Abstract
Strain breeding is much less advanced in the edible and medicinal species Agaricus subrufescens than in Agaricus bisporus, the button mushroom. Both species have a unifactorial system of sexual incompatibility, a mating type locus tightly linked to a centromere, and basidia producing both homokaryotic (n) and heterokaryotic (n + n) spores. In A. bisporus, breeding is mainly based on direct selection among the heterokaryotic offspring and on hybridization between homokaryotic offspring. The parental heterozygosity is highly maintained in the heterokaryotic offspring due to suppression of recombination and preferential pairing in the spores of nuclei, each one per second meiotic divisions; such “non-sister nuclei” heterokaryons are fertile. In A. subrufescens, recent studies revealed that recombination is not suppressed and that nuclei from the same second meiotic division can also be paired in a spore that give rise to a “sister nuclei” heterokaryon in which the nuclei bear the same mating type allele. The objective of the present work was to investigate the potential function of the different categories of spores in A. subrufescens and their possible use in a genetic breeding program. Using eight co-dominant molecular markers, we found that half of the offspring of the A. subrufescens strain WC837 were heterokaryotic, one quarter of them being sister nuclei heterokaryons. These heterokaryons were infertile and behaved like homokaryons, being even able to cross between each other. In contrast, non-sister nuclei heterokaryons could fruit but inconsistently due to inbreeding depression. Potential roles of these two categories of heterokaryons in nature and consequences for strain breeding are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Agaricus subrufescens Peck is a species of medicinal and nutritional interest mainly cultivated in the Americas and in Asia. Even though A. subrufescens and Agaricus bisporus, the button mushroom, belong to the distinctly different Agaricus sections Arvenses and Bivelares, respectively, both are secondary decomposers and can be cultivated on similar substrates (Largeteau et al. 2011; Llarena-Hernández et al. 2013). A. subrufescens is, to our knowledge, the Agaricus species having the broadest climatic and geographical range (Peterson et al. 2000; Kerrigan 2005; Wisitrassameewong et al. 2012a, b; Parra 2013; Gui et al. 2014). Interfertility between isolates from South America, Europe, and Asia has been recently demonstrated (Thongklang et al. 2014), indicating that a broad genetic base is available for the genetic improvement of this species.
Fungal species in the phylum Basidiomycota may sexually reproduce in heterothallic, homothallic, or pseudohomothallic life cycles. In the heterothallic life cycle, the four meiospores produced by each basidium give rise to infertile homokaryotic mycelia [n]. Plasmogamy between sexually compatible homokaryons restores a heterokaryon [n + n] which is fertile since it can fruit and produce spores. In contrast, in pseudohomothallism, two postmeiotic nuclei are paired in each spore. Such spores generally give rise to fertile heterokaryons. Amphithallism refers to the production of both homokaryotic and heterokaryotic spores in the same sporocarp, thus encompassing both heterothallism and pseudohomothallism (Lange 1952; Kennedy and Burnett 1956; Kühner 1977). Amphithallism is not rare. For instance, of approximately 500 species of Agaricales, about 8 % were considered amphithallic (Lamoure 1989). Thongklang et al. (2014) showed that A. subrufescens, like A. bisporus var. bisporus, has a multiallelic unifactorial (bipolar) system of sexual incompatibility with a single mating type locus (MAT) tightly linked to a centromere and an amphithallic life cycle. However, amphithallism in A. subrufescens differed from A. bisporus var. bisporus on three key points: (i) basidia were tetrasporic although 40 to 75 % of the offspring were heterokaryotic; (ii) a fraction reaching 23 and 19 % of the heterokaryoting offspring of the two studied parental strains WC887 and CA487, respectively, were homoallelic at the mating type locus; (iii) the rate of crossovers was much higher than in A. bisporus var. bisporus. The implications of these three key points are detailed in the three following paragraphs, and a theoretical model of the sporogenesis in A. subrufescens is proposed in the Fig. 1a.
Amphithallic tetrasporic random model: a Different categories of spores showing expected ratio of sister nuclei heterokaryons and non-sister nuclei heterokaryons. The two nuclei are horizontally arranged in the spores to indicate they are identical. b Different types of crosses tested between the different types of single spore isolates, showing expected positive and negative mating reactions
The first point indicates that postmeiotic mitosis must occur in basidia as in Mycocalia denudata in which eight postmeiotic nuclei migrate into four spores (Burnett and Boulter 1963). Postmeiotic mitosis also occurs in A. bisporus but in the spores or during the migration of the nuclei through the sterigmata and rarely in the basidia (Kamzolkina et al. 2006); consequently, only four postmeiotic nuclei migrate into the two to four spores of each basidium. The basidium spore number is determined by a major locus BSN linked to MAT (Imbernon et al. 1996). Amphithallism is preponderantly pseudohomothallic in A. bisporus var. bisporus and preponderantly heterothallic in A. bisporus var. burnettii correlatively to their basidial phenotypes which are highly bisporic and tetrasporic, respectively (Raper et al. 1972; Callac et al. 1993; Kerrigan et al. 1994). In A. subrufescens, Thongklang et al. (2014) observed that the ratio of heterokaryotic spores was also quite variable but genetic determinants are unknown and might be completely different. This variable ratio of heterokaryotic spores indicated that the rate of postmeiotic mitoses occurring in basidia must be also variable. Two cases are represented in the model of Fig. 1a. In the basidium on the left side, the postmeiotic mitosis occurs in the four spores. Therefore, homokaryotic spores represented in Fig. 1a are binucleate. However, in A. subrufescens, it is still unknown if the two identical daughter nuclei remain in the mature spores as in A. bisporus (Kamzolkina et al. 2006) or if one of the two nuclei passes in the basidium making the spores uninucleate at maturity. In each of the three other basidia illustrated in the model, postmeiotic mitoses occur in the basidia and the eight postmeiotic nuclei are randomly paired in the four spores.
The second point indicates that postmeiotic nuclei bearing different mating type alleles are not preferentially paired in the same spore as this occurs in A. bisporus but that nuclei migrate at random as in Coprinopsis scobicola (syn. Coprinus bilanatus), a bisporic bifactorial (tetrapolar) species (Kemp 1974) with two mating type loci tightly linked to their respective centromeres (Elliott and Challen 1983; Kemp 1985). For this species, Challen and Elliott (1989) proposed a random bisporic model with a category of heterokaryotic spores called “sister nuclei” progeny because they receive nuclei arising from the same second meiotic division and bearing the same mating type alleles. In A. bisporus, such sister nuclei heterokaryons are infrequent (Summerbell et al. 1989); the spatial position of the spindles of the second meiotic divisions (Evans 1959) and/or their asynchrony (Kamzolkina et al. 2006) could explain the preferential migration in the same spore of two non-sister nuclei, each one per second meiotic divisions. In A. subrufescens, Thongklang et al. (2014) reported the following three primary categories of single spore isolates that are represented in the theoretical tetrasporic random model of Fig. 1a:
-
Non-sister nuclei heterokaryons (also called NSNPP for non-sister nuclear pair progeny heterokaryons): in A. subrufescens as in A. bisporus, the mating type locus MAT is linked to a centromere; therefore, virtually, all these non-sister nuclei heterokaryons are fertile (reproductively competent) sexual (intramixis) heterokaryons. These are the familiar heterokaryons of the pseudohomothallic life cycle allowing uniparental reproduction.
-
Sister nuclei heterokaryons (also called SNPP for sister nuclear pair progeny heterokaryons): these are “unconventional” heterokaryons having a pair of “sister” nuclei arising from the same second meiotic division and therefore having highly homoallelic genotypes in tightly centromeric-linked regions. They are theoretically not reproductively competent although they can be heteroallelic in regions distal to the centromeres. Sister nuclei heterokaryons had not been previously considered in classical concepts of amphithallism.
