Abstract
Aptamers are synthetic DNA recognition elements which form unique conformations that enable them to bind specifically to their targets. In the present study, an attempt was made to standardize a new modified combinatorial method comprising of Ni-NTA affinity Systematic Evolution of Ligands by Exponential Enrichment (SELEX; based on affinity between His tag protein and Ni-NTA), membrane SELEX (based on immobilization of protein on nitrocellulose membrane), and microtiter plate based SELEX (to monitor affinity and to enrich the selected aptamers) for protein targets. For experimental evaluation, staphylococcal interotoxin B was the molecule chosen. The new combinatorial method enhanced selection ability up to 51.20 % in comparison with individual conventional procedures. Employing this method following six rounds of selection, high-affinity aptamers with very different properties could be obtained with a dissociation constant (K d) value as low as 34.72 ± 25.09 nM. The optimal aptamers could be employed in fluorescence binding assay, enzyme-linked oligonucleotide assays, and aptamer-based Western blot assay for characterization and detection. These results pave a potential path without using of any robotics for high-throughput generation of aptamers with advantages in terms of rapidity, simplicity, and ease in handling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
New-generation affinity molecules, namely, aptamers, have became a very attractive class of targeting molecules in many fields of application like medicine (as diagnostic, vaccine, and therapeutic agents), pharmaceutics (for drug discovery and validation), and environmental and food analytics (as biological recognition elements) (Jayasena 1999; Rajendran and Ellington 2002; Wilson and Szostak 1999; Cox et al. 2002a, b; Keefe et al. 2010; Baines and Colas 2006; Becker and Becker 2006; Kärkkäinen et al. 2011). Traditionally reported affinity molecules, i.e., single-chain antibodies (scFv) (Steinhauer et al. 2002), peptides displayed on protein domain scaffolded surfaces (Li 2000), and peptides and peptoids (synthetic peptides) (Humet et al. 2003), have drawbacks in terms of stability in varying conditions (ionic strength, temperature, and pH) and low affinity, making them ineffective for detecting proteins under many conditions (Phizicky et al. 2003). In a quest of finding ligand with less complexity and improved functionality, small-length (20–80 mer) single-stranded nucleic acids, i.e., aptamers, have been proposed. Aptamers can be consistently synthesized through PCR, and it can fold into unique three-dimensional structures in the presence of a target with high affinity and specificity (Tuerk and Gold 1990; Ellington and Szostak 1990). Aptamers are generated by a process that combines combinatorial chemistry with in vitro evolution, known as Systematic Evolution of Ligands by Exponential Enrichment (SELEX), following the incubation of a protein with a library of DNA or RNA sequences, typically 1014 molecules in complexity. Finally, the protein–DNA complexes are isolated, the DNA is amplified, and the process is repeated until the sample is enriched with sequences that display high affinity for the protein of interest (Ellington and Szostak 1992).
SELEX appears to be technically a simple process, but small alterations to protocol can have large impact on the success of generating aptamers. The increasing number of aptamer publications over the years describing their selection and application shows the high interest in this research field. One of the challenges in this area is the optimization of the methodology to get new aptamers with outstanding binding abilities for a certain target. Moreover, there is no standardized SELEX protocol for universal application reported to date. Perhaps this explains why, 13 years since its first citation in the literature, only approximately 40 protein-detecting aptamer sequences have been published and very few have been well characterized and utilized (Stoltenburg et al. 2007). The designing and the selection process of specific aptamer is very complex and time consuming that limits its practical applications (Cox et al. 2002a, b; Murphy et al. 2003). Further, no proper method is available for checking the binding affinity and determination of the presence of specific aptamer for target after every round of SELEX. All these limitations necessitate evaluation and adaptation of novel SELEX procedures to popularize the aptamer-based target detection technology.
In this report, we describe an improved simplified combinatorial SELEX protocol for selection of DNA aptamer against a protein target, i.e., enterotoxin B of Staphylococcus aureus, a biowarfare agent [McBride et al. 2003; Homola et al. 2002]. The procedure allowed reliable synthesis and detection of aptamer of high affinity and specificity. Such methodology is a good alternative of current aptamer synthesis protocols and can be implemented for synthesis of aptamers against different targets.
Methods
Reagents and materials
The single-stranded DNA (ssDNA) library and the primers were obtained from Xcelris Bioscience (Ahmadabad). Unless otherwise mentioned, the following chemicals were obtained from respective manufacturers: Luria–Bertani (LB) broth, LB agar, Baird–Parker agar base, and egg yolk (EY) tellurite enrichment from Himedia (India); trizma base, potassium phosphate dibasic, glycine, sodium hydroxide, ethylenediaminetetraacetic acid (EDTA), Dulbecco’s phosphate-buffered saline with CaCl2 and MgCl2 (DPBS), bovine serum albumin (BSA), albumin, skimmed milk powder, and all PCR reagents from Sigma-Aldrich (India); PCR Purification Kit, Plasmid Extraction Kit, and Gel Extraction Kit from Qiagen (India); TMB/H2O2 from Bangalore Genie (India); TOPO TA Cloning Kit and Streptavidin Conjugate from Invitrogen (India); and 0.22 μm nitro cellulose membrane from Millipore (India). All the solutions were prepared with ultra-high-purity water (Milli-Q).
