Abstract
The genome of the amphotericin producer Streptomyces nodosus was sequenced. A single scaffold of 7,714,110 bp was obtained. Biosynthetic genes were identified for several natural products including polyketides, peptides, siderophores and terpenes. The majority of these clusters specified known compounds. Most were silent or expressed at low levels and unlikely to compete with amphotericin production. Biosynthesis of a skyllamycin analogue was activated by introducing expression plasmids containing either a gene for a LuxR transcriptional regulator or genes for synthesis of the acyl moiety of the lipopeptide. In an attempt to boost amphotericin production, genes for acyl CoA carboxylases, a phosphopantetheinyl transferase and the AmphRIV transcriptional activator were overexpressed, and the effects on yields were investigated. This study provides the groundwork for metabolic engineering of S. nodosus strains to produce high yields of amphotericin analogues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many antibiotics and other drugs are synthesised by micro-organisms (Koehn and Carter 2005). There is a need for a continuous stream of new anti-infectives to counteract the inevitable emergence of resistance in pathogenic microbes. However, few new antibiotics have been identified since the 1960s, suggesting that the supply of easily accessible bioactive natural products is limited (Li and Vederas 2009; Worthington and Melander 2013). Several approaches have been used in an effort to discover micro-organisms that synthesise new antibiotics. High throughput methods have been used to culture larger numbers of potential producer organisms, and more sensitive bioassays have been devised to improve detection of hit compounds (Wang et al. 2006). New producers have also been obtained by sampling of previously unexplored environments and by investigating bacterial symbionts of higher organisms (Wilson et al. 2014). A recent breakthrough has led to laboratory cultivation of soil bacteria that were previously thought to be unculturable (Ling et al. 2015). Re-discoveries of known compounds can be identified and disregarded at an early stage in the screening process (Koehn and Carter 2005; Fair and Tor 2014). As well as direct screening, genome sequencing has revealed that actinomycetes and other bacteria contain silent biosynthetic genes for natural products.
The genomes of three important antibiotic producers Streptomyces coelicolor, Streptomyces avermitilis and Saccharopolyspora erythraea were determined by the Sanger dideoxy sequencing method (Bentley et al. 2002; Ikeda et al. 2003; Oliynyk et al. 2007). Since then, next-generation technologies have allowed rapid and affordable sequencing of many actinomycete genomes. Bioinformatic methods are available for rapid identification of gene clusters for production of metabolite classes for which biosynthesis is relatively well understood (Weber et al. 2015). Some cryptic gene clusters have been activated by changing growth conditions or by overproduction of transcriptional activators (Scherlach and Hertweck 2009; Laureti et al. 2011). Genome sequences are also valuable for unravelling new biosynthetic pathways (Rackham et al. 2010) and for metabolic engineering to increase yields of specific secondary metabolites.
Streptomyces nodosus is the only known producer of amphotericin B (Fig. 1), a medically important antifungal antibiotic. This drug is the most effective treatment for life-threatening systemic mycoses, but has severe side effects. A few less toxic analogues have been obtained by engineering of biosynthetic genes (Stephens et al. 2012; De Poire et al. 2013). Medicinal chemists have also generated promising derivatives by modifying amphotericin B isolated from fermentation cultures of the producer organism (Yamamoto et al. 2015; Davis et al. 2015; Volmer et al. 2010). In this study, we analysed the genome of S. nodosus. The aim was to provide a reference sequence for this industrially important micro-organism. The sequence was mined for biosynthetic genes for other natural products. This was done to identify gene clusters that might specify potentially valuable compounds, as well as pathways that might divert precursors away from amphotericin biosynthesis in producer strains. In addition, genes that affect amphotericin yield were identified. We carried out the first experiments aimed at increasing production of the less haemolytic 16-descarboxyl-16-methyl-analogues (Carmody et al. 2005). Genes for acyl CoA carboxylases, a phosphopantetheinyl transferase and a transcriptional activator were overexpressed and the effects on yields of these amphotericins were investigated.
Materials and methods
Bacterial strains and growth conditions
The S. nodosus wild type strain was ATCC 14899. This organism has also been deposited as strain IMD2693 in culture collection WDCM227 held at University College Dublin. S. nodosus ∆amphNM and S. nodosus ∆amphI were from our laboratory collection. Strains were grown on TS medium or Streptomyces medium. Protoplast transformation was carried out as described (Kieser et al. 2000). Escherichia coli TG1 was used as a host for plasmid constructions. Agar diffusion assays for antibacterial activity were carried out using Bacillus subtilis as an indicator organism.
Genome sequencing
Total cellular DNA was isolated as described previously (Caffrey et al. 2001) and further purified using a QIAGEN 500/G column. The genome was sequenced by MWG Biotech (Eurofins Genomics, Ebersberg, Germany) using a 454 Life Sciences FLX sequencer. Initially, 130 large contigs were obtained. Paired-end techniques were used to assemble these into a single scaffold of 7.7 Mb DNA representing the entire genome. The draft sequence contained several gaps. The largest of these was 11 kb. This region was cloned from a cosmid library (Caffrey et al. 2001). Positive clones were identified by PCR with primers matching sequences adjacent to the gap. The remaining gaps were less than 3 kb. The regions containing these gaps were amplified by PCR and sequenced directly by the dideoxy method. The current version of the sequence contains 96 gaps, of which 16 are estimated at between 100 and 400 nucleotides in length, the rest are estimated as tens of nucleotides. Filling of these gaps is still in progress. None of the gaps is in a natural product biosynthetic gene cluster. The sequence has been deposited in the GenBank database with accession number CP009313. Automated annotation is available online. Biosynthetic gene clusters were identified using antiSMASH (Weber et al. 2015) and were manually curated.
