting in pET-aur1PR3 and pET-aur1PR4, respectively. Both constructs were verified by nucleotide sequencing. An E. coli BL21 (DE3) pLysS host strain was transformed with these plasmids and then grown in LB medium containing 30 μg chloramphenicol ml−1 and 40 μg kanamycin ml−1 at 30 °C to an OD600 of 0.5. Expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and the cells were harvested after 3 h by centrifugation at 12,000×g for 10 min and washed with ice-cold 0.9 % (w/v) NaCl. Cell lysis and native purification of His-tagged SagR on His-Tag Bind resin (Novagen) were carried out as directed by the manufacturer. The eluted proteins were dialysed overnight at 4 °C against a storage buffer (12.5 mM Tris-HCl pH 7.9, 60 mM KCl, 1 mM EDTA, 1 mM DTT, 50 % (v/v) glycerol), cleared by centrifugation at 30,000×g for 10 min and stored at −20 °C. The overproduction and purity of the eluted proteins were confirmed by SDS-PAGE (Laemmli 1970). A prominent, 32-kDa band was identified after IPTG induction in both cases and isolated using His-Tag Bind resin (Fig. S2). This mass corresponds to the expected M r of either His-tagged Aur1PR3 (31,999) or Aur1PR4 (33,315). Protein concentration was determined according to Bradford (1976) with BSA as a standard.
Electrophoretic mobility shift assay
A 410-bp 32P-labelled DNA fragment comprising the sa13-sa14 intergenic region (nucleotide positions −303 to +106 relative to the sa13p promoter transcription start point) and a 457-bp fragment containing the sa52-sa53 intergenic region (nucleotide positions −335 to +121 with respect to the sa52p promoter transcription start point) were prepared by PCR amplification as described above using the 5′ end-labelled primer Sa14S1Rev and unlabelled primer Sa14S1Dir and the 5′ end-labelled primer Sa52S1Rev and unlabelled primer Sa52S1Dir (Table 1), respectively. Electrophoretic mobility shift assay (EMSA) with the purified His-tagged Aur1PR3 and Aur1PR4 was performed as described in Novakova et al. (2010). In brief, the 32P-labelled DNA fragments (0.1 ng, approx. 3000 dpm) were incubated with increasing amounts of purified protein for 15 min at 30 °C, and protein-bound and free DNA were resolved on a non-denaturing polyacrylamide gel.
DNase I footprinting
Binding reactions were performed in 30 μl of binding buffer in the same conditions as for EMSA with 2 ng (approx. 25,000 dpm) of 5′ end-labelled DNA fragments and increasing amounts of purified His-tagged Aur1PR3 and Aur1PR4. After incubation for 15 min at 30 °C, the mixture was incubated with DNase I and additionally treated as described in Novakova et al. (2010). The DNA fragments were analysed on 6 % DNA sequencing denaturing gels together with G + A and T + C sequencing ladders derived from the end-labelled fragment (Maxam and Gilbert 1980).
Analysis of auricin production
Spores (5 × 108 cfu) from wild-type and mutant S. aureofaciens CCM3239 strains were inoculated into 50 ml Bennet medium in 250 ml Erlenmeyer flasks, and the cultures were grown on an orbital shaker at 270 rpm and 28 °C. Four-milliliter aliquots were taken from the cultures at various time points, and auricin was extracted by ethyl acetate and analysed by TLC and HPLC as described previously (Kutas et al. 2013). In brief, extracted samples of auricin were dissolved in 100 μl of 96 % (v/v) ethanol and 10 μl aliquots were subjected to TLC analysis on silica gel 60 F254 plates (Merck) with n-butanol saturated with water. Dried TLC plates were analysed by biochromatography with soft nutrient agar (Kieser et al. 2000) containing a fresh Bacillus subtilis culture. The plates were incubated for 16 h at 37 °C and visually screened for growth inhibition zones. HPLC analysis of 10 μl samples was performed on an OmmiSpher 5 C18 column (5 μm, 250 × 4.6 mm, Varian, Lake Forest, CA) using a Shimadzu LC-10A HPLC system with a SPD-20A UV-VIS detector (Shimadzu, Japan) with acetonitrile and 0.5 % (v/v) acetic acid in water as a mobile phase and detection at 254 nm. Elution was carried out with a linear gradient of acetonitrile from 20 to 100 % for 30 min at a flow rate of 1 ml/min. Under these conditions, auricin exhibited a retention time of 8.7 min. Auricin production was confirmed by electrospray ionisation mass spectra (ESI MS) analysis as a molecular ion [M+H] of m/z = 542.2 with detection in the positive mode (Kutas et al. 2013).
Electrospray ionisation mass spectra analysis
Individual peaks after HPLC runs were taken and subjected to ESI mass spectra analysis in positive and negative ionisation. The ESI spectra were taken using a Velos Pro Hybrid Ion Trap-Orbitrap Mass Spectrophotometer (Thermo Scientific). The ions of interest were subjected to MS/MS fragmentation.
Results
Characterisation of d-forosamine genes in the plasmid pSA3239
Preliminary structural analyses of auricin revealed that a single aminodeoxysugar, d-forosamine, is attached to its aglycon moiety through an O-glycosylation (Kormanec et al. 2014; J. Kormanec, unpublished results). High-resolution ESI MS/MS fragmentation of the auricin ion (m/z = 542.20187 [M+H]+) produced only a single dominant signal at m/z = 142.12259 [M+H]+ (Fig. S3), which corresponds to d-forosamine (Lewer et al. 2009). Sequence analysis of the aur1 gene cluster downstream region (Kormanec et al. 2014) revealed four genes, aur1TQSV, which encode proteins very similar to enzymes involved in the initial steps of deoxysugar biosynthesis (Fig. 1, Table 2) (Salas and Méndez 2005). Surprisingly, no aminotransferase or glycosyltransferase genes have been found in regions close to the auricin cluster; however, genes encoding homologs of these enzymes, sa46, sa52, sa53 and sa59, have been found in a region about 40 kb from the core auricin genes (Fig. 1). The sa59 gene encodes a protein with high similarity to NDP-hexose aminotransferases: its highest similarity to Srm8 from the Streptomyces ambofaciens spiramycin gene cluster (77 % identity and 86 % similarity) and to SpnR from the Saccharopolyspora spinosa spinosyn gene cluster (64 % identity and 78 % similarity). Both these genes are involved in the final steps of d-forosamine biosynthesis in their respective pathways (Karray et al. 2007; Hong et al. 2008). The sa52 gene encodes a protein with high similarity to NDP-aminohexose N-dimethyltransferase, and like sa59, its highest similarity was to dimethyltransferases from the d-forosamine biosynthetic pathways of spiramycin (Srm9c) and spinosyn (SpnS). The two final genes, sa46 and sa53, encode proteins with high similarity to glycosyltransferases (Table 2).