-
Homokaryons are haploid cultures that can arise from spores in two different ways. In the traditional scenario, homokaryotic spores receive only one postmeiotic nucleus. However, in amphithallism involving nuclear pairs, some spores may receive two identical nuclei from one postmeiotic division, and these are also homokaryons as shown in Fig. 1a.
The third point is the higher rate of crossovers of A. subrufescens than in A. bisporus var. bisporus. The suppression or not of recombination has important consequences on the potential role of the spores in nature as for their potential use in breeding strategies. To apprehend these consequences, it is useful to keep in mind how the alleles can be paired in the heterokaryotic spores. Considering a parental strain with a heterozygous genotype Aa at a given locus, the “rate of homoallelism” among the heterokaryotic offspring is used to designate the rate of heterokaryotic single spore isolates having the genotypes AA or aa at this locus. This represents the rate of loss of parental heterozygosity at this locus. Different rates of homoallelism have been considered (for the history, see Challen and Elliott 1989) until Langton and Elliott (1980) re-interpreted the consequence of random migration in pseudohomothallism. In a bisporic random model (i.e., four meiotic products migrating at random in two spores), the theoretical rate is 1/3 (Langton and Elliott 1980). In the tetrasporic random model, a 3/7 rate of homoallelism at any locus had been proposed by Langton and Elliott (1980). However, taking in consideration the fact that 1/7 of these spores are true homokaryons since they receive nuclei from the same postmeiotic mitosis as represented in Fig. 1a, the corrected rate of homoallelism at any locus is 1/3 among the heterokaryotic offspring. Applying this rate to the locus MAT which is tightly linked to a centromere, the one third rate of homoallelic heterokaryons at this locus represents the rate of sister nuclei heterokaryons. Fig. 1a shows how the rates of sister and non-sister nuclei heterokaryons can be simply calculated: 2/7 of the spores receive non-sister nuclei, while 4/7 of the spores receive sister nuclei. Finally, at any locus, the rate of sister nuclei heterokaryons is 1/3 (33 % of the heterokaryotic offspring) in both tetrasporic and bisporic random models. In these models, the rate of homoallelism can also be considered among each of the two categories of heterokaryons and, in this case, it depends on the linkage to centromere: for any centromere-unlinked locus, the rate of homoallelism is 33 % whatever the category of considered heterokaryons. However, higher is the linkage to a centromere (until tightly linked as for the MAT locus), higher is the rate of homoallelism among the sister nuclei heterokaryotic offspring (until 100 %), and lower is the rate of homoallelism among the non-sister heterokaryotic offspring (until 0 %). In A. bisporus var. bisporus, the rate of homoallelism is very low at all loci because the whole heterokaryotic offspring is of the non-sister type and the rate of crossover is very low across the whole genome (Kerrigan et al. 1993). In this variety, the parental heterozygosity remains highly conserved even through multiple intramictic generations; in such “pseudohomothallic“ lineage, accumulation of lethal or deleterious recessive alleles may contribute to this conservation. In contrast, such a suppression of recombination was not observed in A. bisporus var. burnettii nor in the linkage map of an intervarietal hybrid (var. bisporus x var. burnettii) except in pericentromeric regions (Callac et al. 1997; Foulongne-Oriol et al. 2010).
The theoretical model in Fig. 1a mainly describes the basidiosporogenesis phase but does not show completely the life cycles until the fruiting phase because the function of the sister nuclei heterokaryotic spores is unknown and the role of the other spores could be multiple. For example, in A. bisporus var. bisporus, heterokaryotic offspring are potentially fertile but, in certain conditions, can cross with homokaryons (phenomenon of Buller; Buller 1931) instead to fruit (Callac et al. 2006). In rare cases, crosses between heterokaryons have been also evidenced (Raper et al. 1972; Xu et al. 1996).
In A. bisporus var. bisporus, genetic improvement of the strains is based on selection among heterokaryotic offspring (intramictic generations) for the past hundred years, and this is possible because the parental heterozygosity is highly conserved (monospore selection, Kerrigan 1993; Moquet et al. 1998) while phenotypical variations are observed. It is due among other reasons to the random segregation of the centromeres during the meiosis which makes that the homologous chromosomes are randomly redistributed in the two non-sister nuclei of the heterokaryons (Sonnenberg et al. 2011). Since the 1970s, selection is also performed among outcrossing generations mainly between homokaryons as such F1 hybrids (Fritsche 1983); backcrosses are also performed but inbreeding depression was reported (Xu 1995). Heterokaryotic offspring are easily obtained in A. bisporus var. bisporus since the pseudohomothallism predominates. In contrast, the recovering of homokaryotic single spore isolates which are required for outcrossing is problematic due to the low rate of homokaryotic offspring and the absence of reliable morphological criteria for homokaryosis since both homokaryons and heterokaryons have plurinucleate cells and lack clamp connections. In A. bisporus var. bisporus, different indirect methods have been used to distinguish among these, such as fruiting tests, mating tests, or mycelial growth rate tests assuming that only homokaryons are infertile, mating competent, and grow more slowly than the heterokaryons, respectively. However, in A. bisporus var. bisporus, these tests are less reliable than methods based on auxotrophic, alloenzymatic, restriction fragment length polymorphism (RFLP), cleaved amplified polymorphic sequences (CAPS), or any co-dominant markers that have been successfully used to directly evidence heteroallelic loci and thus heterokaryosis of single spore isolates (Kerrigan et al. 1994). Because the rate of homoallelism is very low in A. bisporus var. bisporus, homoallelism of two genetically independent markers (i.e., unlinked to each other) is a reliable criterion for homokaryosis. For example, if the rate of homoallellism does not exceed 10 % at each locus, the probability that a heterokaryon should be homoallelic at both loci is 1 %; therefore, offspring homoallelic at both loci are likely homokaryotic. The more the loci are centromere-linked, the more stringent this test is. In A. subrufescens, cultivated strains have been compared but breeding strategies are not reported in literature except the hybridizations performed by Kerrigan (2005) and Thongklang et al. (2014). For developing such strategies in this species, the test for identifying mating-competent single spore isolates has to be reassessed.
The objective of the present work is to evidence the potential functions of the different types of spores in order to better understand what could be their role in nature and how they could be used in a genetic breeding program. The latter point implies (i) to assess the potential interest of the method of direct selection among the heterokaryotic offspring as it is used in A. bisporus and (ii) to find an outcrossing method by determining what types of offspring are mating competent and how to recognize them easily. By using a larger sample and more markers than Thongklang et al. (2014), we expect to identify enough offspring of each of the three types to study them and to investigate how their relative proportions are consistent with a theoretical tetrasporic random model presented above. We used centromere-linked and centromere-unlinked SCAR markers derived from the genome of A. bisporus for categorizing the offspring of A. subrufescens. Then, the behaviors of the three types of spores were compared as this is classically done in mating tests, mycelial growth rate tests, and fruiting tests with a particular interest in sister nuclei heterokaryons of which the behavior has never been formally studied. We found that the rate of sister nuclei heterokaryons is compatible with a tetrasporic random model and that these heterokaryons behave like homokaryons since they can even mate between themselves. Concerning the non-sister nuclei heterokaryons, the rate of homoallelism at centromere-unlinked loci is even higher than predicted in the random model. Therefore, the role as the potential use of such spores is quite different than in A. bisporus. The consequences for breeding strategies in A. subrufescens are discussed.