Construction of expression plasmids
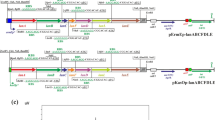
PCR was carried out using Phusion DNA polymerase (New England BioLabs Inc.) and the primers are listed in Table 1. The S. nodosus accA2 gene was amplified with primers PSAf3 and PSAr3. The accB2 gene was amplified with PSAf6 and PSAr6. The two PCR products were digested with Sac I and ligated together. The ligated DNA was cut with NsiI and HindIII and cloned between the Pst I and Hin dIII sites of the pIAGO expression vector (Aguirrezabalaga et al. 2000). The resulting construct was named pIAGO-PSA3-6 (contains accA2 and accB2 and its associated ε gene).
The S. nodosus pccB gene was amplified with primers PSAf2 and PSAr2v2. The DNA was digested with Sac I and ligated to the Sac I-cut PCR-amplified accA2 gene, as in construction of the previous plasmid. The accA2-pccB DNA was digested with Nsi I and Hin dIII and cloned between the Pst I and Hin dIII sites of pIAGO. The resulting construct was named pIAGO-PSA2-3V2 (contains accA2 and pccB with its ε gene).
Construction of the other expression plasmids was straightforward. Biosynthetic genes for the lipopeptide acyl moiety were amplified using primers PS1 and PS4. The Lux, R4, Ppt, and Epim forward and reverse primers were used to amplify the genes for the LuxR protein, AmphRIV, phosphopantetheine transferase, and epimerase. Hin dIII and Bam HI or Bgl II sites were incorporated to allow cloning between the Bam HI and Hin dIII sites of the vector. A list of all the expression plasmids constructed is given in Table 2.
Extraction of lipopeptide
S. nodosus ∆amphI strains transformed with pIAGO-LuxR, pIAGO-SkyPKS or pIAGO were grown for 5 days on fructose-dextrin medium (Caffrey et al. 2001). Cultures were centrifuged to sediment mycelia cells. The supernatant fractions were extracted with ethyl acetate and the pellet fractions were extracted with methanol. The extracts were concentrated by rotary evaporation and analysed by HPLC. Authentic skyllamycin A was provided by Professor Roderich Sussmuth, Technical University of Berlin, Germany.
HPLC and mass spectrometry
HPLC was carried out on a Varian ProStar HPLC system with a ProStar 335 photodiode array detector. All separations were with the use of an Agilent Zorbax SB-C18 (4.6 × 150 mm, 5 μm for analytical and 9.4 × 150 mm, 5 μm for semi-preparative). The solvents used were A = H2O (0.1 % TFA) and B = acetonitrile (0.1 % TFA). The analytical runs were carried out at 1 ml/min and the semi-preparative at 4 ml/min. The gradient was 5 % B for 2 min, 5 to 35 % B over 6 min, 35 to 100 % B over 13 min, 100 % B for 1 min, 100 to 5 % B over 3 min.
Low-resolution mass spectrometry analysis was carried out using a Waters Quattro Micro tandem quadrupole mass spectrometer in both positive and negative ion modes. The ion spectra were determined up to m/z 2000. High resolution measurements were determined using a time-of-flight instrument (Waters Corporation, Micromass Ltd., Manchester, UK).
Analysis of amphotericin production
For accurate comparisons, it was necessary to calculate total polyene yields per gramme dry weight of biomass. To compare polyene yields, S. nodosus ∆amphNM and transformants were grown for 5 days at 30 °C with shaking in Streptomyces medium containing 1 % (w/v) glycerol. To determine dry weights, mycelial cells from 100-ml cultures were sedimented and washed three times with purified water. The pellets were left at 50 °C until the dry weight values were constant.
Polyene yields were estimated by UV-visible spectrophotometry. A sample of each culture was added to an equal volume of butanol and shaken to extract all the polyene into the organic layer. In most cases, a single extraction was sufficient to recover all of the polyene. A320 and A405 were measured to estimate yields of 16-descarboxyl-16-methyl-amphotericin B and 8-deoxy-16-descarboxyl-16-methyl-amphotericin A, with extinction coefficients of 71,500 and 190,000 M−1 cm−1 respectively. These values were combined to obtain the total polyene yield. Triplicate cultures were grown to estimate dry weights and polyene yields.
GC-MS analysis
GC-MS was carried out on organic extracts using a 6890N network GC system, 7683B series injector and 5973 inert mass selective detector, all by Agilent technologies. Samples for derivatisation were incubated with 50 μl N-methyl-N-(trimethylsilyl) trifluoroacetamide at 100 °C for 1 h. Derivatised and non-derivatised samples (1 μl) were injected onto a HP5 MS column. The oven temperature was held at 150 °C for 2 min then raised to 300 °C over 8 min with a run time of 17 min. The operating mass range was from m/z 100–500.