Disruption of d-forosamine biosynthetic genes affects auricin production
To assess the role of these genes in auricin biosynthesis, their chromosomal copies were inactivated using a PCR targeting system for disrupting Streptomyces genes (Gust et al. 2003). None of these mutations affected growth and differentiation (data not shown). The auricin production of these mutants was assessed as described previously (Novakova et al. 2010). Ethyl acetate extracts from the wild-type and disrupted strains were prepared from liquid-grown strains cultivated in Bennet medium and analysed by TLC followed by a bioassay against B. subtilis. In parallel, the level of auricin production was determined by HPLC of these ethyl acetate extracts. The deletion of all four aur1TQSV genes abolished auricin production. No auricin inhibition zone (at Rf = 0.13) was detected by TLC biochromatography. After analysing larger amounts of these extracts, however, several inhibition zones were found which had higher Rf values. These additional zones could also be found when larger amounts of wild-type S. aureofaciens CCM3239 extract were analysed, though they could not be found in extracts from the S. aureofaciens, aur1::tsr strain which lacks the crucial polyketide synthase genes (Novakova et al. 2002) and which was grown under identical conditions (data not shown). This suggests that they arise from auricin intermediates, which are also active against the tested B. subtilis strain, and which share auricin’s instability. A similar phenotype was observed using extracts from the S. aureofaciens Δsa59 mutant (the putative aminotransferase, Fig. 2b), but extracts from the S. aureofaciens Δsa52 mutant (the putative NDP-aminohexose N-dimethyltransferase) still showed a weak inhibition zone with an Rf value close to that of auricin and inhibition zones for the proposed auricin intermediates (Fig. 2b).
a Growth curve of the S. aureofaciens CCM3239 wild-type strain in liquid Bennet medium. The time points used for the TLC analysis of auricin production and transcriptional analysis are indicated by arrows. b Analysis of auricin production in the wild-type and three mutants by biochromatography. Ethyl acetate extracts from each strain grown in Bennet medium to the time points indicated were resolved by TLC and overlaid with B. subtilis as described in the “Materials and methods”. The arrows indicate the inhibition zone corresponding to auricin (Kutas et al. 2013) and deglycosylated auricin intermediates. c HPLC analysis of auricin production by the S. aureofaciens CCM3239 wild-type (WT) and aur1TQSV-, sa59- and sa52-disrupted strains (Δaur1TQSV, Δsa59, Δsa52) grown for 14 h in Bennet medium (details described in the “Materials and methods”). The arrows indicate the positions of peaks for auricin and deglycosylated auricin intermediates
HPLC analysis of the ethyl acetate extracts confirmed that neither the aur1TQSV nor the sa59 mutant produced auricin and that the sa52 mutant produced a small peak with a retention time (Rt) near that of auricin (Fig. 2c). This peak was isolated and high-resolution ESI MS analysis revealed a molecular ion [M+H]+ at m/z 514.1928, which fragmented to a single dominant signal of m/z = 114.098 [M+H]+ (Fig. S4). This mass exactly corresponded to demethylated d-forosamine, thus clearly confirming that sa52 encodes a forosamine N-dimethyltransferase.
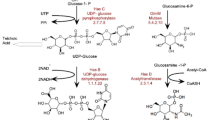
Following HPLC, all peaks were isolated from the wild-type and S. aureofaciens Δaur1TQSV and Δsa59 extracts and investigated for their biological activity using TLC followed by a bioassay against B. subtilis. Two peaks with retention times of 12.5 and 14 min, present in all three strains (Fig. 2c), were active against B. subtilis, giving Rf values similar to the auricin intermediates. High-resolution ESI MS analysis gave molecular ions [M−H]− at m/z 401.09021 and m/z 399.07442, respectively. The corresponding masses of these compounds are close to those of the auricin aglycon without d-forosamine. The sa59 mutant should synthesise NDP-4-keto-2,3,6-trideoxy-d-glucose (Fig. 3) and produce 4-keto-2,3,6-trideoxy-d-glucose-attached auricin derivative. However, no such ion has been identified and MS/MS fragmentation of the ions at m/z 401.09021 and m/z 399.07442 resulted in fragmented ions which do not correspond to the loss of 4-keto-2,3,6-trideoxy-d-glucose (Fig. S5a, b). Therefore, based on these results, it is likely that NDP-4-keto-2,3,6-trideoxy-d-glucose is not recognised by GT and this sugar intermediate is not attached to the auricin aglycon. All these results suggest that these genes have a role in the biosynthesis of d-forosamine. Based on a bioinformatics analysis, a d-forosamine biosynthetic pathway in S. aureofaciens CCM3239 can be proposed (Fig. 3).