Materials and methods
Parental strain and single spore isolates
We used the same strain, WC837, as Thongklang et al. (2014). This strain originated in Brazil, and a subculture is available under the accession number CA454 in the Collection du Germplasm des Agarics à Bordeaux (CGAB), INRA-Bordeaux (http://www6.bordeaux-aquitaine.inra.fr/mycsa_eng/Biological-resources-of-value/The-Agaricus-culture-collection-CGAB).
The strain WC837 was cultivated under standard conditions for sporocarp production as described by Llarena-Hernández et al. (2014). Scoring of basidia of a fresh sporocarp in one- to four-spored classes was performed using light microscopy as described by Callac et al. (1993) and a spore print was obtained from the same sporocarp. For germination, spore suspensions were prepared in a saline solution (NaCl 0.85 %) with trace of Tween 80. Spore counts were estimated using a Malassez cell. One hundred microliters of the spore suspension were spread over 90-mm-diameter Petri dishes on complete yeast medium (Raper et al. 1972). Plates were placed upside down with the addition of grains colonized by A. bisporus mycelium in the lid (a germination stimulus; Rast and Stäuble 1970) for 1 week. After germination, single spore isolates were subcultured on compost extract medium (aqueous extract of pasteurized commercial mushroom compost plus 1 % glucose and 2 % agar).
Choice of informative markers
EST sequences of A. subrufescens identified by Foulongne-Oriol et al. (2014) with putative homologs in the genome of A. bisporus were selected. Information about their physical position on the A. bisporus genome was used to develop informative molecular markers for the distinction between the three expected types of spore among the WC837 single spore isolates (Thongklang et al. 2014). First, we selected A. subrufescens-identified sequences for which A. bisporus homologs were close to MAT and the centromere (RPB2 and PRS088 markers). Secondly, we chose two other A. subrufescens sequences with A. bisporus homologs located on chromosome I in pericentromeric (PRS113 marker) and distal positions (PRS095 marker). Thirdly, four other A. subrufescens sequences (PRS003, PRS016, PRS160, and ITS markers), for which A. bisporus homologs were found in distal positions on chromosomes IV, X, VII, and IX were also used (Table 1).
Method based on homo/heteroallelism to classify single spore isolates in three categories
We used co-dominant single locus markers that were heteroallelic in the parental strain. Heterokaryotic and homokaryotic single spore isolates were first distinguished as follows: single spore isolates heteroallelic at least at one of the loci were unambiguously heterokaryons (either sister or non-sister nuclei heterokaryons). Single spore isolates homoallelic at all loci were regarded as putative homokaryons. This classification was performed with several genetically independent markers (genetically unlinked loci), to avoid misinterpreting a heterokaryon as a homokaryon when it was homoallelic at any of these loci. For each of the genetically independent markers, the frequency of homoallelic single spore isolates was estimated from the experimental data. The probability of misinterpretation of a heterokaryon as a homokaryon was the product of these frequencies for all the markers used. Such a multilocus genotype test has been used in A. bisporus by Kerrigan et al. (1992, 1993, 1994) and in A. subrufescens by Thongklang et al. (2014).
Among the heterokaryotic single spore isolates, non-sister nuclei heterokaryons are expected to be heteroallelic at loci tightly linked to centromeres while sister nuclei heterokaryons are homoallelic. We used markers linked to the MAT locus which is tightly linked to the centromere in A. bisporus (Xu et al. 1993). Homoallelic and heteroallelic single spore isolates at such loci were considered as putative sister nuclei heterokaryons and non-sister nuclei heterokaryons, respectively. The reliability of this method depends on the rates of crossovers between these markers and MAT and thus the centromere. Such rates can be estimated through the analysis of the homokaryotic offspring.
Molecular markers genotyping
CAPS markers were previously used for mapping in A. bisporus (Callac et al. 1997; Foulongne-Oriol et al. 2010) and to study the life cycle of A. subrufescens (Thongklang et al. 2014). CAPS markers exploit heteromorphic positions detected in the sequences; informative loci are heteromorphic and are included in the restriction sites of restriction endonucleases (Table 2). When the restriction site differed from the sequence, derived CAPS (dCAPS) markers were developed (Neff et al. 1998).
DNA extraction was done as described by Zhao et al. (2011). PCR was performed in a 30 μL reaction mixture containing 50 ng genomic DNA, 0.5 μM of each primer, 0.2 mM dNTPs, 2 μg BSA, 1 U Taq DNA polymerase, and 1× incubation buffer. Amplifications were carried out as follows: an initial denaturing step at 94 °C for 5 min, 35 cycles of 94 °C for 1 min, 55 °C for 90 s, 72 °C for 1 min, and a final extension at 72 °C for 5 min. The amplified region was sequenced by Beckman Coulter Genomics Inc. (Takeley, UK). Restriction enzymes recognizing a polymorphic position of the sequence were selected. For the cleavage reaction, 5 μL of the PCR product were digested with 8 U of the appropriate restriction enzyme for 120 min at the optimal incubation temperature recommended by the manufacturer. CAPS products were visualized in 2 % agarose gels, running at 90 V for 90 min. Selected primer sequences and matching restriction enzymes for each marker are listed on Table 2. Loci were named as the sequenced DNA segment followed by the heteromorphic position in the amplified sequence. All the markers are heteroallelic in the parental strain W837. We analyzed all 225 single spore isolates in the same conditions as described above.
Linkage map and Mendelian segregation analyses
To test genetic independence, contingency chi-square tests were performed for all pairwise combinations between the loci used. For the linked loci, their order and genetic distances were computed using MAPMAKER/EXP V3.0b software (Lander et al. 1987). The recombination rate was transformed into map distance (centimorgan cM) using the Kosambi function. For each marker, the hypothesis of the Mendelian allelic segregation ratio of 1:1 was tested among the homokaryotic offspring using chi-square tests.
Mating test and hybridizations
Mating tests were performed according to Kerrigan et al. (1994). A. subrufescens has a unifactorial system of incompatibility: two homokaryons give a positive reaction only when they bear different alleles (Mat-1 and Mat-2) at the mating type locus MAT. The presence of fluffy, vigorous mycelium at the junction zone between the two mycelia characterizes a positive mating reaction (Raper 1976). The test was intended to determine the mating type alleles of the single spore isolates and ultimately to estimate the linkage relationships between MAT and other markers used. A preliminary mating test was performed to identify homokaryotic strains with different mating type alleles. We paired all the homokaryotic single spore isolates and most of the heterokaryons single spore isolates (sister or non-sister nuclei heterokaryons) with four testers previously selected (two Mat-1 and two Mat-2) in duplicate.
Furthermore, mating tests between sister nuclei heterokaryons bearing different mating type genotypes (Mat-1/1 × Mat-2/2) were performed, in triplicate. When positive reactions were observed, putative hybrid mycelia were isolated from the junction and subcultured on a compost-agar medium. Heteroallelism at a marker tightly linked to MAT was used to confirm the hybridization and deductively the restoration of fertility (Mat-1/2).
Moreover, when two sister nuclei heterokaryons crossed, we assumed that the resulting “hybrid” heterokaryon would receive one nucleus of each parental heterokaryon and, therefore, four different hybrids were possible as represented in Fig. 1b. To reveal whether the nuclei randomly assembled in the hybrid, we tested pairs of sister nuclei heterokaryons having the genotypes a1/a2 b1/b1 and a1/a1 b1/b2 at two loci A and B. In this condition, one sister nuclei parent contained the nuclei a1b1 and a2b1 and the other contained the nuclei a1b1 and a1b2. Co-dominant markers at such two loci were used to identify the four possible hybrids which could be homoallelic respectively either at A (a1/a1 b1/b2), at B (a1/a2 b1/b1), at both loci (a1/a2 b1/b2), or at neither (a1/a2 b1/b2). Genotypes of several hybrids isolated from mating between the same two sister nuclei heterokaryons were compared.