Results
Natural product biosynthetic gene clusters in the S. nodosus genome
The genome sequence was mined to identify biosynthetic genes for secondary metabolites. Twenty-four clusters were initially identified using antiSMASH (Table 3). These included five clusters containing polyketide synthase (PKS) genes, and eight containing non-ribosomal peptide synthetase (NRPS) protein genes. There were also clusters capable of synthesising a bacteriocin, ectoine, a lantipeptide, a butyrolactone, and four terpenes. Another three unlinked clusters (7, 10 and 13) function in biosynthesis and transport of an aerobactin-type hydroxamate siderophore (Challis 2005). Most of the clusters showed a high degree of sequence identity with previously characterised clusters.
The amphotericin cluster includes genes for the only modular PKS encoded within the genome. PKS2, PKS3 and PKS4 are likely to be important for the fitness of the producer organism. PKS2 functions in synthesis of hexahydroxyperylenequinone (HPQ) melanin that protects against UV radiation (Funa et al. 2005). PKS3 synthesises Whi spore pigment compounds (Shen et al. 1999), and PKS4 synthesises alkylresorcinol lipids that confer rigidity on the cytoplasmic membrane (Funabashi et al. 2008) PKS5 appears to be capable of synthesising the angucycline urdamycin G (Decker and Haag 1995). Interestingly, the ketosynthase (KS) α gene from this cluster was independently identified by PCR with degenerate primers (Chuck et al. 2006). We previously cloned this region from a cosmid library and generated targeted deletions in the chromosome of S. nodosus ∆amphNM; the deletion mutants did not produce increased yields of 16-descarboxyl-16-methyl-amphotericins (P. Caffrey, unpublished data). An organism closely related to S. nodosus produces saquayamycins (Uchida et al. 1985). However, the fact that angucyclines have not been observed suggests that cluster 5 is silent or expressed at a very low level under growth conditions favourable for amphotericin production.
While the PKS genes are related to previously characterised clusters, some of the NRPSs are novel. More detailed descriptions of new NRPS clusters are presented in the supplementary information. Four of the eight NRPSs (NRPS 1, 2, 5, and 6) contain activation (A) and thiolation (T) domains but not condensation domains; these may form modified amino acids rather than peptide products, or they may function in pathways that have yet to be elucidated (Figs. S1, S2, S3, S4; Tables S1, S2, S3, S4). NRPS3 is closely related to the NRPS for skyllamycins A and B (Pohle et al. 2011). These are 11-residue cyclic depsipeptides modified with a 2-[1-(Z)-propenyl]-cinnamoyl lipid moiety (Fig. 2). The S. nodosus cluster contains a gene (SNOD_28885) for an additional cytochrome P450 (Table S5), suggesting that the product is a skyllamycin analogue with an additional hydroxyl or epoxide group. This cluster is normally silent or expressed at a low level. NRPS4 consists of two multienzyme polypeptides and is related to the pyochelin NRPS of Streptomyces scabiei (Seipke et al. 2011). The gene for the first NRPS4 protein contains a frameshift mutation, which was verified by PCR and re-sequencing. In future work, repair of this frameshift should give a functional assembly line that uses a hydroxybenzoate primer and incorporates and heterocyclises three cysteine residues to thiazoles (Figs. S5 and S6, Table S6). Pyochelin is synthesised in a similar way except that only two cysteines are incorporated.
NRPS7 consists of three NRPS modules and a PKS module housed within two multienzyme polypeptides (Fig. S7, Table S7). NRPS8 is almost identical to the system that synthesises coelichelin in S. coelicolor (Lautru et al., 2005) (Fig. S8, Table S8).
S. nodosus has the genes for production of the terpenoids albaflavenone, geosmin and hopanoids (clusters 9, 11 and 14). Biosynthesis of these compounds has been characterised in other Streptomycetes (Seipke and Loria 2009; Cane and Ikeda 2012; Zhao et al. 2008; Bradley et al. 2010). Cluster 19 contains a putative terpene cyclase with 32 % identity to the pentalenene cyclase of Streptomyces exfoliatus and 31 % identity with avermitilol cyclase SAV_76 of S. avermitilis (Cane and Ikeda 2012). The genome contains genes for all enzymes of the methyl erythritol pathway. These are not clustered.
Cluster 1 includes a gene (SNOD_01550) for a bacteriocin that is 54 % identical to linocin from Brevibacterium linens (Valdes-Stauber and Scherer 1994). Cluster 5 contains three genes specifically required for biosynthesis of ectoine, an osmotic stabiliser (Reshetnikov et al. 2011). Cluster 24 includes a homologue (SNOD_34350) of the gene for the AfsA protein that synthesises A factor from dihydroxyacetone phosphate and a methyl-branched 3-ketoacyl thioester (Kato et al. 2007). The adjacent SNOD_34345 gene encodes a butyrolactone receptor. This suggests that these compounds function in quorum sensing in populations of S. nodosus cells. The lantipeptide encoded by cluster 12 may also function in intercellular communication.