The biosynthesis of d-forosamine in the S. aureofaciens CCM3239 auricin biosynthetic pathway. The homologous enzymes from the spinosyn cluster in Saccharopolyspora spinosa (Hong et al. 2008), the spiramycin cluster in S. ambofaciens (Karray et al. 2007), the landomycin cluster in S. cyanogenus (Westrich et al. 1999) and the urdamycin cluster in S. fradiae (Hoffmeister et al. 2000) are given in brackets under each S. aureofaciens inferred protein product
Two genes encoding putative GTs are involved in the attachment of a d-forosamine to auricin aglycon
A bioinformatics analysis of the region around the core of the auricin gene cluster revealed two genes, sa46 and sa53, encoding proteins similar to GTs (Fig. 1, Table 2). In order to assess their role in auricin biosynthesis, both genes were separately disrupted by the PCR targeting system. These mutations did not affect growth and differentiation. Interestingly, deletion of both genes only partially affected auricin production. The inhibition zones corresponding to auricin in both mutants were smaller than for the wild-type S. aureofaciens CCM3239 strain (Fig. 4a). The quantities of auricin produced were determined by HPLC of the ethyl acetate extracts; in both cases, the deletion caused a 50 % decrease in auricin production relative to the wild-type strain (Fig. 4b). The peaks corresponding to auricin from both mutants were isolated and confirmed by ESI MS. A double S. aureofaciens Δsa46Δsa53 mutant was prepared using the PCR targeting system, and its auricin production was similarly investigated. No auricin was produced by this strain at any time point during cultivation in liquid Bennet medium, and, as for the previously deleted d-forosamine biosynthetic genes, only the inhibition zones of the putative auricin intermediates were seen (Fig. 4a). This suggests, intriguingly, that both the GT sa46 and sa53 genes are in some way involved in the attachment of d-forosamine to the auricin aglycon and that both of them are needed for a sufficient level of GT activity to be observed. Both putative GTs are highly similar to each other with 64 % identity and 77 % similarity (Fig. S6) and so they may therefore be interchangeable in the d-forosamine attachment reaction.
a Analysis of auricin production in the wild-type S. aureofaciens CCM3239 strain (WT) and three mutants by biochromatography. Ethyl acetate extracts from each strain grown in Bennet medium to the time points indicated were resolved by TLC and overlain with B. subtilis as described in the “Materials and methods”. The arrows indicate the inhibition zone corresponding to auricin (Kutas et al. 2013) and deglycosylated auricin intermediates. b HPLC analysis of auricin production by the S. aureofaciens CCM3239 wild-type (WT), sa46- and sa53-disrupted strains (Δsa46, Δsa53) and the Δsa46Δsa53 double mutant grown for 14 h in Bennet medium (details described in the “Materials and methods”). The arrow indicates the position of the peak for auricin
Transcriptional analysis of the d-forosamine genes
The expression of all identified d-forosamine biosynthetic and attachment genes was investigated by S1 nuclease mapping using RNA isolated from the wild-type S. aureofaciens CCM3239 strain at various stages of growth in liquid Bennet medium. Some genes are located in operons (Fig. 1), and therefore, only the first gene of this operon was investigated for the presence of a promoter. Because heptameric direct repeats, similar to the consensus sequence of typical SARP-binding sites (5′-TCGAGXX-3′), could be found at the correct location for SARP regulation, 8 bp upstream of the −10 region (Fig. 5), we also carried out S1 nuclease mapping using RNA from two mutant strains with deletions in SARP family regulator genes which had previously been shown to affect auricin biosynthesis, S. aureofaciens Δaur1PR3 and S. aureofaciens Δaur1PR4 (Novakova et al. 2011b; Rehakova et al. 2013). A single RNA-protected fragment was identified for each gene which corresponded to the indicated promoter (Fig. 6). No RNA-protected fragment was identified using a tRNA control (Fig. 6, lane C). As an internal check of RNA quality, S1 nuclease mappings were also performed using the same RNA samples but with a probe fragment specific for the hrdBp2 promoter, which is expressed fairly constantly during growth and differentiation (Kormanec and Farkasovsky 1993). RNA-protected fragments of similar intensities corresponding to the hrdBp2 promoter were identified in all RNA samples. In the wild-type S. aureofaciens CCM3239 strain, all promoters were inactive during exponential phase (9 h) and were induced at the beginning of stationary phase (12 h), correlating with the onset of auricin production. The level of messenger RNA (mRNA) then decreased during late stationary phase for all promoters (16 h). The level of mRNA for some promoters was partially reduced in both Δaur1PR3 and Δaur1PR4 mutants (sa14p), or more in the Δaur1PR3 mutant (sa52p, sa53p). As already mentioned, the upstream region of all promoters contained two or three heptameric direct repeats similar to the consensus sequence of typical SARP-binding sites with 4- or 15-bp spacers, located exactly 8 bp upstream of the −10 region of promoters (Fig. 5) (Arias et al. 1999; Sheldon et al. 2002; Tanaka et al. 2007; Wietzorreck and Bibb 1997). These results indicate that the promoters are partially, and in some cases also differentially, dependent on both of the SARP family auricin-specific regulatory genes, aur1PR3 and aur1PR4.