Mycelial growth rate test
Agar plugs of 4 mm diameter were transferred to Petri dishes (90 mm diameter) containing 1 % malt agar medium and incubated at 28 °C for 15 days. Colony diameters were measured on two perpendicular axes. These data were used to score the rate of the mycelial growth with two replicate plates for each single spore isolate in a completely randomized experimental design. Mycelial growth rate was compared between each type of isolate using variance analysis and post hoc Duncan’s test applied for multiple means comparison.
Fruiting test
Strains were cultivated in plastic trays filled with 8 kg of conventional compost under standard conditions of cultivation as described by Llarena-Hernández et al. (2014). Forty-five plastic trays were inoculated with the parental strain WC837, homokaryons (four strains, two replicates), non-sister nuclei heterokaryons (nine strains, two replicates), and sister nuclei heterokaryons (nine strains, two replicates).
Results
Classification of the single spore isolates: homokaryons, non-sister nuclei heterokaryons, and sister nuclei heterokaryons
Almost all basidia of the parental sporocarp were tetrasporic: bisporic basidia were not detected and trisporic basidia were rare (less than 1 %). After incubation for 14 days, the rate of germination was estimated as 10 % for a density of approximatively 1000 spores spread on agar medium in a 90-mm-diameter Petri dish. Three hundred and forty germinating spores were isolated from the same spore print. However, many of them stopped growing and only 225 single spore isolates exhibited sufficient mycelial growth to be used.
Single spore isolates were classified based on homo/heteroallelism at eight CAPS markers (Tables 3 and S1 in the Supplementary Material). Among 225 single spore isolates, 49.8 % (112/225) were heteroallelic at least at one of the eight CAPS markers used and were therefore confirmed heterokaryons. The remaining single spore isolates were putative homokaryons with a high level of confidence that we estimated by using the rates of homoallelism among the heterokaryotic offspring at five unlinked loci (PRS088:248, PRS095:266, PRS003:212, PRS016:1, and PRS160:134). The probability that a heterokaryon should be homoallelic at all five loci and therefore misinterpreted as a homokaryon is the product of these five rates: p = 31/112 × 57/111 × 55/111 × 58/110 × 58/111 = 0.019. In other respects, p is also the estimated proportion of undetected heterokaryons p = (T − O)/T where T is the unknown total number of heterokaryons and O is the observed number of heterokaryons. Using these five markers, 111 heterokaryons were detected (O = 111). Finally, by estimation, we would expect T = 111/(1 − p) = 113 heterokaryons. Since we found one supplementary heterokaryon which was heteroallelic at one of the three remaining loci used (single spore isolate WC837-290 was heteroallelic only at locus PRS113:158), the expected total number of heterokaryons is only one more than the 112 confirmed heterokaryons that we found. The final percentage of expected and found heterokaryons are 50.2 % (113/125) and 49.8 %, respectively, i.e., both close to 50 %.
Among the heterokaryons, 21 % (24/112) were likely sister nuclei heterokaryons because they were homoallelic at both loci RPB2:715 and PRS088:248 tightly linked to the centromere and the MAT locus, while 67 % (75/112) were likely non-sister nuclei heterokaryons because they were heteroallelic at these two loci. Twelve percent (13/112) of the heterokaryons that were homoallelic at only one of these two loci remained unclassified. Possibly, following a crossover between RPB2:715 and PRS088:248, they received both one recombinant and one non-recombinant nucleus; theoretically, they could be either sister nuclei heterokaryons or non-sister nuclei heterokaryons with equal probability. Without these 13 unclassified heterokaryotic single spore isolates, the rate of sister nuclei heterokaryons was estimated as 24 % (24/99) among the heterokaryons. Taking them into account with the half considered as sister nuclei heterokaryons, this rate can be estimated to 27 % (30.5/112). This rate is relatively close to the rate of 33 % expected in the theoretical tetrasporic random model represented in Fig. 1a.
Mating tests between single spore isolates and tester homokaryons
Mating tests were performed between tester homokaryons bearing Mat-1 or Mat-2 alleles and single spore isolates of the three categories with expected positive reactions as such indicated in Fig. 1b. In all the types of mating tests (homokaryon × homokaryon, homokaryon × sister nuclei heterokaryon, and homokaryon × non-sister nuclei heterokaryon), positive and negative mating reactions were observed (Fig. 2). In positive reactions, more or less fluffy mycelium emerged from one or several points on the junction line between the two mycelia. The new putative hybrid mycelium often formed a new circular and vigorous colony overlapping its parent mycelia. Negative reactions were characterized by the absence of fluffy mycelium in the contact zone; moreover, the contact zone remained often poorly invaded, sometimes exhibiting a solid white line (Fig. 2d, f). With two replicates for each mating test, it was not rare to observe a positive reaction in only one of the two replicates. False negative reactions are common in A. bisporus (Kerrigan et al. 1994) as in some species of Agaricus section Arvenses to which A. subrufescens belongs (Calvo-Bado et al. 2000).
Positive and negative reactions in mating tests: a Positive between homokaryons (WC837-309, WC837-145). b Positive between sister nuclei heterokaryons (WC837-142, WC837-18). c, d Between sister nuclei heterokaryons and homokaryon: positive (WC837-142, WC837-20) and negative (WC837-142, WC837-145), respectively. e, f Between non-sister nuclei heterokaryons and homokaryons: positive (WC837-97, WC837-145) and negative (WC837-121, WC837-20), respectively
In mating tests between four tester homokaryons and the 113 putative homokaryons identified with the CAPS markers, positive mating reactions were obtained for 75 % of them (Table 4) giving an unbalanced segregation ratio of 59 Mat-1:26 Mat-2. These two alleles co-segregated completely with those of PRS088:271 (0 recombinants) and almost completely with those of RPB2:715 (2 recombinants).
In mating tests between four tester homokaryons and 66 putative non-sister nuclei heterokaryons heteroallelic at the MAT locus, positive reactions were observed for 20 of them (30 %): 13 had a positive reaction with tester(s) Mat-1, six with tester(s) Mat-2, and only one with two testers bearing either Mat-1 or Mat-2 allele. Such positive mating reactions between homokaryons and heterokaryons (the Buller phenomenon) are not rare in Basidiomycota (Buller 1931; Quintanilha 1937; Raper et al. 1972; Callac et al. 2006). Considering that all these heterokaryons should theoretically mate with all tester homokaryons regardless their mating type alleles (Fig. 1b), the success rate is finally about only 16 %.
In mating tests between four tester homokaryons and 20 putative sister nuclei heterokaryons (homoallelic at the MAT locus), positive reactions were observed for 16 of them (80 %). The data completely agreed with the expected homoallelic genotypes Mat-1/1 and Mat-2/2 of these sister nuclei heterokaryons which gave positive reactions with Mat-2 and Mat-1 tester strains, respectively. Finally, in this limited sample, there was no evidence of recombination between the tightly linked loci MAT, RPB:175 and PRS088:248: 11 had the genotype Mat-1/1 Rpb2:175-1/1 PRS088:248-1/1 and five had the genotype Mat-2/2 Rpb2:715-2/2 PRS088:248-2/2. It is noteworthy that this unbalanced ratio 11 (Mat-1/1)/5 (Mat-2/2) = 2.20 is similar to the ratio 59 (Mat-1)/26 (Mat-2) = 2.27 observed among the homokaryons. It was expected that sister nuclei heterokaryons would behave like homokaryons in mating tests but this had previously never been formally shown.