Bacterial cultures and growth conditions
For routine microbiological work, S. aureus strain ATCC14458 and Escherichia coli strain BL21 (DE3) were grown in Baird–Parker agar base with egg yolk (EY) tellurite enrichment and LB medium and incubated at 28 and 37 °C, respectively. Cultures were maintained on its respective agar media and stored at 4 °C for short-term preservation, and for long-term preservation, cultures were stored at −80 °C in 15 % glycerol.
Design of aptamer library and primer
A 76-bp oligonucleotide single-stranded DNA library was constructed, consisting of a 40-nt randomized region flanked by 17- and 19-nt-long primer binding regions at the end. The naive library was resuspended in DNA storage buffer (10 mM Tris–HCl, pH 7.4 ± 0.2) to a concentration of 100 μM. The primers for this study were designed by using gene runner software version 3.05 (http://generunner.net/). The primers were resuspended in DNA storage buffer to a concentration of 10 μM. The library and the primers were stored at −20 °C until further use. The gene sequences of the library and the primers (normal, biotin-labeled, and FITC-labeled) are listed in Table 1.
Amplification of single-stranded DNA library
In order to generate the ssDNA template, asymmetric polymerase chain reaction (PCR) (Berezovski et al. 2006) was performed in a PCR reaction (40 μl) containing 4 μl of 10× PCR buffer, 3.2 μl of 1.6 mM MgCl2, 2 μl of 2.0 mM dNTPs, 2 U μl−1 of 0.4 μl Taq DNA polymerase, 1 μl of the template library, 27.4 μl of dH2O, and different ratios of forward to reverse primer (1.1:0.9,1.2:0.8, 1.3:0.7, 1.4:0.6, 1.5:0.5, 1.6:0.4, 1.7:0.3, 1.8:0.2, 1.9:0.1, 2.0:0) to obtain desired amplification. PCR was performed (Biorad, India) with the following protocol: initial denaturation at 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing at 56 °C for 45 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. Finally, PCR amplicons were separated by 2.5 % agarose gel electrophoresis, and PCR product of desired size (76 bp) was eluted from the gel using the Qiagen MiniElute Gel Extraction Kit following manufacturer’s protocol.
Cloning, expression, and purification of seb gene
seb encoding full mature peptide was amplified by PCR from the genome of S. aureus strain ATCC14458. The gene along with vector pET22b was digested with NcoI/XhoI restriction enzymes and ligated in-frame with N terminal pelB signal peptide and C terminal His6-tag of pET22b vector (Novagen, Germany). E. coli BL21 (DE3) (Invitrogen, Bangalore, India) transformed with recombinant plasmids were selected on LB agar supplemented with 100 μg ml−1 ampicillin and further screened by PCR (Studier et al. 1990). The recombinant clones were inoculated into LB broth and grown until A600 reaches 0.6. Cells were then induced with isopropyl β-D-1-thio galactopyranoside (IPTG) to a final concentration of 1 mM and pelleted after 5 h of induction. Expression of recombinant proteins was examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The localization of expressed recombinant proteins in E. coli BL21 (DE3) was determined by sequential extraction of proteins from periplasmic space, cytosol, and inclusion bodies (Gupta et al. 1999) and purified under native condition by immobilized metal affinity chromatography using Ni-NTA slurry (Qiagen, Germany) as per manufacturer’s protocol. The purified protein was dialyzed against binding buffer (50 mM Na2HPO4, pH 7.2 ± 0.2, 150 mM NaCl, 2 mM MgCl2) and stored at −80 °C. Homogeneity of purified protein was determined by SDS-PAGE, and the protein content was quantified by Bradford method (Sambrook et al. 1989).
In vitro selection of aptamers
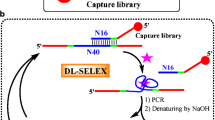
The SELEX protocol for the enrichment of aptamer pool against target was done in the following order: Ni-NTA affinity SELEX, nitrocellulose membrane SELEX, and microtiter plate based SELEX, considered as a single round of SELEX (Fig. 1). The SELEX round was performed until the affinity of selected pool enriched to optimum level. Counter SELEX was introduced in this protocol after second and fourth round of selection. All SELEX rounds and counter SELEX round were performed by keeping target and aptamer concentration constant, where bound and unbound aptamer pooled DNA was purified and suspended in the same volume of water, and concentration was measured by Nano Drop-2000 (Thermo Scientific, India). For comparison of the efficiency of the combinatorial SELEX protocol, all three SELEX methods were performed individually with equal number of SELEX rounds, and affinity of the selected aptamers was also determined. The details of protocol are as described below.
Ni-NTA affinity SELEX
Preparation of aptamer library and elimination of aptamers specific to Ni-NTA Sepharose
Two microliters of Aptamer library (240 ng μl−1) was resuspended in 30 μl of 20 mM Tris–HCl, pH 7.4 ± 0.2, in a microcentrifuge tube, heated at 94 °C for 10 min, rapidly cooled in ice for 15 min, and kept at room temperature for 10 min in 500 μl of selection buffer (2.5 mM CaCl2, 5 mM MgCl2 in Dulbecco’s phosphate-buffered saline, pH 7.4 ± 0.2). Finally, it was passed through a Ni-NTA column (Qiagen, India) pre-washed with selection buffer. The flow-through was collected and used as a library for selection of aptamers against the targets.