Comparison of the heptameric direct repeats (black highlighted) of the promoters governing expression of the d-forosamine genes identified in the auricin cluster or its flanking regions to the previously identified sa13p promoter, which is dependent upon the SARP family auricin-specific regulators Aur1PR3 and Aur1PR4 (Rehakova et al. 2013), and to the binding sites of the ActII-ORF4 SARP regulator in the actVI-ORF1p promoter (Arias et al. 1999). The putative −10 box and transcription start points are underlined
High-resolution S1 nuclease mapping of the transcription start points for the sa14p, sa46p, sa52p and sa53p promoters in the S. aureofaciens CCM3239 wild-type (wt), S. aureofaciens aur1PR3-disrupted (Δaur1PR3) and S. aureofaciens aur1PR4-disrupted (Δaur1PR4) strains. The 5′-labelled DNA fragments were hybridised with RNA isolated from cultures grown in liquid Bennet medium to the time points indicated (these corresponded to different growth phases). E. coli tRNA was used as a control (lane C). The control S1 nuclease mapping experiment (bottom panel) used the same RNA samples and a DNA probe for the hrdBp2 promoter (Kormanec and Farkasovsky 1993). The RNA-protected DNA fragments were analysed on DNA sequencing gels together with G + A (lane A) and T + C (lane T) sequencing ladders derived from the end-labelled fragments (Maxam and Gilbert 1980). Thin horizontal arrows indicate the positions of RNA-protected fragments, and thick bent vertical arrows indicate the nucleotide corresponding to the transcription start point. Before assigning the transcription start point, 1.5 nucleotides were subtracted from the length of the protected fragment to account for the difference in the 3′ ends resulting from S1 nuclease digestion and chemical sequencing reactions. In the control experiment, the same RNA preparations were hybridised in parallel with all of the probes. All S1 nuclease mapping experiments were performed twice with independent sets of RNA, giving similar results
Both auricin-specific SARP regulators, Aur1PR3 and Aur1PR4, bind the d-forosamine promoters
The transcriptional analysis indicated that the d-forosamine promoters are partially dependent upon both the aur1PR3 and aur1PR4 genes. To investigate whether this dependence is direct, both genes were overexpressed in the E. coli T7 RNA polymerase expression system, and the ability of purified N-terminally His-tagged Aur1PR3 and Aur1PR4 to bind the sa13p, sa14p, sa52p and sa53p promoter regions was tested by EMSA using a 410-bp fragment containing the sa13-sa14 intergenic region (nucleotide positions −303 to +106 with respect to the sa13p promoter transcription start point) and a 457-bp fragment containing the sa52-sa53 intergenic region (nucleotide positions −335 to +121 with respect to the sa52p promoter transcription start point). A single retarded band was clearly visible for each promoter fragment with increasing concentration of both Aur1PR3 and Aur1PR4 (Fig. 7). The specificity of the interaction was demonstrated by the competitive binding of the unlabelled fragment (Fig. 7, lane 5). These results indicated that both Aur1PR3 and Aur1PR4 are capable of binding to the sa13-sa14 and the sa52-sa53 intergenic regions.
Detection of Aur1PR3 and Aur1PR4-DNA complexed by EMSA using 5′ end-labelled DNA fragments comprising the sa13–sa14 and the sa52–sa53 intergenic regions with increasing amounts of purified His-tagged Aur1PR3 and Aur1PR4. Lane 1, a labelled DNA fragment in the absence of protein; lanes 2 to 5, 1, 3, 5 and 5 μg, respectively, of the purified His-tagged protein indicated. Addition of 200 ng of the unlabelled corresponding DNA fragment was used to demonstrate Aur1PR3 and Aur1PR4 binding specificity (lane 5). The arrows indicate the free DNA fragment and the shifted fragment corresponding to the proposed complex. All binding experiments were performed twice with independent sets of protein samples, giving similar results
To map the Aur1PR3 and Aur1PR4 binding sites within these regions, a DNase I footprinting assay was performed using the same 32P-labelled sa13-sa14 and sa52-sa53 intergenic DNA fragments. Two clearly separated regions covering the sa13p, sa14p, sa52p and sa53p promoters were protected in each DNA fragment. In each case, the protected regions contained the heptameric direct repeats which are similar to the consensus sequence of the SARP-binding sites (Fig. 8). The protected DNA fragments were identical for both Aur1PR3 and Aur1PR4 proteins. These results indicate that both these auricin-specific SARP family regulators bind to the promoters in identical regions covering the SARP-binding heptameric direct repeats to directly activate the expression of the directed genes.
a DNaseI footprinting of Aur1PR3 and Aur1PR4 binding to the 5′ end-labelled DNA fragments comprising the sa13-sa14 (left) and the sa52-sa53 (right) intergenic regions. The vertical bar indicates the positions of the binding sites. The numbering is relative to the transcription start point of the given promoter. Lane 1 is without Aur1PR3 or Aur1PR4. Lanes 2 and 3 contain 30 and 60 μg, respectively, of purified His-tagged Aur1PR3 and Aur1PR4. Lanes A and T contain G + A and C + T sequencing ladders (Maxam and Gilbert 1980). All binding experiments were performed twice with independent sets of protein samples, giving similar results. b The nucleotide sequence of the S. aureofaciens CCM3239 sa13–sa14 and sa52–sa53 intergenic regions. The transcription start points of the promoters are indicated by bent arrows. The putative −10 box of the promoters is in bold and underlined. The heptameric direct repeats similar to the consensus SARP-binding sites are in bold only. The nucleotides that were protected from DNase I by Aur1PR3 or Aur1PR4 binding are shaded. The nucleotide sequence shown is part of the aur1 cluster that can be found in the GenBank/EMBL/DDBJ databases under accession number KJ396772
Discussion
Sugar moieties of glycosylated antibiotics are frequently crucial for their biological activities. Preliminary structural characterisation of auricin revealed that it is a unique angucycline antibiotic with a single aminodeoxysugar, d-forosamine (Kormanec et al. 2014; Fig. S1). The present work was therefore aimed at characterising the genes encoding biosynthetic enzymes for this aminodeoxysugar and its attachment to the auricin aglycon. Although several antibiotics have been found which contain this aminodeoxysugar (referred in Hong et al. 2008), only two antibiotic gene clusters containing d-forosamine biosynthetic genes, S. ambofaciens spiramycin (Karray et al. 2007) and Saccharoplyspora spinosa spinosyn (Waldron et al. 2001), have been characterised to date, though it should be noted that the biosynthesis of TDP-d-forosamine in the spinosyn pathway strain has been fully elucidated in vitro (Hong et al. 2008).