Single locus segregation ratios
Chi-square value for Mendelian segregation (df = 1, p = 0.05) tests among the homokaryotic offspring are reported in Table 3. The 1:1 segregation was rejected for all the loci of the large linkage group (see next paragraph: MAT, RPB2:715, PRS088:248, PRS113:158, PRS095:266) and for PRS003:212. For the large linkage group, the numbering of the alleles followed the parental chromosomes: it was always allele 2 of each locus that belonged to the same parental chromosome, and that was always the under-represented allele.
Although the number of identified sister nuclei heterokaryons was low, it must be noted that similar unbalanced segregations among the postmeiotic nuclei having migrated in these heterokaryotic spores were observed since the genotype 1/1 was much more frequent than the genotype 2/2 at all five loci of the large linkage group listed above (11:5, 15:8, 16:8, 10:3, and 10:2).
Pairwise segregation ratios, linkage map, and synteny with A. bisporus
Using chi-square contingency tests for nine loci, genetic independence was rejected (P < 0.001, 1 df) for the following eight pairs of loci among the 36 tested pairs (Tables S2 and S3 in the Supplementary Material): ITS and PRS160:134, PRS113:158, and PRS095:266, and all the pairwise combinations between the four loci PRS113:158, PRS088:248, RPB2:715, and MAT. Therefore, there were two linkage groups: a large one including five loci (MAT, RPB2:715, PRS088:248, PRS113:158, PRS095:266) and a small one (ITS:200, PRS160:134).
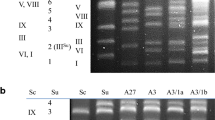
The linkage map obtained by the maximum-likelihood method implemented in Mapmaker (Fig. 3) was consistent with the loci independence tests. By projection on the A. bisporus genome, the genetic order of markers in the large linkage group of A. subrufescens respects the physical order of their respective homologs observed in A. bisporus. Since no recombinant was found between MAT and PRS088:248, these two loci were found collocated. Two recombinants were observed between MAT and RPB2:175. Conversely, the small linkage group did not agree with A. bisporus data. PRS160:134 and ITS:200 loci are linked in A. subrufescens but are unlinked in A. bisporus, since the ITS locus belongs to chromosome IX and PRS160 to chromosome VII.
Linkage map obtained from the segregation analysis at 9 loci in 113 homokaryotic single spore isolates. Values indicated on the left side are the distance intervals in centimorgans (cM) using Kosambi function. Markers including molecular CAPS markers and the phenotypic MAT loci are labelled on the right side
Observed rates of homoallelism and comparison with the theoretical model
The rate of homoallelism among all the heterokaryons is expected to be 33 % at all loci in the theoretical tetrasporic random model represented in Fig. 1a. The observed rates are given in Table 3. It varied from 27 to 31 % for the three centromere-linked loci of the large linkage group (RPB2:715, PRS088:248, and PRS113:158). Lethal or deleterious recessive alleles at centromere-linked loci might explain this little deviation from the random model and also the deviation from Mendelian segregation ratio observed among the homokaryotic offspring. The rate of homoallelism exceeded the 33 % expected in the theoretical model for the five centromere-unlinked markers: it varied from 50 to 53 % for four of them and reached 60 % at the locus ITS:200.
In the theoretical tetrasporic random model, the rate of homoallelism among sister nuclei heterokaryons decreases from 33 to 0 % for tightly centromere-linked loci since heterokaryons inherit of homologous centromeres; in contrast, the rate of homoallelism among non-sister nuclei heterokaryons increases from 33 to 100 % for tightly centromere-linked loci since heterokaryons inherit of heterologous centromeres. In Fig. 4, the observed mean rate of homoallelism has been calculated among 75 non-sister nuclei heterokaryons and 24 sister nuclei heterokaryons from data of Table 3 for centromere-unlinked loci, centromere-linked loci, and for the locus PRS113:158 which is intermediary. Although the rate of homoallelism at centromere-unlinked loci was higher than expected, the observed variations globally agreed with the theoretical model: when the linkage to the centromere or the MAT locus varied from unlinked to tightly linked, the rate of homoallelism increased from 50 to 100 % for the sister nuclei heterokaryons, whereas it decreased from 50 to 0 % for the non-sister nuclei heterokaryons. However, it should be noted that the 13 heterokaryons remaining unclassified were not taken into account in the Fig. 4. Some of them are probably heteroallelic sister nuclei heterokaryons or homoallelic non-sister nuclei heterokaryons at one of the two centromere-linked loci. Consequently, the rates of homoallellism would not be exactly 0 or 100 % for the non-sister and sister nuclei heterokaryons, respectively, at the loci tightly linked to MAT but there would be little variation from these values in agreement with the fact that they are not completely linked to MAT.
Relationships between linkage to centromere and homoallelism among 75 non-sister nuclei heterokaryons and among 24 sister nuclei heterokaryons. Mean rates of homoallelism were calculated for two loci tightly linked to the locus MAT and the centromere, for four centromere-unlinked loci which are genetically independent from each other, and for the locus PRS113:158 which is intermediary
Matings between sister nuclei heterokaryons and confirmation of the crosses
Since sister nuclei heterokaryons behaved as homokaryons in mating tests when confronted to tester homokaryons, we chose some of them to test whether they could cross among themselves. Seven confrontations with three replicates were performed between sister nuclei heterokaryons having not only different mating type genotypes (Mat-1/1 vs. Mat-2/2) but also appropriate genotypes at other loci in order to test whether the nuclei of two mated sister nuclei heterokaryons are randomly paired in the resulting hybrid heterokaryon (see Fig. 1b and materials and methods). In five of the seven confrontations, positive mating reactions were observed (Fig. 2b) and two to four putative hybrids were isolated from different replicate plates or from distinct reactions appearing in the same confrontation plate.
Sixteen putative hybrids isolated from positive mating reactions between sister nuclei heterokaryons were all heterozygous at the PRS088:248 locus (Table 5). This not only confirmed their hybrid status but also indicated that these unconventional crosses between infertile heterokaryons led to fertile heterokaryons since PRS088:248 is tightly linked to the MAT locus. Genotypes at two other loci (only one in one case) were the same for all the hybrids isolated from mating between the same couple of sister nuclei heterokaryons (Table 5). Moreover, except in the case where a single locus was used, these data showed that only one of the four possible pairings between the two nuclei of each “parental” heterokaryon was found. In two cases, there were four identical hybrid genotypes from the same type of mating. If the pairing was random, the probability of finding the same pairing four times would be 4/44 = 0.016. These data demonstrated that the pairings were not random and even suggested that only a single pairing was possible. However the process of pairing of nuclei remains unknown. In fact, such crosses are unconventional selfing and apparent preferential pairing of nuclei could simply result to the presence of deleterious or lethal recessive alleles and/or from unintended selective sampling.
Mycelial growth rate test
The distribution of frequency of colony diameters of 91 single spore isolates (39 homokaryons, 37 non-sister nuclei heterokaryons, and 15 sister nuclei heterokaryons) measured after 15 days of growth is shown in Fig. 5. The mean diameter measured for homokaryons was 34.9 mm and did not differ significantly from the mean diameter for sister nuclei heterokaryons (34.1 mm). The non-sister heterokaryons differed significantly (p < 0.05) from both sister nuclei heterokaryons and homokaryons with a mean diameter of 47.2 mm.
The homokaryotic single spore isolates presented a peak of growth rate in the range of 31–40 mm where 56.4 % (22/39) are grouped. Sister nuclei heterokaryons showed a peak in the range 31–40 mm with 33.3 % of the individuals (5/16) in that range. The non-sister nuclei heterokaryons were present in all ranges represented in the graph, with a peak in the range 51–60 mm containing 37.8 % (9/41) of them.