Activation of skyllamycin-like lipopeptide biosynthesis
Three different skyllamycins have been identified so far (Fig. 2). Skyllamycin A has anti-cancer activity (Pohle et al. 2011) whereas skyllamycins B and C inhibit bacterial biofilms (Navarro et al. 2014). Slight structural changes can alter biological activity. We attempted to activate the S. nodosus cluster encoding a putative skyllamycin analogue. The NRPS genes are preceded by a 10-kb region containing genes for two acyl carrier proteins and multiple discrete KS, ketoreductase (KR), and dehydratase (DH) enzymes that assemble the substituted cinnamoyl lipid moiety. The PKS and NRPS genes are apparently transcribed in the same direction (Fig. S9). The PKS genes were amplified with primers PS4 and PS1 and cloned into pIAGO. The construct pIAGO-SkyPKS was transformed into an amphI mutant of S. nodosus that does not produce amphotericins. We did not detect a substituted cinnamate by GC-MS or HPLC. However, a new metabolite appeared that had a UV-visible absorption spectrum very similar to that of skyllamycin A, but had a slightly earlier retention time (Fig. 3 and Fig. S10). It is possible that the product of the lipid biosynthetic genes activates the cluster, or alternatively, the 11-kb insert causes integration of the construct into the chromosome by homologous recombination. This would place the cluster downstream from the strong ermE* promoter. The luxR (SNOD_28895) gene was also cloned into pIAGO and transformed into S. nodosus ∆amphI. The new putative lipopeptide was again produced. Extracts containing the new compound were not active against B. subtilis. However, even at concentrations of 105 to 140 μM, skyllamycin A only has weak antibacterial activity (Toki et al. 2001).
Skyllamycin A has a mass of 1482.7 (Pohle et al. 2011). The new S. nodosus metabolite was purified by preparative HPLC and analysed by mass spectrometry (Fig. S11). This revealed that the new compound had a mass appropriate for a hydroxylated skyllamycin A ([M + Na]+ = 1521.4). Further work will be required to identify the hydroxylation site and to investigate biological activities.
Genes affecting amphotericin production
The amphotericin cluster has previously been extensively characterised (Caffrey et al. 2001). This contains PKS and cytochrome P450 genes for assembly and modification of the macrolactone core, genes involved in conversion of GDP-α-D-mannose to GDP-D-mycosamine, a mycosamine glycosyltransferase, and export and regulatory genes. Many unlinked genes also contribute to amphotericin production. These include genes for phosphopantetheinylation of PKS acyl carrier protein (ACP) domains, and genes involved in acyl CoA precursor supply. In this study, some of these genes were investigated, with a view to increasing the yields of amphotericin B and its analogues. We have previously shown that phosphomannose isomerase (SNOD_13725) and phosphomannomutase (SNOD_13740) are important for generating GDP-α-D-mannose for mycosamine formation (Nic Lochlainn and Caffrey 2009).
Acyl CoA carboxylases
Polyketides are synthesised from activated acyl units, commonly malonyl CoA and (2S)-methylmalonyl CoA (Marsden et al. 1994). There is evidence that overproduction of acyl CoAs in cells can boost polyketide production (Ryu et al. 2006; Olano et al. 2008). Malonyl CoA is synthesised from acetyl CoA and carbon dioxide in an ATP-requiring reaction catalysed by acetyl CoA carboxylase. Methylmalonyl CoA can be formed by a number of pathways. In one of these, (2S)-methylmalonyl CoA is synthesised by carboxylation of propionyl CoA. Another important pathway involves rearrangement of succinyl CoA to form (2R)-methylmalonyl CoA, which can be epimerised to the 2S stereoisomer by an epimerase (Leadlay and Fuller 1983). Two other pathways to methylmalonyl CoA are known (Li et al. 2004). However, the propionyl CoA carboxylase route appears to be the most important for complex polyketide biosynthesis (Murli et al. 2003).
Acetyl CoA and propionyl CoA carboxylases each contain α and β subunits (Rodriguez and Gramajo 1999). The α subunit contains carboxylase and biotin carrier protein domains. The carboxylase catalyses ATP-dependent carboxylation of the biotin prosthetic group. The β subunit binds acetyl CoA or propionyl CoA and catalyses transfer of the carboxyl group from biotin to C-2 of the acyl group to form the dicarboxylic acyl thioesters. Some acyl CoA carboxylases also have an ε subunit that aids association between α and β subunits.
In S. nodosus the SNOD_20870 accA2 gene encodes acetyl CoA carboxylase α subunit. SNOD_23485 encodes acetyl CoA carboxylase β subunit gene accB2. The SNOD_20890 pccB gene encodes propionyl CoA carboxylase β subunit. The S. nodosus accB2 and pccB genes both have downstream ε subunit genes.
The SNOD_20870, SNOD_23485 and SNOD_20890 genes are homologous to the S. coelicolor SCO4921 accA2, SCO5535 accB and SCO4926 pccB genes, respectively. [There is no S. nodosus counterpart of the non-essential S. coelicolor accA1 gene SCO6271]. These S. coelicolor enzymes have been experimentally characterised. The S. coelicolor AccA2 SCO4921 protein can interact with the SCO5535 β subunit to form malonyl CoA, or with SCO4926 to form (2S)-methylmalonyl CoA (Diacovich et al. 2002). The β subunits must have their cognate ε subunits for good activity in vitro. Overexpression of these enzymes in S. coelicolor gave a sixfold increase in yield of actinorhodin (Ryu et al. 2006).