Analysis of the region downstream from the auricin core biosynthetic genes identified four close tandem putative deoxysugar genes, aur1TQSV (Fig. 1, Table 2), which encode proteins with high similarity to those enzymes involved in the initial steps of the biosynthesis of the intermediate NDP-4-keto-2,3,6,-trideoxy-d-glucose (Fig. 3) (Salas and Méndez 2005). Deletion of these genes resulted in the absence of auricin and the production of deglycosylated auricin intermediates, which still had biological activity against a B. subtilis test strain. These genes must therefore have a role in d-forosamine biosynthesis. Sequence comparison of the inferred protein products of aur1TQSV, with those of known proteins, especially those for d-forosamine biosynthesis from the spinosyn and spiramycin gene clusters, allowed us to identify a potential enzyme for each of the steps of the proposed pathway shown in Fig. 3 (see Table 2 for the list). Interestingly, the inferred protein products of all four aur1TQSV genes are most similar to their counterparts from the angucycline landomycin and urdamycin gene clusters (Fig. 3, Table 2). Both antibiotics contain the deoxysugar l-rhodinose, and the first five steps in the biosynthesis of this deoxysugar are identical to the initial steps of d-forosamine biosynthesis (Hoffmeister et al. 2000; Westrich et al. 1999). The identities and similarities of Aur1TQSV to the landomycin enzymes are the following: LanH 80/88 %, LanQ 85/93 %, LanS 74/83 % and LanT 69/76 %; to the urdamycin enzymes: UrdH 74/85 %, UrdQ 83/91 %, UrdS 69/78 % and UrdT 59/67 %. Identities/similarities of Aur1TQSV to their counterparts from the d-forosamine gene clusters for spinosyn and spiramycin are significantly lower; for spinosyn: Gdh 64/76 %, SpnQ 68/81 %, SpnO 52/63 % and SpnN 53/63 %; and for spiramycin: Srm5 66/77 %, Srm12 72/83 %, Srm7 49/59 % and Srm15c 54/62 %. On the other hand, the two final d-forosamine biosynthetic genes, sa59 and sa52, encoding respectively an NDP-hexose aminotransferase and an NDP-aminohexose N-dimethyltransferase (Table 2, Fig. 3), are most similar to the genes encoding the enzymes for the corresponding final steps in the d-forosamine biosynthesis of spiramycin and spinosyn, which are macrolides rather than angucyclines (Karray et al. 2007; Hong et al. 2008). They are, moreover, located in a region rather distant from the auricin core gene cluster (Fig. 1). Their unusual location, together with their similarity to macrolide rather than angucycline biosynthesis genes, suggests that sa59 and sa52 have a different evolutionary origin from the other genes in the auricin biosynthesis cluster. While the genes for the initial steps of d-forosamine biosynthesis (aur1TQSV) are homologous to those of the deoxysugars of other angucyclines, even those with different deoxysugars (e.g. landomycin and urdamycin), these final two genes were probably transferred to the S. aureofaciens CCM3239 linear plasmid pSA3239 from different d-forosamine biosynthetic clusters by horizontal gene transfer.
Normally, deoxyhexoses are synthesised from glucose-1-phosphate by way of the intermediate 4-keto-6-deoxy-d-glucose. The first step, activation of glucose by addition of NDP, is catalysed by an NDP-glucose synthase. In several antibiotic clusters, the gene for this enzyme is missing and NDP-glucose is acquired from the primary metabolism (Salas and Méndez 2005). This is the case for the biosynthesis of d-forosamine in the Saccharopolyspora spinosa spinosyn antibiotic cluster where the NDP-d-glucose synthase Gtt actually belongs to the primary metabolism and is also important for cell wall biosynthesis (Hong et al. 2008). In the present case, there are two genes, sa14 and sa50, in the vicinity of the aur1 cluster whose likely protein products are similar to NDP-d-glucose synthase (Table 2, Fig. 1), especially to the Saccharoplyspora spinosa gtt gene (Hong et al. 2008). They are also highly similar to each other (86 % identity and 92 % similarity), suggesting that they arose from a single ancestor. This suggests that either of these two proteins may be used for the initial glucose activation, and, in fact, disruption of one of them, sa14, has no effect on auricin biosynthesis (data not shown).
Like the case of NDP-glucose synthase, there are also two genes, sa46 and sa53, encoding glycosyltransferase homologs (Table 2, Fig. 1), even though auricin is modified by only a single deoxyhexose. Phenotypic analysis of mutants of both genes indicated that both of them participate at least partially in the attachment of d-forosamine to auricin aglycon. Deletion of either gene alone caused only a partial decrease in auricin production, and only deletion of both genes resulted in no auricin production. The inferred protein products of these genes, Sa46 and Sa53, are highly similar to each other, with 64 % identity and 77 % similarity (Fig. S2); comparing them to protein sequence databases showed that all other GTs of known function had significantly lower homology (about 30–40 % identity and 40–50 % similarity). The high similarity between Sa46 and Sa53 suggests that either of them may be used to attach d-forosamine to auricin aglycon. Therefore, an explanation of the observed phenotype might be that both enzymes are required for efficient attachment of d-forosamine to the auricin aglycon. This behaviour is uncommon for the glycosylation reaction. Normally, only one GT is found in a biosynthetic cluster for each attached sugar (e.g. for jadomycin, urdamycin). Moreover, in several cases (e.g. landomycin A and saquayamycin Z), the number of GT genes is actually lower than the number of sugars being attached, and some GTs have been described which catalyse the attachment of two sugars to different positions on the chain (Luzhetskyy et al. 2005). On the other hand, similar redundant glycosyltransferase activity has also been previously observed in the biosynthesis of the macrolide antibiotic spiramycin in S. ambofaciens. This molecule has three deoxysugar moieties (including d-forosamine), but an analysis of the spiramycin gene cluster found four genes (srm5, srm29, srm30 and srm38; srm29 is involved in d-forosamine attachment) encoding putative GTs, only three of which are involved in spiramycin biosynthesis. The inactivation of srm30 had no effect on spiramycin biosynthesis, and the role of this gene remains elusive (Nguyen et al. 2010).
