Abstract
Based on natural selection and the survival of the fittest by evolutionary adaption, a smart high-throughput system was developed to select active haloalkane dehalogenase variants from a large mutant library. Only active enzyme variants can hydrolyse toxic halogenated alkanes to promote growth, whereas inactive mutants starve or die due to the toxic compound. With this powerful tool, huge enzyme mutant libraries can be screened within a few days. The selection is done without any artificial substrates that are hard to synthesize and they also resemble typical ones for haloalkane dehalogenases. Three saturation libraries, with a size of more than 106 cells, based on inactive variants of the haloalkane dehalogenases DhaA or DhlA were successfully screened to retrieve active enzymes. The enrichment of the active wild-type enzyme in contrast to the inactive variants was about 340-fold. In addition, this selection approach can be applied for continuous directed evolution experiments for the enrichment of cells expressing adapted haloalkane dehalogenases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Haloalkane dehalogenases (HLD) are enzymes, which can degrade toxic pollutants by cleaving the carbon-halogen bond of halogenated aliphatic compounds (Janssen 2004). They belong to the α/β hydrolase fold family with a core domain bearing the catalytic triad of Asp-His-Asp/Glu and a variable, mostly helical cap domain, which provides essential residues to stabilize the transition state, bind substrates and products and determine the selectivity. Completing the catalytic pentad, two residues (Trp and Trp/Asn) of the latter domain form the halide binding site (Chovancova et al. 2007). The hydrolytic reaction turns (mostly toxic) halogenated alkanes to the corresponding, non-hazardous alcohols. The need for new haloalkane dehalogenases or the engineering of characterized enzymes is high as they are useful for, e.g., detoxification of halide compounds (Bosma et al. 2002) or the production of enantiopure fine chemicals (Prokop et al. 2010; van Leeuwen et al. 2012). The first described haloalkane dehalogenase was in 1985 the DhlA from Xanthobacter autotrophicus GJ10, which is able to hydrolyse 1,2-dichloroethane (Keuning et al. 1985). The majority of the haloalkane dehalogenases were discovered by strain isolation (Kulakova et al. 1997) and in silico sequence comparisons (Jesenska et al. 2005).
One major challenge is that enzymes evolved by nature often lack essential properties like sufficient enantioselectivity, temperature stability (Arnold and Volkov 1999), substrate spectrum (Böttcher et al. 2008) or possess only poor activity (Bornscheuer and Pohl 2001). Directed evolution as a protein engineering method can help to improve the benefits of these enzymes as demonstrated for numerous examples and many enzyme classes (Chen and Arnold 1993; Jochens et al. 2011). The process of engineering leads to mutant libraries, which are constructed by various mutagenesis methods. Afterwards, the obtained variants need to be screened for the new or improved feature. Directed evolution is one option for protein engineering and can be used to generate custom-made enzymes with desired properties for industrial applications (Bornscheuer 2013). In contrast to rational protein design, where an extensive knowledge of the structure and the catalytic mechanism is needed and which only covers a small sequence space, directed evolution can be performed without this information, but it requires suitable high-throughput screening or selection methods (Arnold 1998; Bornscheuer et al. 2012; Kazlauskas 2009).
Selection methods are commonly used in many application areas (Acevedo-Rocha et al. 2014). Usually, auxotrophic strains or antibiotic resistance are the basis for these selection procedures. Unfortunately, the establishment of selection assays for protein engineering is hard because finding of toxic substrates, which fit the individual requirements of the engineering target, is difficult (Schmidt-Dannert and Arnold 1999), so that only a few examples were reported. For instance, auxotrophic E. coli strains have been used to engineer aminotransferases (Yano et al. 1998) or to develop an alteration of the enzyme topology of chorismate mutase (MacBeath et al. 1998). In another work, an alkyltransferase-deficient strain was utilized in a selection method using the mutagen N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) to create functional mutants of human DNA-alkyltransferases (Christians and Loeb 1996). We have used cell sorting by FACS analysis in combination with pseudoenantiomeric substrates to select esterase variants with improved enantioselectivity (Fernández-Álvaro et al. 2011; Reetz et al. 2008). In all procedures, selection of desired mutants was performed through survival of adapted phenotypes to accumulate the fittest variants.