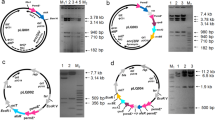
S. nodosus has another acetyl CoA carboxylase β subunit gene accB1 SNOD_29345 which is linked to the urdamycin cluster and lacks an associated ε subunit gene. Expression plasmids were constructed for the S. nodosus acetyl and propionyl CoA carboxylases, as shown in Fig. 4.
The mutase/epimerase pathway for generation of 2S methylmalonyl CoA
The S. nodosus genome contains genes for three methylmalonyl CoA mutases. SNOD_20515 and SNOD_27715 each encode α2 homodimeric enzymes, whereas SNOD_29730 plus SNOD_29735 encode an αβ heterodimer. SNOD_26810 appears to encode an isobutyryl CoA mutase. The S. nodosus methylmalonyl CoA mutase enzymes should be capable of synthesising (2R)-methylmalonyl CoA from succinyl CoA. However, there is no obvious gene for a methylmalonyl CoA epimerase that can interconvert (2R)- and (2S)- methylmalonyl CoA. Possibly, this activity is present, but the gene cannot be identified by sequence homology. Some of these epimerases are incorrectly annotated as glyoxylases (Gross et al. 2006). A gene for this enzyme had been identified in a preliminary draft genome sequence of an aromatic heptaene producer, Actinoplanes caeruleus (Stephens et al. 2013). The GenBank accession number for the epimerase region is KT374297. This gene was amplified and cloned into pIAGO and the construct was transformed into S. nodosus. The aim was to investigate whether overexpression of the epimerase would increase yields of amphotericins and analogues.
Phosphopantetheine transferases
The ACPs and peptidyl carrier proteins (PCPs) involved in fatty acid, polyketide and non-ribosomal peptide biosynthesis are post-translationally modified with phosphopantetheine prosthetic groups. There are three 4′-phosphopantetheine transferases encoded in the S. nodosus genome. These were assigned functions based on size, conserved motifs, and homology to known phosphopantetheinyl transferases (Beld et al. 2014) One, SNOD_19940 protein, is likely to be involved in modifying discrete ACPs involved in fatty acid biosynthesis. Another, SNOD_28340 protein, is likely to be involved in modifying ACP domains in NRPSs and PKSs. The third, SNOD_29350 protein, is encoded within the urdamycin cluster and is probably dedicated to ACPs involved in biosynthesis of this aromatic polyketide (Wang et al. 2001). Jiang et al. (2013) found that overproduction of a Sfp-type phosphopantetheinyl transferase in Streptomyces chattanoogensis L10 gave a 40 % increase in production of pimaricin. The SNOD28340 gene for an Sfp-type phosphopantetheine transferase was amplified and cloned into pIAGO.
AmphRIV
Regulation of nystatin and pimaricin biosynthesis has been thoroughly investigated (Sekurova et al. 2004; Santos-Alberturas et al. 2011, 2012). From these studies, it is clear that AmphRIV and homologues are pathway-specific regulators that activate transcription of polyene biosynthetic genes. The amphRIV gene was also cloned into pIAGO.
Effects of expression plasmids on polyene yields
Transformations were carried out to introduce the five expression plasmids and the empty pIAGO vector into the S. nodosus ∆amphNM mutant. Transformants were grown and total polyene yields per g dry weight of biomass were determined.
In strain ∆amphNM, neither the acetyl CoA carboxylase (AccA2-AccB2) nor the propionyl CoA carboxylase (AccA2-PccB) gave a significant increase in yield (Fig. 5). The epimerase and phosphopantetheine transferase also made little difference. Overexpression of the amphRIV gene gave a fourfold increase in yield as compared to the untransformed strain. This indicates that the levels of PKS proteins are the limiting factor in amphotericin production.
Discussion
The S. nodosus genome was sequenced for two reasons: to search for biosynthetic gene clusters for potentially valuable natural products and to enable attempts to increase yields of amphotericin B and analogues. The genome was found to contain 24 biosynthetic gene clusters for natural products. Of these, 18 are apparently capable of specifying known metabolites or closely related compounds. This is not surprising as rediscovery is common in Streptomyces species (Koehn and Carter 2005). It is becoming clear that rare non-streptomycete actinomycetes are a more promising source of new chemical entities (Jose and Jebukumar 2013). Of the clusters that specify unknown products, four contain NRPS adenylylation and thiolation domains but no condensation domains and are likely to activate and modify single amino acid residues. NRPS7 appears to synthesise a peptide-ketide hybrid compound from three amino acids and an organic acid. These enzymes may be amenable to overexpression and characterisation in vitro, which may assist identification of their products. NRPS3 and NRPS4 are of interest because they have the potential to synthesise analogues of skyllamycins and pyochelins. Skyllamycins are important because they are active against some cancers and against bacterial biofilms. Skyllamycin A inhibits mitosis of certain tumour cells by interfering with the platelet-derived growth factor signalling pathway (Tori et al. 2001; Pohle et al. 2011). It also has weak antibiotic activity against B. subtilis and Staphylococcus aureus but not against Gram-negative bacteria or fungi. Skyllamycins B and C are non-antibiotic inhibitors of biofilms caused by the opportunistic human pathogen Pseudomonas aeruginosa (Navarro et al. 2014). Skyllamycin B is capable of dispersing biofilms and may be important in combination therapies for treating respiratory tract infections in cystic fibrosis patients (Navarro et al. 2014). Slight structural changes have profound effects on biological activity. We have shown that S. nodosus can be activated to produce a new skyllamycin analogue. This compound will be investigated further in future work.