Behavior of single spore homokaryotic and heterokaryotic isolates (non-sister nuclei and sister nuclei heterokaryons) in fruiting tests
The parental strain fruited normally. The homokaryons were unable to invade the culture substrate and were replaced by competitors or contaminants. Among the non-sister nuclei heterokaryons, four failed to adequately invade the substrate, two invaded the substrate but did not fruit, and three fruited but only two of them produced mature sporocarps. Among the sister nuclei heterokaryons, three did not adequately invade the substrate and among the six remaining, two fruited locally as a result of hybridization with uncontrolled inoculum (confirmed by sequencing, data not shown) but only one of these reached maturity, two others produced balls of mycelium (popcorn-like), and two formed both immature fruiting bodies and balls of mycelium which also resulted from uncontrolled hybridizations. Finally, as expected, only the parental strain and non-sister nuclei heterokaryons were fertile but the latter poorly fructified. The sister nuclei heterokaryons were more vigorous than the homokaryons in invading the substrate and appeared to easily cross with uncontrolled inoculum from the environment.
Discussion
Different categories of spores and agreement with the theoretical tetrasporic random model
To analyze the offspring of the strain of A. subrufescens WC837, we used a larger offspring (225 vs. 94) and more markers (eight vs. three) than Thongklang et al. (2014) in order to perform more accurate assessments and to get enough single spore isolates of each category to study them. Incidentally, synteny with A. bisporus was evidenced for a linkage group of five loci including MAT; however, another smaller unexpected linkage group was found: the rDNA (ITS locus) was linked to a locus of which the putative homolog sequence is not located on the same chromosome in the genome of A. bisporus. Finally, using four independent loci or linkage groups of loci, we found that 50 % of the single spore isolates were heterokaryotic and among them, the percentage of sister nuclei heterokaryons was estimated as 24–27 %. These values are of the same order as the values estimated in the previous offspring of the same parent (Thongklang et al. 2014): 40 % of single spore isolates were heterokaryotic and 23 % of them were sister nuclei heterokaryons. These rates of sister nuclei heterokaryons are lower than the 33 % expected in the theoretical random model. The presence of deleterious or lethal alleles in centromeric regions could simply explain such a deficit in sister nuclei heterokaryons. The presence of such alleles in one of the two homologous parental chromosomes bearing the large linkage group of five loci could similarly explain the significantly unbalanced allelic segregation found at these loci among the homokaryotic offspring (deficit in alleles 2) and correlatively among the heterokaryotic offspring (deficit in genotypes 2/2). However, in this case, the dis-equilibrium affects similarly both centromere-linked and unlinked loci and thus does not provide a demonstrative explanation for the deficit in sister nuclei heterokaryons.
Another rate, i.e., that of homoallellism at centromere-unlinked loci among the heterokaryotic offspring, can be used to test the theoretical random model in which it is expected to also be 33 %. We found that this rate of homoallelism was higher than expected and consistently close to 50 % or even higher at one locus. Thongklang et al. (2014) found similar frequencies but did not highlight these results, which in their study were not significant because of the small number of centromere-unlinked markers (two vs. four) and heterokaryons used (39 vs. 113 in the present study).
As did Thongklang et al. (2014), we conclude that recombination is not suppressed and that the process of migration of nuclei into the spores clearly differs from the non-random process known in A. bisporus. The theoretical tetrasporic random model is the simplest model that can explain our data, but it remains to clarify either how the observed rate of homoallelism among the heterokaryotic offspring might provide an overestimate of the rate theoretically expected among the heterokaryotic spores or how the processes of meiosis or sporogenesis might really differ from the theoretical random model.
Homoallelism, inbreeding depression, and consequences for the use of non-sister nuclei heterokaryons
Whatever the reason of the relatively high rate of homoallelism of 50 % resulting from the intramictic process at centromere-unlinked loci in A. subrufescens, it represents a rate of loss of parental heterozygosity similar to the rate of 50 % that could be expected in a classical selfing. However, in agreement with the theoretical model, this rate at centromere-linked loci tends to 100 % for the sister nuclei heterokaryons and to 0 % for the non-sister nuclei heterokaryons (Fig. 4). It is expected that, correlatively, the inbreeding depression in these two categories of heterokaryons should be respectively higher and lower than in a classical selfing. This difference between the two categories of heterokaryons was confirmed by the significantly greater mycelium growth rate on malt agar medium and the higher ability to invade the culture substrate for the non-sister nuclei heterokaryons. However, these potentially fertile heterokaryons remained less vigorous than the parental strain to invade the compost and some of them even failed to fruit. Thongklang et al. (2014) performed some fruiting tests with heterokaryotic offspring from their Brazilian-French hybrid strains and also observed that these poorly fruited compared to the parental hybrid. While it is feasible in A. bisporus var. bisporus (Moquet et al. 1998), because of the inbreeding depression, it is unlikely to obtain non-sister nuclei heterokaryotic single spore isolates that would be as performant as the parent in term of yield but phenotypically different.
Spawn producers who fear that competitors applied such a method to quickly get new cultivars from their protected commercial hybrid strains are presently trying to obtain recognition of the heterokaryotic offspring of A. bisporus var. bisporus as essentially derived varieties (EDV) as defined by the International Union for the protection of new varieties of plants (UPOV) convention (Sonnenberg et al. 2011). Such a procedure should not be required in A. subrufescens, at least for varieties derived from strains studied by us and/or Thongklang et al. (2014).
Properties of the sister nuclei heterokaryons and consequences for their usefulness in cross-breeding
The sister nuclei heterokaryons represented 24–27 %, i.e., about one quarter of the heterokaryotic single spore isolates and one eighth of the total analyzed offspring. Their mycelial growth rate was on average significantly lower than that of the non-sister nuclei heterokaryons and similar to that of the homokaryons on malt agar medium. We used this medium because it is more standard than the compost extract agar medium that we used for the mating tests. Xu (1995) reported that detection of inbreeding depression in A. bisporus varied depending of the culture medium for criteria of fitness as the mycelium growth rate or the interactions between mycelia. Although we did not formally test certain differences between the homokaryons and the sister nuclei heterokaryons, we noted that the latter generally faster grew on compost extract agar medium and gave more visible positive reactions in mating tests (often not easily detected between homokaryons). In these tests, 80 % gave positive reactions with tester homokaryons and thus interacted as well as or possibly better than the homokaryons. In fruiting test, although the tested sister nuclei heterokaryons were infertile like the homokaryons, they better invaded the culture substrate.
The sister nuclei heterokaryons easily crossed also among themselves, but always the same pairs of nuclei were found in the resulting fertile heterokaryotic hybrids among the four possible pairings. This is not surprising for at least two reasons: first, in this particular case of selfing, lethal or deleterious recessive alleles inherited by the sister heterokaryons make certain pairing unviable; second, competition may occur between nuclei or between the new formed heterokaryons; moreover, the more they are vigorous, the greater are their chances of being isolated. In conclusion, sister nuclei heterokaryons are mating-competent heterokaryons behaving like homokaryons. They are possibly even more competent to cross and to survive in the wild due to their heterozygosity. However, they differ from the homokaryons on a major point: they can transmit not only recessive deleterious or lethal alleles but also advantageous alleles that could be on linked loci and which could not be found in viable homokaryotic single spore isolates. Such a linkage has been shown through the Buller phenomenon in A. bisporus, for a locus involved in disease resistance (Callac et al. 2008).