Haloalkane dehalogenases such as DhaA (Newman et al. 1999) or DhlA (Janssen et al. 1989) became of interest for protein engineering experiments (Chaloupkova et al. 2003; Pavlova et al. 2009). The lack of easy to use high-throughput assays turned out to be a major hindrance as screening for active or enantioselective haloalkane dehalogenase variants could only be achieved with low throughput using a pH assay (Holloway et al. 1997) or gas chromatographic (GC) analysis (van Leeuwen et al. 2012). For the pH assay, the proton released during the reaction causes a pH shift indicated by phenol red, with the disadvantage of only a modest sensitivity and the drawback that often false-positive variants are detected, especially when crude cell lysate is used. Alternatives like tryptophane quenching (Tang et al. 2003) or the use of isothermal titration calorimetry (Arnold and Volkov 1999) provide a hint of substrate binding and are thus extremely sensitive, but cannot be used for the analysis of mutant libraries as purified enzyme is required. One selection system for dehalogenases was established, but this is restricted to Pseudomonas strains able to grow on long chain alcohols released (Kazlauskas 2009) and was also limited to a small number of substrates (Kawasaki et al. 1995). We here describe a concept for the selection of active haloalkane dehalogenase variants, which can be derived from directed evolution approaches or metagenome libraries using typical substrates of this enzyme class. This method is based on growth of E. coli strains expressing the haloalkane dehalogenase variants in the presence of toxic substrates. Active (desired) variants will detoxify the substrate and hence turn it into a carbon source, thus providing a growth advantage for the E. coli bearing the gene encoding the desired enzyme mutant. To verify the selection method, different libraries of two non-functional haloalkane dehalogenase enzymes—by mutating the essential catalytic residues—were created and subsequently selected for rescued haloalkane dehalogenase activity in the growth assay.
Materials and methods
Template preparation and plasmid construction
The genes encoding DhlA (GenBank AAA88691.1) and EchA (Nardini et al. 1999) (GenBank CAA73331.1) were cloned in the pJOE plasmid (Altenbuchner 1988) with restriction enzymes NdeI and HindIII. The DhlA gene was ligated into plasmid pET11a (Novagen, Germany) via NdeI and BamHI and the DhaA gene, cloned into plasmid pET21b, was a kind gift from the group of Prof. Jiří Damborský from Masaryk University, Brno, Czech Republic (Dvorak et al. 2014). The CIF gene (GenBank ABJ12172.1) was ordered commercially (GenScript, USA) and subcloned in vector pET28a with NcoI and XhoI.
Site directed and saturation mutagenesis
Oligonucleotides for the mutation of the DhlA gene were GGTCGTTCAGGCGTGGGGCGG and CTGTACGAAAGCGCCAGCGTC for the substitutions D124A and H289A, respectively. For saturation mutagenesis, the underlined nucleotides were replaced by NNK (CTGGTCGTACAGNNKTGGGGCGGA) or NDT (CTGTACGAAAHNGCCAGCGTC), respectively. N represents all possible nucleotides, K only thymine and guanine and D adenine, guanine and thymine. Plasmid-depending primers, flanking the multi-cloning site of pET, were TAATACGACTCACTATAGGG (T7) and CTAGTTATTGCTCAGCGGTG (pET-RP). These oligonucleotides were used for sequencing or PCR. The respective mutants of DhaA (CATCCACGCGTGGGGCTCAGC, D106A or GGAGGTACGCCAATCCCGG, H272A) were created analogously with the following saturation of the nucleophile (CATCCACNNKTGGGGCTCAGC, A106NNK) and/or the catalytic base (CCTGGAGGTAAHNCAATCCC, A272NDT).
To create the alanine mutants, the genes were amplified in a PCR: For the nucleophile pET-RP and for H289A T7-primer was used in the reaction with the corresponding primer. The mixture with 50 ng of each primer, 100 ng template, 2.5 U PfuPlus! and 0.2 mM NTPs in the appropriate buffer was incubated at 95 °C for 2 min, then for 35 cycles at 95 °C for 30 s, 57 °C for 60 s and 72 °C for 1.5 min. The final elongation was at 72 °C for 5 min. The PCR product was the template for a MEGAWHOP reaction (Miyazaki 2003). The product was mixed with the template plasmid, and the subsequent reaction was performed at 68 °C for 5 min, then at 95 °C for 1 min, 10 cycles at 95 °C for 30 s, 55 °C for 30 s and 68 °C for 7 min and 14 cycles at 95 °C for 30 s, 55 °C for 30 s and 68 °C for 11 min. Afterwards, the original plasmid was digested with DpnI for 2 h at 37 °C, followed by an inactivation step at 80 °C for 20 min. The megaprimer with the mismatch for saturation mutagenesis was also amplified in a PCR and a MEGAWHOP. All genes were finally transformed into TOP10 cells. For experiments with BL21 (DE3) cells, the plasmids of an overnight culture from the freshly transformed TOP10 cells were isolated and transformed into BL21 (DE3).