Repair of the frameshift in the SNOD_29570 gene for the first NRPS4 protein may result in synthesis of a novel siderophore related to pyochelin. This will be investigated in future work. Pyochelin siderophores are important virulence factors in P. aeruginosa. Pyochelin analogues have been synthesised that exert antibiotic activity because they block iron uptake (Mislin et al. 2006). Siderophores have also been conjugated to drugs to improve penetration of the outer membranes of Gram-negative pathogens (Starr et al. 2014).
The polyketide macrolactone of amphotericin B is assembled from activated acyl precursors, 16 acetyl and three propionyl units. Overproduction of AmphRIV gave a fourfold increase in yield in the ΔamphN strain. This agrees with findings of other groups who have investigated the effects of AmphRIV homologues on production of other polyenes (Sekurova et al. 2004; Santos-Alberturas et al. 2011, 2012). Elevation of AmphRIV is likely to increase transcription of PKS and late genes. The cellular levels of PKS proteins appear to be limiting, because overexpression of acetyl and propionyl CoA carboxylase genes made little difference to polyene production. Overexpression of acetyl CoA carboxylase significantly increased yields of actinorhodin in S. coelicolor (Ryu et al. 2006), but the small discrete enzymes of a type II PKS are more rapidly translated than the largest hexamodular amphotericin PKS proteins. These considerations suggest that with aromatic polyketide biosynthesis, precursor supply may be the limiting factor whereas with polyene biosynthesis, it is the abundance of the PKS catalyst. While Jiang et al. (2013) found that overproduction of phosphopantetheine transferase gave a modest increase in pimaricin production, this approach did not increase the yield of amphotericin. This indicates that the basal level of SNOD_28340 phosphopantetheine transferase is sufficient for post-translational modification of all 19 ACP domains in the amphotericin PKS.
Further work should lead to greater increases in yields of amphotericins and analogues. With new methods for disruption of chromosomal genes (Cobb et al. 2015), this can be achieved more efficiently. The availability of a reference sequence will also allow identification of mutations that increase yields in high producer strains generated by traditional strain improvement techniques. These methods involve radiation or chemical mutagenesis, followed by screening for mutants that give higher yields. This approach is laborious but genome sequencing of improved strains can give insights into secondary metabolism by revealing which genes are affected in high producers (Wang et al. 2014). The provision of a reference sequence is essential for this approach.
This study reports the first sequencing and analysis of the S. nodosus genome. The work has led to isolation of a new skyllamycin analogue, and has provided the basis for further metabolic engineering to improve amphotericin yields.
References
Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Brana AF, Mendez C, Salas JA (2000) Identification and expression of genes involved in biosynthesis of L-oleandrose and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob Agents Chemother 44:1266–1275
Beld J, Sonnenschein EC, Vickery CE, Noel JP, Burkart MD (2014) The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep 2014(31):61–108
Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bradley AS, Pearson A, Sáenz JP, Marx CJ (2010) Adenosylhopane: the first intermediate in hopanoid side chain biosynthesis. Org Geochem 41:1075–1081
Caffrey P, Lynch S, Flood E, Finnan S, Oliynyk M (2001) Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem. Biol. 8:713–723
Cane DE, Ikeda H (2012) Exploration and mining of the bacterial terpenome. Acc Chem Res 45:463–472
Carmody M, Murphy B, Byrne B, Power P, Rai D, Rawlings BJ, Caffrey P (2005) Biosynthesis of amphotericin derivatives lacking exocyclic carboxyl groups. J. Biol. Chem. 280:34420–34426
Challis GL (2005) A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6(4):601–611
Chuck J-A, Dunn C, Facultad F, Nakazone C, Nikodinovic J, Barrow KD (2006) Amplification of DNA encoding entire type I polyketide synthase domains and linkers from Streptomyces species. Curr Microbiol 53:89–94
Cobb RE, Wang Y, Zhao H (2015) High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol 4(6):723–728
Davis SA, Vincent BM, Endo MM, Whitesell L, Marchillo K, Andes DR, Lindquist S, Burke MD (2015) Nontoxic antimicrobials that evade drug resistance. Nature Chem Biol 11(7):481–487
Decker H, Haag S (1995) Cloning and characterization of a polyketide synthase gene from Streptomyces fradiae Tü2717, which carries the genes for biosynthesis of the angucycline antibiotic. J Bacteriol 177(21):6126–6136
De Poire E, Stephens N, Rawlings BJ, Caffrey P (2013) Engineered biosynthesis of disaccharide-modified polyene macrolides. Appl Environ Microbiol 79(19):6156–6159