Binding of target to Ni-NTA
Four hundred microliters of Ni-NTA sepharose beads was washed three times with equal amount of binding buffer (50 mM Na2HPO4, pH 7.2 ± 0.2, 150 mM NaCl, 2 mM MgCl2, 0.05 % Tween-20). It was separated by centrifugation and incubated with SEB protein (1 mg ml−1) for 30 min at room temperature for binding. Unbound SEB proteins were removed from the bead-bound SEB following three times washing with 400 μl of binding buffer. Finally, it was centrifuged at 6000 rpm for 20 s and used for further study.
Binding and recovery of aptamers specific to the target
For the selection of SEB aptamer, the pre-prepared aptamer library was transferred to the bead-bound SEB and incubated for 45 min for binding of aptamer to the SEB protein. After incubation, the SEB-DNA complex was washed three times with 900 μl of binding buffer to remove unbound aptamer sequences and then centrifuged.
For recovery of the SEB-selected aptamers, 200 μl of 20 mM Tris–HCl (pH 7.4 ± 0.2) was added to the pellet, heated for 10 min at 85 °C, vortexed for 1 min, and centrifuged at 10,000 rpm for 10 min, and the supernatant was transferred to a fresh tube. This step was repeated and supernatants were combined. The aptamer DNA from the supernatant was recovered by PCR purification kit and dissolved in 30 μl of distilled water, and the concentration was measured by Nanodrop-2000 (Thermo Scientific, India). Finally, the DNA was amplified with earlier standardized PCR conditions. This DNA was used as library for further round of selection. Two microliters of aptamer library (240 ng μl−1) was resuspended in 30 μl of 20 mM Tris–HCl, pH 7.4 ± 0.2, in a microcentrifuge tube, heated at 94 °C for 10 min, and then rapidly cooled in ice for 15 min.
Enhancing specificity of the selected aptamer pool by nitrocellulose membrane SELEX
Purified rSEB protein (1 mg ml−1) was immobilized on a nitrocellulose membrane and was blocked with 3 % bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Then, the denatured ssDNA library was dissolved in 500 μl of binding buffer and incubated with the immobilized proteins on the membrane at room temperature for 1 h. The nitrocellulose membrane was washed with 1 ml of binding buffer for five times and cut into pieces. One hundred microliters of elution buffer (7 M urea, 0.5 M NH4COOH, 7 mM SDS, 1 mM EDTA, pH 8.0 ± 0.2) was added to the pieces, heated at 94 °C for 10 min, and centrifuged at 7500 rpm for 10 min at 4 °C, and the supernatant was collected. The ssDNA was extracted by chloroform and ethanol precipitation. The precipitated ssDNA was suspended in 30 μl of de-ionized water, and the concentration was measured by Nanodrop-2000 (Thermo Scientific, India). Bound aptamer pool was amplified separately using three different primer pairs, i.e., normal forward and normal reverse, biotin-forward and normal reverse, and FITC-forward and normal reverse, respectively, for next round of selection and affinity determination. PCR amplification was done with previously standardized conditions. Two microliter of each pool aptamer library (240 ng μl−1) was resuspended in 30 μl of 20 mM Tris–HCl, pH 7.4 ± 0.2, in a microcentrifuge tube, heated at 94 °C for 10 min, and then rapidly cooled in ice for 15 min.
Affinity determination and enhancing aptamer pool specificity by microtiter plate based SELEX
SEB protein (1 mg ml-1) was coated onto a microtiter plate (Nunc) in 100 μl of carbonate-bicarbonate buffer (pH 9.6 ± 0.2) and incubated at room temperature for 60 min. The plate was washed three times with binding buffer. Unbound sites were blocked with 3 % BSA and incubated for 1 h, followed by thorough washing. Thirty microliters of each pre-denatured aptamer pool (240 ng μl−1) was added to each well of the plate and incubated for 60 min at room temperature. After thorough washing, 100 μl of boiling water was added wells to break protein–DNA interaction. The solution was transferred to a fresh 1.5-ml tube. Biotin-labeled pool was used in enzyme-linked oligonucleotide assay (ELONA) for checking affinity of the pool. Fluorescence intensity was checked for FITC-labeled aptamer pool by Infinite M1000 spectrophotometer (TECAN, India). Normal aptamer pool was recovered by PCR purification kit and dissolved in 30 μl of distilled water, and the concentration was measured by Nanodrop-2000 (Thermo Scientific, India). The DNA was amplified with earlier standardized PCR conditions. This aptamer pool was used as library for further selection. Two microliters of aptamer library (240 ng μl−1) was resuspended in 30 μl of 20 mM Tris–HCl, pH 7.4 ± 0.2, in a microcentrifuge tube, heated at 94 °C for 10 min, and then rapidly cooled in ice for 15 min and was further proceeded for next SELEX rounds to remove aptamer having cross-reactivity with closely related and other proteins. Binding affinity in each round was also monitored by fluorescence microscopy (Nikon, India) by using drop of FITC-labeled aptamer pool targets bound on glass slide.