Typically, GTs attach NDP-activated sugars, normally TDP-conjugated ones, to acceptor substrates. They are classified into more than 90 families, of which family 1 (GT1) is the most common glycosylator of secondary metabolites (Coutinho et al. 2003). Despite their significant sequence diversity, GTs show great structural similarity. They fall into two structural superfamilies termed GT-A and GT-B. Most of the GTs involved in secondary metabolite biosynthesis belong to the GT-B family, which is characterised by two similar Rossmann-like domains, an N-terminal domain which provides the acceptor-binding site and a C-terminal domain which provides the donor-binding site. These two domains are connected by a linker region and the active site is located between them. A phylogenetic analysis of GTs showed that GTs from strains producing similar compounds tend to be related (Luzhetskyy et al. 2005); consequently, Sa46 and Sa53 were compared with several GTs which modify angucycline antibiotics and with two GTs which are known to transfer d-forosamine to macrolide antibiotics (Fig. S2). (The crystal structure of one of these SpnP has recently been published (Isiorho et al. 2014)). This comparison showed that, like other GTs involved in the glycosylation of secondary metabolites, both Sa46 and Sa53 belong to the GT1 family and likely have a GT-B fold. Significant similarities were found in both the N-terminal acceptor-binding sites and the C-terminal donor-binding sites in all aligned GTs, but no residues specific for the angucycline acceptor or the d-forosamine donor could be identified (Fig. S2). SpnP was found to possess a three-helix motif (H-X-R-X5-D-X5-R-X12–20-D-P-X3-W-L-X12–18-E-X4-G) predictive of GTs which require an auxiliary protein (Isiorho et al. 2014). This motif is not present in either Sa46 or Sa53 but was found in Srm29, which does require an auxiliary protein (Nguyen et al. 2010). It therefore seems likely that neither Sa46 nor Sa53 requires an auxiliary protein. Moreover, there is no homolog gene encoding an auxiliary protein in the vicinity of the auricin cluster.
An analysis of the expression of the d-forosamine biosynthetic and attachment genes revealed interesting features. The four initial d-forosamine biosynthetic genes, aur1TQSV, are located on the large operon aur1A-aur1U together with the core auricin biosynthetic genes (Fig. 1), whose transcription is controlled by the aur1Ap promoter (Novakova et al. 2005, 2011a). This promoter is under the direct control of the auricin-specific activator Aur1P (Novakova et al. 2005). In contrast, the other d-forosamine specific genes are located in regions rather distant from the core auricin biosynthetic genes (Fig. 1). They appear to be grouped into two putative operons each of whose expression is governed by a single promoter which is activated at the beginning of stationary phase and which is differentially dependent upon two SARP family auricin-specific activators, Aur1PR3 (Novakova et al. 2011b) and Aur1PR4 (Rehakova et al. 2013). The sa46p promoter likely directs the expression of an operon containing the sa46, sa47, sa48, sa49 and sa50 genes (Fig. 1); of these, sa50 appears to be a gene for the first d-forosamine biosynthetic enzyme, NDP-d-glucose synthase (Table 2). Similarly, based on its location and close proximity (Fig. 1), the sa53p promoter may also direct a putative operon containing the genes sa53, sa54, sa55, sa56, sa57, sa58 and sa59; the last of these encodes an NDP-hexose aminotransferase. The remaining genes in this putative operon, interestingly enough, encode homologs of primary metabolism proteins for methionine and S-adenosylmethionine biosynthesis (Table 2). We have also previously identified the sa13p promoter, which directs the expression of a putative operon located in a different part of the plasmid, containing the four putative auricin tailoring genes sa13, sa12, sa11 and sa10 (Fig. 1). This promoter is also differentially dependent upon the same two SARP family regulators (Rehakova et al. 2013). Finally, in the present work, we also identified the divergently located promoter sa14p, which directs the sa14 gene encoding an additional putative NDP-glucose synthase, and which is also at least partially dependent upon Aur1PR3 and Aur1PR4. All of these promoters contain heptameric direct repeats similar to the consensus sequence of typical SARP-binding sites which are located in the correct places (Fig. 5) (Arias et al. 1999; Sheldon et al. 2002; Tanaka et al. 2007; Wietzorreck and Bibb 1997). Furthermore, binding studies with four selected promoters confirmed that there is a direct interaction between both SARP family regulators and these regions, indicating that these genes are most likely direct targets of both Aur1PR3 and Aur1PR4. These results show that the mechanism which regulates auricin production has at least two levels of auricin-specific regulation. The biosynthetic core enzymes, together with the d-forosamine initial biosynthetic genes aur1TQSV, are governed by the auricin-specific activator Aur1P, which is under the negative control of the TetR-family negative regulator Aur1R (Kormanec et al. 2014). The auricin tailoring enzymes, on the other hand, are directed by two SARP family regulators that are under the control of both Aur1P and Aur1R (Kormanec et al. 2014). The key question is whether these SARP regulators function cooperatively or independently in the activation of these genes. This question is currently under investigation.
This abundance of SARP family regulators is fairly unusual in the activation of antibiotic clusters, which are normally activated by a single regulatory gene whose promoter is regulated by a complex signal transduction pathway that includes many global regulators; the best studied example is probably the SARP, ActII-ORF4 (Liu et al. 2013; van Wezel and McDowall 2011). However, there are an increasing number of cases in which several pathway-specific activators are involved in the regulation of antibiotic clusters. For example, two SARP family regulators, TylS and TylT, and three additional regulators, TylR, TylU and TylQ, are involved in the regulation of the Streptomyces fradiae tylosin cluster, though only TylS and TylR are essential for tylosin production (Cundliffe 2008). Likewise, the kinamycin alp cluster of S. ambofaciens contains five regulatory genes, including three SARP family regulators, but only one, AlpV, has been found to be involved in kinamycin regulation (Bunet et al. 2011). Two SARP family regulators, SrrY and SrrZ, are also involved in the regulation of lankamycin production in Streptomyces rochei (Suzuki et al. 2010), and two SARP family regulators, NanR1 and NanR2, are essential for nanchangmycin production in Streptomyces nanchangensis. These latter are thought to form a heterodimer to bind and activate their cognate genes (Yu et al. 2012). It can be seen, therefore, that many SARP homologs vary in their regulatory mechanisms and interplay, which may be specific for a given antibiotic gene cluster.