Possible roles of the heterokaryons in nature
As Billiard et al. (2011, 2012) conjectured, we can wonder whether pseudohomothallism evolved to favor intramixis or to achieve universal mating compatibility (i.e., for example, non-sister nuclei heterokaryons can potentially cross via the Buller phenomenon with homokaryons bearing any mating types). The non-sister nuclei heterokaryons poorly fruited and poorly crossed in selfing and rarely with testers homokaryons bearing different mating types. Probably, this was partly due to deleterious or lethal recessive alleles, maybe more particularly due to those located in centromeric regions since sister nuclei heterokaryons crossed much more easily with the same testers. However, the sister nuclei heterokaryons are both unable to fruit and unable to achieve universal mating compatibility. In fact, we believe they simply have a role of sentinel mycelia more vigorous than the homokaryons and waiting for crossing. This hypothesis would agree with the geographical and climatic distribution range of this opportunistic but rarely abundant species, which is one of the widest in the genus.
Breeding strategies for A. subrufescens
Our results highlight certain consequences for breeding strategies in A. subrufescens. Direct selection among non-sister nuclei heterokaryons does not seem a promising method due to the inbreeding depression and, therefore, efforts would focus on outcrossing. This is also the opportunity to exploit genetic resources available at a large geographical range. Depending on the objective and the context, a simple strategy of treating mating-competent sister nuclei heterokaryons as homokaryons may be appropriate for the two following reasons: first, this can avoid selection against alleles of interest that could be present in sister nuclei heterokaryons but absent in homokaryons; secondly, this is faster and cheaper because four or five centromere-unlinked markers are necessary to identify the sister nuclei heterokaryons, while one or two markers including a centromere-linked one are sufficient to identify and discard almost all non-sister nuclei heterokaryons. Such markers as RPB2 and PRS088 in the present study or even better the marker MIP in Thongklang et al. (2014) might be used. Although the reproductive strategy varies among the different strains of A. subrufescens studied by Thongklang et al. (2014), this method should be efficient in all cases. In contrast, other methods based on mycelial growth rate tests, mating tests, or fruiting tests did not appear efficient to reliably identify the sister nuclei heterokaryons.
Finally, we propose a simple method of hybridization for an amphithallic species in which recombination is not suppressed and the postmeiotic nuclei migrate (almost) at random into the spores. This method does not require identifying all the types of spores. Our study is complementary to the recent work of Thongklang et al. (2014) on the interfertility between geographically distant specimens of A. subrufescens, and together, they respond to the questions of what to cross and how to cross them. This contributes to facilitate the genetic improvement of strains of A. subrufescens and other amphithallic species.
References
Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, Giraud T (2011) Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev Camb Philos Soc 86:421–442. doi:10.1111/j.1469-185X.2010.00153.x
Billiard S, López-Villavicencio M, Hood ME, Giraud T (2012) Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J Evol Biol 25:1020–1038. doi:10.1111/j.1420-9101.2012.02495.x
Buller AHR (1931) Research on fungi, vol IV. Longmans, Green and Co, London
Burnett JH, Boulter ME (1963) The mating systems of fungi II. Mating systems of the gasteromycetes, Mycocalia denudata and M. duriaeana. New Phytol 62:217–236. doi:10.1111/j.1469-8137.1963.tb06328.x
Callac P, Billette C, Imbernon M, Kerrigan RW (1993) Morphological, genetic, and interfertility analyses reveal a novel, tetrasporic variety of Agaricus bisporus from the Sonoran desert of California. Mycologia 85:835–851. doi:10.2307/3760617
Callac P, Desmerger C, Kerrigan RW, Imbernon M (1997) Conservation of genetic linkage with map expansion in distantly related crosses of Agaricus bisporus. FEMS Microbiol Lett 146:235–240. doi:10.1016/S0378-1097(96)00482-X
Callac P, Spataro C, Caille A, Imbernon M (2006) Evidence for outcrossing via the Buller phenomenon in a substrate simultaneously inoculated with spores and mycelium of Agaricus bisporus. Appl Environ Microbiol 72:2366–2372. doi:10.1128/AEM.72.4.2366-2372.2006
Callac P, Imbernon M, Savoie JM (2008) Outcrossing via the Buller phenomenon in a substrate simultaneously inoculated with spores and mycelium of Agaricus bisporus creates variability for agronomic traits. In: Lelley JI, Buswell JA (eds) Proceedings of the 6th International Conference on Mushroom Biology and Mushroom Products. GAMU, Krefeld, Germany p 113–119. http://wsmbmp.org/Previous_Conference_6.html
Calvo-Bado L, Noble R, Challen M, Dobrovin-Pennington A, Elliott T (2000) Sexuality and genetic identity in the Agaricus Section Arvenses. Appl Environ Microbiol 66:728–734. doi:10.1128/AEM.66.2.728-734.2000
Challen MP, Elliott TJ (1989) Segregation of genetic markers in the 2-spored secondarily homothallic basidiomycete Coprinus bilanatus. Theor Appl Genet 78:601–607. doi:10.1007/BF00290848
Elliott TJ, Challen MP (1983) Genetic ratios in secondarily homothallic basidiomycetes. Exp Mycol 7:170–174. doi:10.1016/0147-5975(83)90060-9
Evans HJ (1959) Nuclear behaviour in the cultivated mushroom. Chromosoma 10:115–135. doi:10.1007/BF00396566
Foulongne-Oriol M, Spataro C, Cathalot V, Monllor S, Savoie JM (2010) An expanded genetic linkage map of Agaricus bisporus based on AFLP, SSR and CAPS markers sheds light on the recombination behaviour of the species. Fungal Genet Biol 47:226–236. doi:10.1016/j.fgb.2009.12.003
Foulongne-Oriol M, Lapalu M, Ferandon C, Spataro C, Ferrer N, Amslem J, Savoie JM (2014) The first set of expressed sequence tags (EST) from the medicinal mushroom Agaricus subrufescens delivers resource for gene discovery and marker development. Appl Microbiol Biotechnol 98:7879–7892. doi:10.1007/s00253-014-5844-y
Fritsche G (1983) Breeding Agaricus bisporus at the mushroom experimental station, Horst. Mushroom J 122:49–53
Gui Y, Zhu GS, Callac P, Hyde KD, Parra LA, Chen J, Yang TJ, Huang WB, Gong GL, Liu ZY (2014) Agaricus section Arvenses: three new species in highland subtropical Southwest China. Fungal Biol 119:79–94. doi:10.1016/j.funbio.2014.10.005
Imbernon M, Callac P, Gasqui P, Kerrigan RW, Velcko AJ Jr (1996) BSN, the primary determinant of basidial spore number and reproductive mode in Agaricus bisporus, maps to chromosome I. Mycologia 88:749–761. doi:10.2307/3760970
Kamzolkina OV, Volkova VN, Kozlova MV, Pancheva EV, Dyakov YT, Callac P (2006) Karyological evidence for meiosis in the three different types of life cycles existing in Agaricus bisporus. Mycologia 98:763–770. doi:10.3852/mycologia.98.5.763