Counter SELEX by nitrocellulose membrane method
Purified rSEC, rSEA, skimmed milk and BSA, albumin (1 mg ml−1) were immobilized on a nitrocellulose membrane. The membrane was blocked with 3 % bovine serum albumin (BSA) in phosphate-buffered saline (PBS). The denatured ssDNA library (240 ng μl−1) was dissolved in 500 μl of binding buffer and incubated with the immobilized proteins on the membrane at room temperature for 1 h. Afterward, the nitrocellulose membrane was washed with 1 ml of binding buffer for five times; all wash solutions were collected in a single vial and recovered by PCR purification kit and dissolved in 30 μl of distilled water, and the concentration was measured by Nanodrop-2000 (Thermo Scientific, India). Finally, the DNA was amplified and used as library for further round of SELEX.
TOPO cloning, sequencing, and analyzing for identification of consensus aptamer sequences
The selected aptamer pools from sixth round of combinatorial SELEX was amplified with standardized PCR conditions using normal aptamer forward and reverse primers with Taq DNA polymerase and subsequently cloned by using TOPO TA cloning Kit (Invitrogen/Life Technologies, India) as per manufacturer’s protocol. The ligated vectors were transformed into One Shot Top10 chemical competent E. coli cells (provided with TOPO TA cloning Kit). Positive transformants were analyzed by colony PCR using a combination of a vector-specific primer (M13 forward and reverse primer) and aptamer-specific primer (normal aptamer forward and reverse primer, Table 1). The plasmid DNA of positive clones were isolated using the Gene Elute Plasmid Miniprep Kit (Qiagen, India), and the inserted aptamer DNA of each clone was sequenced by custom sequencing facility provided by Xceleris, Bangalore. The obtained sequences were analyzed and aligned by using the web-based tool ClustalW provided by the EBI web server (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Larkin et al. 2007; Thompson et al. 1994). The selected ssDNA molecules were subjected to secondary structure prediction using the M-fold software (http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form) at 26 °C in 150 mM (Na+) and 1 mM (Mg++) for determination of free energy (Zuker 1989, 2003).
Aptamer binding assay
Fluorescence-labeled ssDNA binding assay was performed to monitor the enrichment of each SELEX round and to evaluate binding affinity of aptamer. In brief, fluorescence-labeled ssDNA was thermally denatured in 200 μl of selection buffer and incubated in the dark with SEB-coated 96-well microtiter plates (Nunc) at 37 °C for 1 h on a shaking platform. After incubation, unbound ssDNA was collected and SEB-bound ssDNA was eluted by adding 200 μl of pre-heated selection buffer. The fluorescence intensity of unbound and eluted ssDNA was measured following excitation at 492 nm and emission at 532 nm using Infinite M1000 spectrophotometer (TECAN, India). All binding assays were repeated three times. To monitor the enrichment of each SELEX round and to evaluate binding affinity of aptamer, fluorescence intensity of ssDNA aptamer (200 nM) with SEB (20 μM) was measured.
For evaluation of cross-reactivity among different related toxins and other proteins, the binding assays were repeated using rSEA, rSEC, BSA, skimmed milk, and albumin.
Determination of K d of selected aptamers
To determine the binding kinetics of a selected aptamer, the binding assay was performed as described earlier in “Determination of specificity of SEB2 aptamer by ELONA” section but with increasing amounts of FITC-labeled ssDNA aptamer (0 to 350 nM), and a constant amount of SEB (20 μM) for each assay was used. To calculate the dissociation constants (K d) of the aptamers, the aptamer quantity bound to targets was plotted and data points were fitted to the equation Y = B max X / (K d + X) via non-linear regression analysis, using GraphPad Prism 6.
Enzyme linked oligonucleotide assay (ELONA)
ELONA was performed to demonstrate the affinity and specificity of aptamer sequences to SEB protein with modifications. One hundred microliters of rSEB protein (50 μg ml−1) in carbonate–bicarbonate buffer (pH 9.6 ± 0.2) was coated on microtiter plate (Nunc). The unbound sites were blocked by overnight incubation with 3 % bovine serum albumin (BSA) in phosphate-buffered saline (PBS). Each well was washed thoroughly with PBS-T followed by 45-min incubation with pre-denatured 100 μl biotinylated SEB2 aptamers (1 ng μl−1). The unbound biotin aptamers were removed by three times washing with 1.5 % Tween-20 in binding buffer. Finally, 100 μl of a 1:2500 dilution of streptavidin-HRP conjugate solution was added and mixed properly. Following 30-min incubation at room temperature on a shaking platform, the plate was washed three times with binding buffer and developed using TMB/H2O2 (Bangalore Genie) according to the manufacturer’s instructions. The reaction was inhibited with the addition of 150 μl of 1 M H2SO4. Protein-bound aptamer–streptavidin-HRP conjugate complex was quantified by determining the absorbance at 490 nm using Infinite M1000 spectrophotometer (TECAN, India). For evaluation of cross-reactivity, among different related and other proteins, the binding assays were repeated using rSEA, rSEC, BSA, skimmed milk, and albumin.