The current results allow our model of auricin regulation, presented in Kormanec et al. (2014) and Mingyar et al. (2015), to be extended (Fig. 9). The most important auricin-specific regulator, Aur1P, directs the expression of the core auricin biosynthetic genes along with the initial d-forosamine biosynthetic genes (aur1TQSV) from the aur1Ap promoter. The expression of Aur1P itself is directly and negatively regulated by the TetR-family negative regulator Aur1R. The genes for both of these regulators are induced after entry into stationary phase by the SagA-SagR γ-butyrolactone autoregulator-receptor system (Mingyar et al. 2015). Once the core genes of the cluster have begun expression, some basal level of auricin biosynthesis may begin, but it is limited because Aur1R inhibits the expression of the Aur1PR3 activator. Transcription of the auricin-specific activators remains blocked by Aur1R until the cells are fully prepared for auricin production, perhaps by permitting the expression of auricin resistance genes, for example. Once a suitable threshold level of auricin intermediates has built up, they bind to Aur1R, unblocking repression and resulting in the increased production of both Aur1P and Aur1PR3 activators. Aur1P then directly binds and activates the gene for Aur1PR4, another activator. All three activators then induce the expression of all auricin biosynthetic genes: Aur1P directly (and probably also Aur1PR3 indirectly) induces the expression of the core auricin biosynthetic genes together with the initial d-forosamine biosynthetic genes; meanwhile, both Aur1PR3 and Aur1PR4 induce the expression of other auricin tailoring genes, including the final d-forosamine biosynthetic and attachment genes. The effect of this activity is to dramatically increase the biosynthesis of auricin to its peak level. This high level of production is not maintained for long however. When a high enough level of auricin intermediates binds to Aur1P, it prevents it from further functioning as a transcriptional activator, resulting in a decreased expression of all auricin biosynthetic genes.
In this complex regulatory scheme, several auricin-specific regulators serve as checkpoints to control the biosynthesis of this compound. Aur1P mainly directs the genes for the biosynthesis of the aglycone, the initial d-forosamine biosynthetic genes, and auricin resistance. The auricin intermediates thereby produced are likely not as toxic to the cells as the final glycosylated product. The glycosylation and tailoring genes responsible for finishing auricin synthesis are, in turn, under the control of two additional SARP family positive regulators, Aur1PR3 and Aur1PR4, which are regulated by two independent branches through Aur1P and Aur1R. It may also be that the glycosylation reaction may be connected with the rapid export of auricin from the cell. An interesting question concerns the differential role of the two SARP regulators in controlling the expression of these tailoring genes. One possibility is that they may form a heterologous dimer, as NanR1 and NanR2 in regulating the nanchangmycin gene cluster (Yu et al. 2012); however, our binding studies showed that they are also able to bind the promoter regions separately. Further studies will therefore be needed to investigate this interesting feature of auricin regulation.
In conclusion, we characterised d-forosamine specific genes essential for the biosynthesis of this sugar. Interestingly, although auricin contains a single deoxyhexose, two GT genes were found to participate in the attachment of d-forosamine to the auricin aglycon. Two auricin-specific regulators belonging to the SARP family, Aur1PR3 and Aur1PR4, have been found to directly regulate the expression of the genes which are responsible for d-forosamine biosynthesis and attachment.
References
Arias P, Fernandez-Moreno MA, Malpartida F (1999) Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol 181:6958–6968
Ausubel FM, Brent R, Kingston RE, Moore DO, Seidman JS, Smith JA, Struhl K (1995) Current protocols in molecular biology. Wiley, New York
Bibb MJ (2005) Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol 8:208–215
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bunet R, Song L, Mendes MV, Corre C, Hotel L, Rouhier N, Framboisier X, Leblond P, Challis GL, Aigle B (2011) Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J Bacteriol 193:1142–1153
Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328:307–317
Cundliffe E (2008) Control of tylosin biosynthesis in Streptomyces fradiae. J Microbiol Biotechnol 18:1485–1491
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 18:1541–1548
Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A (2007) Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep 24:162–190
Hoffmeister D, Ichinose K, Domann S, Faust B, Trefzer A, Drager G, Kirsching A, Fischer C, Kunzel E, Bearden DW, Rohr J, Bechthold A (2000) The NDP-sugar co-substrate concentration and enzyme expression level influence the substrate specificity of glycosyltransferases: cloning and characterization of deoxysugar biosynthetic genes of the urdamycin biosynthetic gene cluster. Chem Biol 7:821–831
Hong L, Zhao Z, Melancon CE III, Zhang H, Liu H (2008) In vitro characterization of the enzymes involved in TDP-forosamine biosynthesis in the spinosyn pathway of Saccharopolyspora spinosa. J Am Chem Soc 130:4954–4967
Horinouchi S, Hara O, Beppu T (1983) Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol 155:1238–1248
Isiorho EA, Jeon B-S, Kim NH, Liu H-W, Keatinge-Clay AT (2014) Structural studies of the spinosyn forosaminyltransferase, SpnP. Biochemistry 53:4292–4301
Karray F, Darbon E, Oestreicher N, Dominguez H, Tuphile K, Gagnat J, Blondelet-Rouault M-H, Gerbaud C, Pernodet J-L (2007) Organization of the biosynthetic gene cluster for the macrolide antibiotic spiramycin in Streptomyces ambofaciens. Microbiology-SGM 153:4111–4122
Kharel MK, Pahari P, Shephard MD, Tibrewal N, Nybo SE, Shaaban KA, Rohr J (2012) Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat Prod Rep 29:264–325
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Kormanec J (2001) Analyzing the developmental expression of sigma factors with S1-nuclease mapping. In: Chein CH (ed) Nuclease methods and protocols. Methods in molecular biology 160. Humana, Totowa, pp 481–494