Diacovich L, Peirú S, Kurth D, Rodríguez E, Podestá F, Khosla C, Gramajo H (2002) Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J Biol Chem 277:31228–31236
Fair RJ, Tor Y (2014) Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem 6:25–64
Funa N, Funabashi M (2005) Biosynthesis of hexahydroxyperylenequinone melanin via oxidative aryl coupling by cytochrome P-450 in Streptomyces griseus. J Bacteriol 187(23):8149–8155
Funabashi M, Funa N, Horinouchi S (2008) Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 283(20):13983–13991
Gross F, Ring MW, Perlova O, Fu J, Schneider S, Gerth K, Kuhlmann S, Stewart AF, Zhang Y, Muller R (2006) Metabolic engineering of Pseudomonas putida for methylmalonyl-CoA biosynthesis to enable complex heterologous secondary metabolite formation. Chem. Biol. 13:1253–1264
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Jiang H, Wang Y-Y, Ran X-X, Fan W-M, Jiang X-H, Guan W-J, Li Y-Q (2013) Improvement of natamycin production by engineering of phosphopantetheinyl transferases in Streptomyces chattanoogensis L10. Appl Environ Microbiol 79:3346–3354
Jose PA, Jebakumar SRD (2013) Non-streptomycete actinomycetes nourish the current microbial antibiotic drug discovery. Front Microbiol 4:240
Kato J-Y, Funa N, Watanabe H, Ohnishi Y, Horinouchi S (2007) Biosynthesis of gamma-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Nat. Acad. Sci. USA 104:2378–2383
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich United Kingdom
Koehn FE, Carter GT (2005) The evolving role of natural products in drug discovery. Nat Rev Drug Discov 4:206–220
Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B (2011) Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA 108:6258–6263
Lautru S, Deeth RJ, Bailey LM, Challis GL (2005) Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nature Chem Biol 1:265–269
Leadlay PF, Fuller JQ (1983) Proton transfer in methylmalonyl-CoA epimerase from Propionibacterium shermanii. Studies with specifically tritiated (2R)-methylmalonyl-CoA as substrate. Biochem J 213(3):635–642
Li C, Florova G, Akopiants K, Reynolds KA (2004) Crotonyl-coenzyme a reductase provides methylmalonyl-CoA precursors for monensin biosynthesis by Streptomyces cinnamonensis in an oil-based extended fermentation. Microbiology 150:3463–3472
Li JW, Vederas JC (2009) Drug discovery and natural products: end of an era or an endless frontier? Science 325(5937):161–5
Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones MJ, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459
Marsden AFA, Caffrey P, Aparicio JF, Loughran MS, Staunton J, Leadlay PF (1994) Stereospecific acyl transfers on the erythromycin-producing polyketide synthase. Science 263:378–380
Mislin GLA, Hoegy F, Cobessi D, Poole K, Rognan D, Schalk IJ (2006) Binding properties of pyochelin and structurally related molecules to FptA of Pseudomonas aeruginosa. J Mol Biol 357:1437–1448
Murli S, Kennedy J, Dayem LC, Carney JR, Kealey JT (2003) Metabolic engineering of Escherichia coli for improved 6-deoxyerythronolide B production. J Ind Microbiol Biotechnol 30:500–509
Navarro G, Cheng AT, Peach KC, Bray WM, Bernan VS, Yildiz FH, Linington RG (2014) Image-based 384-well high-throughput screening method for the discovery of skyllamycins a to C as biofilm inhibitors and inducers of biofilm detachment in Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:1092–1099
Nic Lochlainn L, Caffrey P (2009) Phosphomannose isomerase and phosphomannomutase gene disruptions in Streptomyces nodosus: impact on amphotericin biosynthesis and implications for glycosylation engineering. Metab Eng 11(1):40–47
Olano C, Lombo F, Mendez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292
Oliynyk M, Samborskyy M, Lester JB, Mironenko TA, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nature Biotechnol. 25:447–453
Pohle S, Appelt C, Roux M, Fiedler H-P, Süssmuth RD (2011) Biosynthetic gene cluster of the non-ribosomally synthesized cyclodepsipeptide skyllamycin: deciphering unprecedented ways of unusual hydroxylation reactions. J Am Chem Soc 133(16):6194–6205
Rackham EJ, Grüschow S, Ragab AE, Dickens S, Goss RJM (2010) Pacidamycin biosynthesis: identification and heterologous expression of the first uridyl peptide antibiotic gene cluster. Chembiochem 11(12):1700–1709
Reshetnikov AS, Khmelenina VN, Mustakhimov II, Trotsenko YA (2011) Genes and enzymes of ectoine biosynthesis in halotolerant methanotrophs. Meth. Enzymol. 495:15–30
Rodríguez E, Gramajo H (1999) Genetic and biochemical characterization of the α and β components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2). Microbiology 145:3109–3119
Ryu Y-G, Butler MJ, Chater KF, Lee KJ (2006) Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl Environ Microbiol 72(11):7132–7139
Santos-Aberturas J, Vicente CV, Guerra SM, Payero TD, Martín JF, Aparicio JF (2011) Molecular control of polyene macrolide biosynthesis: direct binding of the regulator PimM to eight promoters of pimaricin genes and identification of binding boxes. J. Biol. Chem. 286:9150–9161