Aptamer-based Western blot analysis
Protein samples were separated in 15 % polyacrylamide gels under denaturing conditions using the mini-Protean 3 system (Bio-Rad, India). The proteins were electro-blotted onto nitrocellulose membrane (Millipore, India). It was blocked by overnight incubation with 5 % BSA in PBS-T and then probed with biotinylated aptamer diluted to 1 μg ml−1 in 5 ml of PBS-T for 2 h at room temperature with rotation. The blots were washed three times with 10 ml of PBS-T and then probed with streptavidin–HRP conjugate diluted to 1:2500 in PBS-T. The blots were washed, and the positive bands were detected by immersing it in a developing solution of TMB/H2O2 (Bangalore Genie) at room temperature for 5 min. The enzyme reaction was terminated by washing the blot in water. For evaluation of cross-reactivity in presence of different related and other proteins, the assay was repeated using rSEA, rSEC, BSA, and albumin.
Statistical analysis
All experiments were repeated independently three times with similar conditions. Results were presented as the mean value ± standard deviation (SD). Statistical differences between treatments were analyzed by univariate (ANOVA) and Tukey’s test assuming P value (P < 0.05, P < 0.01, and P < 0.001) presented using GraphPad Prism 6.
Results
Asymmetric PCR
In this study, asymmetric PCR was performed for amplification of aptamer library. The number of PCR cycles was also optimized to avoid over-amplification, which is evidenced by mis-annealed products. Asymmetric PCR preferentially increased the target ssDNA and decreased the primer dimers as well as non-specific amplification (Supplementary Fig. S2). Optimization reactions showed a sharp band of expected base size (agarose gel analysis) with 0.45 pmol template DNA and primer ratio at 1.6:0.4 (forward/reverse) after 30 cycles.
Cloning, expression, and purification of SEB protein
The rSEB amplicons were cloned into E. coli BL21 (DE3) using pET22b vector, and the integrity of cloned genes in recombinant plasmid pET22b-rSEB was confirmed by sequencing analysis. The rSEB chimeric gene was of 719 bp in length, and E. coli host cells harboring the recombinant plasmid pET22b-rSEB, and the nucleotides which are on in silico translation matched with amino acid sequences of the selected SEB mature peptide (Gene Bank accession M11118.1). E. coli host cells harboring the recombinant plasmid pET22B-rSEB expressed proteins with apparent molecular weight of 28.37 kDa, upon 5-h induction with 1 mM IPTG at 37 °C. The molecular mass of expressed recombinant proteins as seen in 12 % SDS-PAGE correlated with the estimated molecular mass (Supplementary Fig. S1a). The soluble recombinant proteins were purified from 500 ml induced cultures by immobilized metal affinity chromatography using Ni-NTA column under native conditions (Supplementary Fig. S1b). The presence of C terminal His6-tag in polypeptide was confirmed by Western blot analysis (Supplementary Fig. S1c).
In vitro selection of ssDNA aptamers
Six rounds of in vitro selection were used to isolate aptamers that could favorably bind to SEB. The enrichment of target-specific aptamers was monitored during the selection process. An increasing amount of binding ratio (Fig. 2a), fluorescence intensity (Fig. 2b), and enzyme-linked oligonucleotide assay absorbance (Fig. 2c) was observed with increasing number of selection rounds and then remained nearly in a steady state in the fifth and sixth rounds. This suggested that SEB binding aptamers with good binding affinity were enriched, and the binding ability of selection pools plateaued after about the fifth round of selection. The fluorescence was observed on the toxin-immobilized glass slide after washing. The glass slide was washed with extensive amounts of binding buffer. Presence of highly localized fluorescent dots and the absence of background smear provide an indication of successful selection (Supplementary Fig. S3). The stringency of the selection was controlled by adjusting the target protein concentrations, the incubation times, and the washes during the course of optimizing our protocol; we have observed enrichment of sequences that were not unique to target protein and, therefore, we have incorporated counter-selection after the second and fourth round against closely related and other proteins to prevent enrichment of aptamers showing affinity to proteins other than the target. Binding percentage was increased up to 51.20 % after the sixth round of selection which proves the efficiency of the newly described protocol compared to other methods. However, by following individual conventional SELEX, even after six rounds of selection, the binding percentage of Ni-NTA affinity SELEX, membrane SELEX, and microtiter SELEX was as low as 8, 6.95, and 9.50 %, respectively (Fig. 3a). Binding ratio in combinatorial SELEX with increasing number of selection rounds is also higher compared to individual SELEX protocol used here (Fig. 3b).