Kemp RFO (1974) Bifactorial incompatibility in the two-spored basidiomycetes Coprinus sassii and C. bilanatus. Trans Br Mycol Soc 62:547–555. doi:10.1016/S0007-1536(74)80066-5
Kemp RFO (1985) Gene segregation in the 2-spored basidiomycete, Coprinus bilanatus. Heredity 54:391–395
Kennedy ME, Burnett JH (1956) Amphithallism in fungi. Nature 177:882–883. doi:10.1038/177882a0
Kerrigan RW (1993) New prospects for Agaricus bisporus strain improvement. Rep Tottori Mycol Inst 31:188–200
Kerrigan RW (2005) Agaricus subrufescens, a cultivated edible and medicinal mushroom, and its synonyms. Mycologia 100:876–892. doi:10.3852/mycologia.97.1.12
Kerrigan RW, Baller LM, Horgen PA, Anderson JB (1992) Strategies for the efficient recovery of Agaricus bisporus homokaryons. Mycologia 84:575–579. doi:10.2307/3760324
Kerrigan RW, Royer JC, Baller LM, Kohli Y, Horgen PA, Anderson JB (1993) Meiotic behavior and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics 133:225–236
Kerrigan RW, Imbernon M, Callac P, Billette C, Olivier JM (1994) The heterothallic life cycle of Agaricus bisporus var. burnettii, and the inheritance of its tetrasporic trait. Exp Mycol 18:193–210. doi:10.1006/emyc.1994.1020
Kühner R (1977) Variation of nuclear behavior in the homobasidiomycetes. Trans Br Mycol Soc 68:1–16. doi:10.1016/S0007-1536(77)80145-9
Lamoure D (1989) Indices of useful information for incompatibility tests in basidiomycetes. V.—Agaricales sensu lato. Cryptogam Mycol 10:41–80
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181. doi:10.1016/0888-7543(87)90010-3
Lange M (1952) Species concepts in the genus Coprinus. Dansk Bot Arkiv 14:1–140
Langton FA, Elliott TJ (1980) Genetics of secondarily homothallic basidiomycetes. Heredity 45:99–106. doi:10.1038/hdy.1980.53
Largeteau ML, Llarena-Hernández RC, Regnault-Roger C, Savoie JM (2011) The medicinal Agaricus mushroom cultivated in Brazil: biology, cultivation and non-medicinal valorization. Appl Microbiol Biotechnol 92:897–907. doi:10.1007/s00253-011-3630-7
Llarena-Hernández RC, Largeteau ML, Farnet AM, Foulongne-Oriol M, Minvielle N, Regnault-Roger C, Savoie JM (2013) Potential of European wild strains of Agaricus subrufescens for productivity and quality on wheat straw based compost. World J Microbiol Biotechnol 29:1243–1253. doi:10.1007/s11274-013-1287-3
Llarena-Hernández RC, Largeteau ML, Ferrer N, Regnault-Roger C, Savoie JM (2014) Optimization of the cultivation conditions for mushroom production with European wild strains of Agaricus subrufescens and Brazilian cultivars. J Sci Food Agric 94:77–84. doi:10.1002/jsfa.6200
Moquet F, Guedes-Lafargue MR, Mamoun M, Olivier JM (1998) Selfreproduction induced variability in agronomic traits for a wild Agaricus bisporus. Mycologia 90:806–812
Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagy LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, Bailey AM, Billette C, Coutinho PM, Deakin G, Doddapaneni H, Floudas D, Grimwood J, Hildén K, Kües U, LaButti KM, Lapidus A, Lindquist EA, Lucas SM, Murat C, Riley RW, Salamov AA, Schmutz J, Subramanian V, Wösten HAB, Xu J, Eastwood DC, Foster GD, Sonnenberg ASM, Cullen D, de Vries RP, Lundell T, Hibbett DS, Henrissat B, Burton KS, Kerrigan RW, Challen MP, Grigoriev IV, Martin F (2012) Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci U S A 109:17501–17506. doi:10.1073/pnas.1206847109
Neff MM, Neff JD, Chory J, Pepper AE (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14:387–392. doi:10.1046/j.1365-313X.1998.00124.x
Parra LA (2013) Agaricus L. Allopsalliota Nauta & Bas. (Parte II). Candusso, Alassio
Peterson KR, Desjardin DE, Hemmes DE (2000) Agaricales of the Hawaiian Islands. 6. Agaricaceae I: Agaricaceae: Agaricus and Melanophyllum. Sydowia 52:204–257
Quintanilha A (1937) Contribution à l’étude génétique du phénomène de Buller. C R Hebd Seances Acad Sci 205:745–747
Raper CA (1976) Sexuality and life cycle of the edible, wild Agaricus bitorquis. Microbiology 95:54–66. doi:10.1099/00221287-95-1-54
Raper CA, Raper JR, Miller RE (1972) Genetic analysis of the life cycle of Agaricus bisporus. Mycologia 64:1088–1117
Rast D, Stäuble EJ (1970) On the mode of action of isovaleric acid in stimulating the germination of Agaricus bisporus spores. New Phytol 69:557–566. doi:10.1111/j.1469-8137.1970.tb07608.x
Sonnenberg ASM, Baars JJP, Hendrickx PM, Lavrijssen B, Gao W, Weijn A, Mes JJ (2011) Breeding and strain protection in the button mushroom Agaricus bisporus. In: Savoie JM, Foulongne-Oriol M, Largeteau Largeteau M, Barroso G (eds) Proceedings of the 7th International Conference on Mushroom Biology and Mushroom products, vol 1. INRA Bordeaux, France, p 7–15. http://wsmbmp.org/Previous_Conference_7.html
Summerbell RC, Castle AJ, Horgen PA, Anderson JB (1989) Inheritance of restriction fragment length polymorphisms in Agaricus brunnescens. Genetics 123:293–300
Thongklang N, Hoang E, Estrada AER, Sysouphanthong P, Moinard M, Hyde KD, Kerrigan RW, Foulongne-Oriol M, Callac P (2014) Evidence for amphithallism and broad geographical hybridization potential among Agaricus subrufescens isolates from Brazil, France and Thailand. Fungal Biol 188:1013–1024. doi:10.1016/j.funbio.2014.10.004
Wisitrassameewong K, Karunarathna SC, Thongklang N, Zhao R, Callac P, Chukeatirote E, Bahkali AH, Hyde KD (2012a) Agaricus subrufescens: new to Thailand. Chiang Mai J Sci 39:131–146
Wisitrassameewong K, Karunarathna SC, Thongklang N, Zhao R, Callac P, Moukha S, Ferandon C, Chukeatirote E, Hyde KD (2012b) Agaricus subrufescens: a review. Saudi J Biol Sci 19:131–146. doi:10.1016/j.sjbs.2012.01.003
Xu J (1995) Analysis of inbreeding depression in Agaricus bisporus. Genetics 141:137–145
Xu J, Kerrigan RW, Horgen PA, Anderson JB (1993) Localization of the mating type gene in Agaricus bisporus. Appl Environ Microbiol 59:3044–3049
Xu J, Horgen PA, Anderson JB (1996) Somatic recombination in the cultivated mushroom Agaricus bisporus. Mycol Res 100:188–192. doi:10.1016/S0953-7562(96)80119-5
Zhao R, Karunarathna S, Raspe O, Parra LA, Guinberteau J, Moinard M, De Kesel A, Barroso G, Courtecuisse R, Hyde KD, Guelly AK, Desjardin DE, Callac P (2011) Major clades in tropical Agaricus. Fungal Divers 51:279–296. doi:10.1007/s13225-011-0136-7
Acknowledgments
Manuela Brito gratefully acknowledges the Brazilian agencies “Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior” (CAPES) and “Programa de Doutorado Sanduiche no Exterior” (PDSE-CAPES 13006-12-6) for financial support. This work was financially supported by a research project funded by a bilateral cooperation between Mexico (project 115790 CONACYT) and France (ANR-09-BLAN-0391-01). We would like to thank Mrs Heather Yoell for her very helpful assistance in English.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 183 kb)
Rights and permissions
About this article
Cite this article
Rocha de Brito, M., Foulongne-Oriol, M., Moinard, M. et al. Spore behaviors reveal a category of mating-competent infertile heterokaryons in the offspring of the medicinal fungus Agaricus subrufescens . Appl Microbiol Biotechnol 100, 781–796 (2016). https://doi.org/10.1007/s00253-015-7070-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7070-7