Santos-Aberturas J, Vicente CM, Payero TD, Martín-Sánchez L, Cañibano C, Martín JF, Aparicio JF (2012) Hierarchical control on polyene macrolide biosynthesis: PimR modulates pimaricin production via the PAS-LuxR transcriptional activator PimM. PLoS One 7(6):e38536
Scherlach K, Hertweck C (2009) Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem 7:1753–1760
Sekurova ON, Brautaset T, Sletta H, Borgos SEF, Jakobsen ØM, Ellingsen TE, Strøm AR, Valla S, Zotchev SB (2004) In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J Bacteriol 186:1345–1354
Seipke RF, Loria R (2009) Hopanoids are not essential for growth of Streptomyces scabies 87–22. J Bacteriol 191:5216–5223
Seipke RF, Song L, Bicz J, Laskaris P, Yaxley AM, Challis GL, Loria R (2011) The plant pathogen Streptomyces scabies 87–22 has a functional pyochelin biosynthetic pathway that is regulated by TetR- and AfsR-family proteins. Microbiology 157:2681–2693
Shen Y, Yoon P, Yu TW, Floss HG, Hopwood D, Moore BS (1999) Ectopic expression of the minimal whiE polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes. Proc Natl Acad Sci USA 96:3622–3627
Starr J, Brown MF, Aschenbrenner L, Caspers N, Che Y, Gerstenberger BS, Huband M, Knafels JD, Lemmon MM, Li C, McCurdy SP, McElroy E, Rauckhorst MR, Tomaras AP, Young JA, Zaniewski RP, Shanmugasundaram V, Han S (2014) Siderophore receptor-mediated uptake of lactivicin analogues in Gram-negative bacteria. J Med Chem 57(9):3845–3855
Stephens N, Rawlings BJ, Caffrey P (2012) Streptomyces nodosus host strains optimized for polyene glycosylation engineering. Biosci Biotechnol Biochem 76(2):384–387
Stephens N, Rawlings BJ, Caffrey P (2013) Versatility of enzymes catalyzing late steps in polyene 67-121C biosynthesis. Biosci Biotechnol Biochem 77(4):880–883
Tori S, Agatsuma T, Ochiai K, Saitoh Y, Ando K, Nakanishill S, Lokker NA, Giese NA, Matsuda Y (2001) RP-1776, a novel cyclic peptide produced by Streptomyces sp.9 inhibits the binding of PDGF to the extracellular domain of its receptor. J. Antibiotics 54(5):405–414
Uchida T, Imoto M, Watanabe Y, Miura K, Dobashi T, Matsuda N, Sawa T, Naganawa H, Hamada M, Takeuchi T, Umezawa H (1985) Saquayamycins, new aquayamycin-group antibiotics. J. Antibiotics 38(9):1171–1181
Valdés-Stauber N, Scherer S (1994) Isolation and characterization of linocin M18, a bacteriocin produced by Brevibacterium linens. Appl Environ Microbiol 60:3809–3814
Volmer A, Szpilman AM, Carreira EM (2010) Synthesis and biological evaluation of amphotericin B derivatives. Nat Prod Rep 27:1329–1349
Wang F-Q, Zhong J, Zhao Y, Xiao J, Liu J, Dai M, Zheng G, Zhang L, Yu J, Wu J (2014) Duan B (2014) genome sequencing of high-penicillin producing industrial strain of Penicillium chrysogenum. BMC Genomics 15(Suppl 1):S11
Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB (2006) Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441(7091):358–361
Wang L, McVey J, Vining LC (2001) Cloning and functional analysis of a phosphopantetheinyl transferase superfamily gene associated with jadomycin biosynthesis in Streptomyces venezuelae ISP5230. Microbiology 147(Pt 6):1535–1545
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 30 — A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. doi:10.1093/nar/gkv437
Wilson MC, Mori T, Ruckert C, Uria AR, Helf MJ, Takada K, Gernert C, Steffens UAE, Heycke N, Schmitt S, Rinke C, Helfrich EJN, Brachmann AO, Gurgui C, Wakimoto T, Kracht M, Crusemann M, Hentschel U, Abe I, Matsunaga S, Kalinowski J, Haruko Takeyama H, Piel J (2014) An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506:58–62
Worthington RJ, Melander C (2013) Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31(3):177–84
Yamamoto T, Umegawa Y, Tsuchikawa H, Matsumori N, Hanashima S, Murata M, Haser R, Rawlings BJ, Caffrey P (2015) Role of polyol moiety of amphotericin B in ion channel formation and sterol selectivity in bilayer membrane. Bioorg Med Chem 23(17):5782–5788
Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE (2008) Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2). J. Biol. Chem. 283:8183–8189
Acknowledgments
We thank Niamh Stephens for carrying out some of the initial curation of the genome sequence and Kevin Conboy, UCD School of Chemistry, for MS analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was conducted with the financial support of Science Foundation Ireland, grant number 09/RFP/GEN2132. Paul Sweeney declares that he has no conflict of interest. Cormac Murphy declares that he has no conflict of interest. Patrick Caffrey declares that he has no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 422 kb)
Rights and permissions
About this article
Cite this article
Sweeney, P., Murphy, C.D. & Caffrey, P. Exploiting the genome sequence of Streptomyces nodosus for enhanced antibiotic production. Appl Microbiol Biotechnol 100, 1285–1295 (2016). https://doi.org/10.1007/s00253-015-7060-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7060-9