a Enrichment of SEB-specific aptamers during the combinatorial SELEX. Comparative enrichment of binding percentage of SEB-specific aptamers during the combinatorial SELEX. The bar graph shows a significant increasing eluted aptamer concentration in each selection round and nearly kept a steady state from rounds 5 and 6. Counter selection step (CS) was introduced to remove ssDNA non-specifically bound to the related toxin (after each round, DNA was purified and dissolved in 30 μl of water). b Comparative enrichment of fluorescence intensity during combinatorial SELEX. Comparative enrichment of fluorescence intensity of bound and unbound SEB-specific aptamers during the combinatorial SELEX. The bar graph shows significant increasing bound aptamer pool fluorescence intensity in each selection round of SELEX and remained nearly a steady state in rounds 5 and 6. Counter selection step (CS) was introduced to remove ssDNA non-specifically bound to the related toxin (after each round, DNA was purified and dissolved in 30 μl of water). The experiment was repeated for three times. c Monitoring of SEB-specific aptamers during the combinatorial SELEX by enzyme-linked oligonucleotide assay (ELONA). Enrichment of SEB-specific aptamers during the combinatorial SELEX in enzyme-linked oligonucleotide assay (ELONA). The bar graph shows a significant increasing signal over background observed in each selection round and remained nearly a steady state in the fifth round and sixth round. Counter selection step (CS) was introduced to remove ssDNA non-specifically bound to the related toxins. The experiment was repeated for three times
a Comparative binding percentage of SEB-specific aptamers during combinatorial SELEX and individual conventional SELEX. The bar graph shows the binding percentage of ssDNA aptamer pool in each selection round. Counter selection step (CS) was introduced to remove ssDNA non-specifically bound to the related toxin (after each round, DNA was purified and dissolved in 30 μl of water). The result shows that the binding percentage is significantly higher in combinatorial SELEX in comparison to individual SELEX method in each round. b Comparison enrichment of SEB-specific aptamers during combinatorial SELEX and individual conventional SELEX. The bar graph shows the amounts of ssDNA eluted from each selection round. Counter selection step (CS) was introduced to remove ssDNA non-specifically bound to the related toxin (after each round, DNA purified by DNA purification kit and dissolved in 30 μl of water). The result shows that the binding percentage is significantly higher in combinatorial SELEX in comparison to individual SELEX method in each round
Cloning and sequence analysis of aptamer pools
Aptamer pools from the sixth round of SELEX displayed the highest affinity for the target protein. Hence, after cloning and transformation of the selected aptamer pool, 39 E. coli clones were further examined. Of these, 28 positive transformants were determined, and the contained plasmid DNA was prepared for sequencing the inserted aptamer DNA of each clone. The 76-bp sequence in each aptamer was unambiguously identified. Multiple sequence alignments using ClustalW and phylogenetic analysis revealed that four groups of sequences were enriched modestly and accomplished according to their sequence similarity (Fig. 4).
The first group is the one with the largest number of aptamers comprising of 12 sequences (4 identical sequences). Group 2 with its eight sequences (two identical sequences) is much more homogeneous than group 1. Group 3 consist of five sequences (two identical sequences). Group 4 consist of three sequences (data not shown). The secondary structure analysis of these candidate aptamers was assessed using the Mfold software which yielded one potential secondary structure in the empirical condition of selection (Fig. 5 and Table 2). As it can be observed, all sequences show complex typical stem–loop structure. We anticipate that the stem–loop region might play a particularly crucial role during aptamer–epitope interactions, thus leading to higher selectivity in these aptamers. We take into consideration the Gibbs free energy (dG) value and two representatives from each group of aptamers having the lowest Gibbs free energy (dG) considered for characterization (Table 2). Multiple sequences alignment (Fig. 4), Jalview (Supplementary Fig. S4), and phylogenetic analysis (Supplementary Fig. S5) of these selected eight sequences share homology between 60 and 96 % and few nucleotide conserved sequences.
Aptamer binding assay
The binding affinity of the candidate aptamers was measured using fluorescein isothiocyanate (FITC)-labeled aptamers. After binding analysis of each aptamer, the specificity of aptamers was assessed toward closely related and other proteins (Fig. 6). It was observed that the fluorescence intensity of SEB1, SEB2, and SEB 27 aptamers was higher toward SEB protein. It is noteworthy that the random DNA sequence library pool did not show any significant binding toward the target molecule. This result suggested that the binding of identified aptamers was sequence specific and not due to non-specific polyanionic effect.
Fluorescence intensity of selected aptamers. Binding assays with the individual SEB aptamer to test their ability to bind to SEB protein. In each assay, proteins were incubated with 250 pmol fluorescence-labeled aptamer in a volume of 200 μl according to the SELEX conditions. The unselected, fluorescence-labeled ssDNA library (BLANK) was used as the negative control. After several washing steps, the bound aptamers were eluted by heat treatment and quantified by fluorescence analysis. The results are shown as a bar graph. The experiment was repeated for three times
Determination of K d values
K d values of selected aptamers were determined by performing fluorescence-based affinity assays. The quantity of the targets in each binding assay was consistently maintained using the same volume of toxin suspensions. Binding curves were plotted using the relative fluorescence intensity (the difference between the fluorescence intensity of the 5׳-FITC-labeled aptamers measured from SEB-coated microtiter well and uncoated well) against the gradient concentration (0–350 nM). The experimental aptamers showed varying degrees of affinity for the SEB (Fig. 7). From the results (Table. 2), we concluded that SEB1 and SEB 2 and SEB 27 aptamers are able to detect SEB protein in a concentration-dependent manner with K d = 49.55 ± 11.33, 36.34 ± 11.23, and 34.72 ± 25.09 nM, respectively. Hence, SEB2 aptamer was considered for further characterization and development of aptamer-based detection assay.
Binding kinetics of selected aptamer sequences. Determination of the dissociation constants (K d) for selected aptamer sequences with increasing amounts FITC-labeled ssDNA aptamer (0 to 350 nM) and a constant amount of SEB (20 μM) for each assay. On the basis of the amount of aptamers eluted, saturation curves were obtained and the dissociation constants (K d) were calculated by non-linear regression analysis
Determination of specificity of SEB2 aptamer by ELONA
Enzyme-linked oligonucleotide assay (ELONA) was performed for determination of specificity of SEB2 aptamer. Results obtained from ELONA assay in order to prioritize the SEB2 aptamers for further characterization revealed highest affinity and specificity of SEB2 aptamer for the purified native rSEB toxin (confirmed by kinetic studies), and no cross-reactivity/spurious signals were observed with exerted closely related toxins and negative control (BSA, skimmed milk, and albumin) (Fig. 8 and Supplementary Fig. S6). Employing the colorimetric detection system (TMB/H2O2) for streptavidin peroxidase conjugated to a significant (100×) signal over background was observed with purified native rSEB toxin.