Kormanec J, Farkasovsky M (1993) Differential expression of principal sigma factor homologues of Streptomyces aureofaciens correlates with the developmental stage. Nucleic Acids Res 21:3647–3652
Kormanec J, Novakova R, Mingyar E, Feckova L (2014) Intriguing properties of the angucycline antibiotic auricin and complex regulation of its biosynthesis. Appl Microbiol Biotechnol 98:45–60
Kutas P, Feckova L, Rehakova A, Novakova R, Homerova D, Mingyar E, Rezuchova B, Sevcikova B (2013) Strict control of auricin production in Streptomyces aureofaciens CCM 3239 involves a feedback mechanism. Appl Microbiol Biotechnol 97:2413–2421
Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680–685
Lewer P, Hahn DR, Karr LL, Duebelbeis DO, Gilbert JR, Crouse GD, Worden T, Sparks TC, McKamey P, Edwards R, Graupner PR (2009) Discovery of the butenyl-spinosyn insecticides: novel macrolides from the new bacterial strain Saccharopolyspora pogona. Bioorg Med Chem 17:4185–4196
Liu G, Chater KF, Chandra G, Niu G, Tan H (2013) Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143
Luzhetskyy A, Vente A, Bechthold A (2005) Glycosyltransferases involved in the biosynthesis of biologically active natural products that contain oligosaccharides. Mol BioSyst 1:117–126
Martin JF, Liras P (2010) Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol 13:263–273
Maxam AM, Gilbert W (1980) Sequencing end-labelled DNA with base specific chemical cleavages. Methods Enzymol 65:449–560
Mingyar E, Feckova L, Novakova R, Bekeova C, Kormanec J (2015) A γ-butyrolactone autoregulator-receptor system involved in the regulation of auricin production in Streptomyces aureofaciens CCM 3239. Appl Microbiol Biotechnol 99:309–325
Nguyen HC, Karray F, Lautru S, Gagnat J, Lebrihi A, Huynh TDH, Pernodet J-L (2010) Glycosylation steps during spiramycin biosynthesis in Streptomyces ambofaciens: involvement of three glycosyltransferases and their interplay with two auxiliary proteins. Antimicrob Agents Chemother 54:2830–2839
Novakova R, Bistakova J, Homerova D, Rezuchova B, Kormanec J (2002) Cloning and characterization of a polyketide synthase gene cluster involved in biosynthesis of a proposed angucycline-like polyketide auricin in Streptomyces aureofaciens CCM3239. Gene 297:197–208
Novakova R, Homerova D, Feckova L, Kormanec J (2005) Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology-SGM 151:2693–2706
Novakova R, Kutas P, Feckova L, Kormanec J (2010) The role of the TetR-family transcriptional regulator Aur1R in negative regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology-SGM 156:2374–2383
Novakova R, Rehakova A, Feckova L, Kutas P, Knirschova R, Kormanec J (2011a) Genetic manipulation of pathway regulation for overproduction of angucycline-like antibiotic auricin in Streptomyces aureofaciens CCM 3239. Folia Microbiol 56:278–282
Novakova R, Rehakova A, Kutas P, Feckova L, Kormanec J (2011b) The role of two SARP-family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology-SGM 157:1629–1639
Novakova R, Knirchova R, Farkasovsky M, Feckova L, Rehakova A, Mingyar E, Kormanec J (2013) The gene cluster aur1 for the angucycline antibiotic auricin is located on a large linear plasmid pSA3239 in Streptomyces aureofaciens CCM 3239. FEMS Microbiol Lett 342:130–137
Rehakova A, Novakova R, Feckova L, Mingyar E, Kormanec J (2013) A gene determining a new member of the SARP family contributes to transcription of genes for the synthesis of the angucycline polyketide auricin in Streptomyces aureofaciens CCM3239. FEMS Microbiol Lett 346:45–55
Salas JA, Méndez C (2005) Biosynthesis pathways for deoxysugars in antibiotic-producing actinomycetes: isolation, characterization and generation of novel glycosylated derivatives. J Mol Microbiol Biotechnol 9:77–85
Sheldon PJ, Busarow SB, Hutchinson CR (2002) Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol Microbiol 44:449–460
Suzuki T, Mochizuki S, Yamamoto S, Arakawa K, Kinashi H (2010) Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci Biotechnol Biochem 74:819–827
Tanaka A, Takano Y, Ohnishi Y, Horinouchi S (2007) AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369:322–333
van Wezel GP, McDowall KJ (2011) The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep 28:1311–1333
Waldron C, Matsushima P, Rosteck PR Jr, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz R (2001) Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem Biol 8:487–499
Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A (1999) Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol Lett 170:381–387
Wietzorreck A, Bibb M (1997) A novel family of proteins that regulates antibiotic production in streptomycetes appear to contain an OmpR-like DNA-binding fold. Mol Microbiol 25:1181–1184
Yu Q, Du A, Liu T, Deng A, He X (2012) The biosynthesis of the polyether antibiotic nanchangmycin is controlled by two pathway-specific transcriptional activators. Arch Microbiol 194:415–426
Acknowledgments
We are grateful to Dr. Bertold Gust (John Innes Centre, Norwich, UK) for kindly providing all the plasmids and strains used in the PCR targeting system; the system itself was supplied by Plant Bioscience Ltd. (Norwich, UK). This work was supported by the Slovak Research and Development Agency under contract No. APVV-0203-11. The research leading to these results received funding from the European Commission’s Seventh Framework Programme (FP7/2007-2013) under the grant agreement STREPSYNTH (project No. 613877). This work was co-funded by the Slovak Research and Development Agency under contract No. DO7RP-0037-12.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animal preformed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 748 kb)
Rights and permissions
About this article
Cite this article
Bekeova, C., Rehakova, A., Feckova, L. et al. Characterisation of the genes involved in the biosynthesis and attachment of the aminodeoxysugar d-forosamine in the auricin gene cluster of Streptomyces aureofaciens CCM3239. Appl Microbiol Biotechnol 100, 3177–3195 (2016). https://doi.org/10.1007/s00253-015-7214-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7214-9