Specificity of SEB 2 aptamer by ELONA. Determination of specificity of SEB2 aptamer using enzyme-linked oligonucleotide assay. Results show little or no cross-reactivity observed with exerted closely related toxins and negative control (BSA, skimmed milk, albumin). We employed the colorimetric detection system (TMB/H2O2) for streptavidin peroxidase conjugated to a significant (100×) signal over background observed with purified native rSEB toxin
Specificity determinations of the SEB2 aptamer using protein blot analysis
The results of the enzyme-linked assay suggested that the SEB2 aptamer exhibited specificity for SEB protein. In order to verify the specificity and determination of the ability to detect the denatured form of SEB protein, as well as to investigate further the potential uses of the aptamer, protein blot analysis was performed. The SEB2 aptamer was indeed able to bind to both native and denatured SEB protein on the blot, and no cross-reactivities in terms of chemiluminescent signal intensity were observed with exerted closely related toxins and negative control (BSA, albumin) (Supplementary Fig. S7). The performance of aptamer was similar to the anti-6X His antibody (Sigma, India) in terms of chemiluminescent signal intensity and specificity (Supplementary Fig. S1c).
Discussion
Antibodies with high affinity and specificity are still at the heart of many pathogen diagnostic systems, but their predominance has been challenged by various nucleic-acid-based methods. Although most of these approaches aim at identification of pathogens via determination of their nucleic acid composition, since the advent of aptamers, oligonucleotides could be exploited for direct detection of the protein components of a given microbe. In this report, we presented straightforward selection protocol and application of the nuclease resistance protein–specific DNA aptamers. The SELEX procedure used comprises Ni-NTA affinity SELEX (based on affinity between His-tag and Ni-NTA), membrane SELEX (based on adherence of protein on nitrocellulose membrane), and microtiter plate based SELEX (to monitor affinity and to enrich the selected aptamers) for individually purified protein targets. Additionally, we have utilized the advantage of the highly specific and strong streptavidin–biotin interaction for several approaches for detection. Single-stranded DNA generated by employing asymmetric PCR after each round of selection reduced the additional single-strand separation. Most of the published protocols describe relatively a complex method for selection and enrichment. Moreover, for checking the binding affinity and determination of presence of specific aptamer for target after every round of SELEX, a defined proper method is also not available. In the presently described SELEX protocol, this aspect appears to get overruled by the fluorescence binding assay step that determines the affinity after every round of SELEX. More so, the functional versatility of aptamers could also be analyzed by the tested assays such as enzyme-linked oligonucleotide assay and Western blot analysis wherein biotinylated aptamers were employed. Supremacy of this combinatorial method was proven adequately as it reduced the number of selection rounds and provided improved selection efficiency of more than 51.20 % higher than what was achieved with conventional individual selection method. Once subjected to ClustalW and phylogenetic analysis, the selected aptamer sequences revealed a very high homology. The selected aptamers also had the affinity which was comparable to what was achieved in earlier published reports, and it could well recognize the target protein with an affinity constant in very low nanomolar range (Liu et al. 2013).
By this combinatorial method, one round of SELEX can be completed within 9 h and needs only six rounds of selection to obtain high-affinity aptamers. On the other hand, the reported SELEX protocol might take 5 h or less for completion of each round and definitely needs more than 15 rounds for achieving the desired results of obtaining high-affinity and specific aptamers following the manual operation. However, automated aptamer acquisition platform with a throughput of 120 aptamers/month for eight proteins has been described in the literature (Cox et al. 2002a, b). The presently developed combinatorial SELEX protocol also has the capability of high-throughput yield without the need of any automation and robotics thereby making it more cost-effective in terms of utility. Another high-throughput SELEX protocol using 96-well microtiter plates has been described but still is robotic dependent (Drolet et al. 1999). This particular protocol relied on hydrophobic immobilization of proteins on microtiter plates, and the authors commented that the four proteins tested adhered to the wells with varying efficiency, thereby relatively making it difficult to control the amount of protein needed for each experiment.
The key virtue of the proposed method also lies in its ability to be easily modified according to the target and condition, little to no changes to the selection conditions, or the library design, but the approach may definitely require a certain level of standardization to customize the SELEX process. This SELEX protocol has the additional flexibility to introduce the desired change for selection of aptamer not only based on His-tag protein targets but also for any purified protein targets. Although the described method amply demonstrated for obtaining aptamers from DNA library, the same protocol should be applicable for obtaining RNA aptamers from RNA libraries as well.
However, it is adequately proven that the newly described combinatorial SELEX protocol has edge over the existing methods with a test example of making use of rSEB as a target protein. The procedure needs to be evaluated on other target protein molecules of varied type and size along with incorporating few basal modifications to suit the optimal desired application. Probably, only then can the real virtues of the proposed method be merited for a possible universal application.
The findings of combinatorial SELEX method presented here provide an optimized and improved method for successful enrichment of tight-binding aptamers within a SELEX library for generating ssDNA aptamers for protein targets without the need of expensive robotics. The protocol described here is very efficient compared to the traditional SELEX which takes more time and reagents. The SELEX process provides assessment and characterization of the progress of each SELEX round selection by direct comparison between the background elution and the target binding and monitors the enrichment of target-specific aptamer during selection by use of FITC-labeled ssDNA and biotin-labeled ssDNA aptamers. The configuration of the combinatorial SELEX process has the additional flexibility to introduce the desired change for selection of aptamer not only based on His-tag protein targets but also for any purified protein targets. We have provided methodological background for successful selection of aptamers for SEB toxin of S. aureus (a biowarfare agent). The selected aptamers had the affinity which was comparable to what was achieved in earlier published reports, and it could well recognize the target protein with an affinity constant in very low nanomolar range proves protocol efficiency.
References
Baines IC, Colas P (2006) Peptide aptamers as guides for small-molecule drug discovery. Drug Discov Today 11(7):334–341
Becker KC, Becker RC (2006) Nucleic acid aptamers as adjuncts to vaccine development. Curr Opin Mol Ther 8(2):122–129
Berezovski MV, Musheev MU, Drabovich AP, Jitkova JV, Krylov SN (2006) Non-SELEX: selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat Protoc 1(3):1359–1369
Cox J, Rajendran M, Riedel T, Davidson EA, Sooter LJ, Bayer TS, Ellington AD (2002a) Automated acquisition of aptamer sequences. Comb Chem High Throughput Screen 5(4):289–299
Cox JC, Hayhurst A, Hesselberth J, Bayer TS, Georgiou G, Ellington AD (2002b) Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res 30(20):e108
Drolet DW, Jenison RD, Smith DE, Pratt D, Hicke BJ (1999) A high throughput platform for systematic evolution of ligands by exponential enrichment (SELEX [TM]). Comb Chem High Throughput Screening 2:271–278
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Ellington AD, Szostak JW (1992) Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures
Gupta P, Waheed SM, Bhatnagar R (1999) Expression and purification of the recombinant protective antigen of Bacillus anthracis. Protein Expr Purif 16(3):369–376
Homola J, Dostalek J, Chen S, Rasooly A, Jiang S, Yee SS (2002) Spectral surface plasmon resonance biosensor for detection of staphylococcal enterotoxin B in milk. Int J Food Microbiol 75(1):61–69
Humet M, Carbonell T, Masip I, Sánchez-Baeza F, Mora P, Cantón E, Messeguer A (2003) A positional scanning combinatorial library of peptoids as a source of biological active molecules: identification of antimicrobials. J Comb Chem 5(5):597–605
Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45(9):1628–1650
Kärkkäinen RM, Drasbek MR, McDowall I, Smith CJ, Young NW, Bonwick GA (2011) Aptamers for safety and quality assurance in the food industry: detection of pathogens. Int J Food Sci Technol 46(3):445–454
Keefe AD, Pai S, Ellington A (2010) Aptamers as therapeutics. Nat Rev Drug Discov 9(7):537–550
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Li M (2000) Applications of display technology in protein analysis. Nat Biotechnol 18(12):1251–1256
Liu A, Zhang Y, Chen W, Wang X, Chen F (2013) Gold nanoparticle-based colorimetric detection of staphylococcal enterotoxin B using ssDNA aptamers. Eur Food Res Technol 237(3):323–329
McBride MT, Gammon S, Pitesky M, O’Brien TW, Smith T, Aldrich J, Venkateswaran KS (2003) Multiplexed liquid arrays for simultaneous detection of simulants of biological warfare agents. Anal Chem 75(8):1924–1930
Murphy MB, Fuller ST, Richardson PM, Doyle SA (2003) An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res 31(18):e110
Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S (2003) Protein analysis on a proteomic scale. Nature 422(6928):208–215
Rajendran M, Ellington AD (2002) Selecting nucleic acids for biosensor applications. Comb Chem High Throughput Screen 5(4):263–270
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning (Vol. 1, No. 7.58). Cold spring harbor laboratory press, New York
Steinhauer C, Wingren C, Hager AC, Borrebaeck CA (2002) Single framework recombinant antibody fragments designed for protein chip applications. Biotechniques 33:38–45
Stoltenburg R, Reinemann C, Strehlitz B (2007) SELEX—a (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 24(4):381–403
Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185:60–89
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968):505–510
Wilson DS, Szostak JW (1999) In vitro selection of functional nucleic acids. Annu Rev Biochem 68(1):611–647
Zuker M (1989) On finding all suboptimal foldings of an RNA molecule. Science 244(4900):48–52
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415
Acknowledgments
The first author is indebted to Department of Science and Technology (DST, New Delhi, India), for financial assistance through INSPIRE program. The third author is indebted to Department of Science and Technology (DST, New Delhi, India), for financial assistance through Women Scientist-A (WOS-A). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 604 kb)
Rights and permissions
About this article
Cite this article
Mondal, B., Ramlal, S., Lavu, P.S.R. et al. A combinatorial systematic evolution of ligands by exponential enrichment method for selection of aptamer against protein targets. Appl Microbiol Biotechnol 99, 9791–9803 (2015). https://doi.org/10.1007/s00253-015-6858-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6858-9