Plasmid-DNA of colonies was isolated with the innuPREP Plasmid Mini Kit (Analytic Jena, Germany) and sequenced (Eurofins Genomics GmbH, Germany).
Cultivation conditions
The E. coli strains BL21 (DE3) and TOP10 were cultivated in Luria-Bertani (LB) medium with 10 g/l NaCl, 10 g/l tryptone and 5 g/l yeast extract at pH 7.5 with 100 μg/ml ampicillin or 50 μg/ml kanamycin. For agar plates, 15 g/l agarose was added to the LB medium. The M9 minimal medium for the toxicity tests and the selection assay was composed of 50 mM NaH2PO4, 22 mM KH2PO4, 8.5 mM NaCl, 20 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 6 μM thiamine hydrochloride and 4 μM biotin at pH 6.6 with the respective antibiotic.
Toxicity tests
The haloalkane dehalogenases and epoxide hydrolases were freshly transformed into TOP10 or BL21 (DE3) E. coli cells. The OD600 of 30 ml LB media in shaking flasks was set to 0.05 with an overnight culture of the cells, and they were induced with a final concentration of 0.2 % L-rhamnose or 0.1 mM IPTG. After overnight growth and expression at 20 °C, cells were harvested (4500 g, 20 min at 4 °C) and set to an OD600 of 10 after washing twice with an appropriate volume of selection media without supplementation. An amount of 5 ml selection media in culture tubes was inoculated for cultivation with 50 μl washed cells (final OD600 of 0.1). Toxic substrates were added to their final concentrations with 2 % acetonitrile and 0.1–0.4 % glucose. The OD600 was measured regularly to determine the change in growth between the different cell cultures and substrate concentrations. The growth ratio was calculated as OD600 values compared to a similar selection culture without toxin. This way, the inhibition effect concerning the growth of the cells could be traced.
Selection assay
An overnight culture was inoculated with a freshly transformed DhaA or DhlA library in BL21 (DE3) cells. A 30 ml LB culture was set to an OD600 of 0.05 with these cells. At an OD600 of 0.4, the culture was induced with L-rhamnose or IPTG (final concentrations 0.2 % or 0.1 mM, respectively) and cultivated further at 20 °C for 14 h. Afterward, the OD600 was set to 10 in M9 media, and the cells were washed twice with not supplemented selection media. A total of 300 μl of the final suspension was used to inoculate the 30-ml selection approaches. The selection media contained 30 ml M9 media with 100 μg/ml ampicillin/50 μg/ml kanamycin, 0.02 % L-rhamnose/0.1 mM IPTG, 20 mM toxic substrate (in acetonitrile, to a final concentration of 2 % acetonitrile) and 0.1–0.4 % glucose, depending on the substrate. The final OD600 was set to 0.1. The culture was incubated 24 h at 37 °C at 180 rpm shaking in an incubator. A dilution was plated onto agar plates, and the complete selection approach was centrifuged at 4 °C for 20 min and 2000g. The whole pellet was washed with M9 media, and a new round of selection started by inoculating a freshly prepared selection media with the whole pellet. The colonies on the plates were counted to determine the cfu/ml.
Results
Selection of substrates and toxicity tests
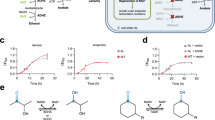
In a toxicity test, pre-induced cultures expressing the haloalkane dehalogenases DhaA or DhlA were incubated for 24 h with different substrates (Fig. 1) in concentrations from 0 to 50 mM.
For selection, the toxic substrates are converted to harmless alcohols and in case of substrates 1 and 2 to glycerol, a growth substrate. Preliminary tests were performed with 0.2 % glucose in selection media for substrates 3–7 and without glucose for substrates 1 and 2 (Fig. 2) using DhaA or DhlA as representatives of haloalkane dehalogenases and the two epoxide hydrolases EchA (Nardini et al. 1999) or CIF (Bahl et al. 2010) as negative controls in growth experiments (Fig. 2). After determining the proper concentration range of the haloalkanes (usually ~15–25 mM), the glucose concentration was also varied from 0.1 to 0.4 %. Only the combinations of substrate/glucose inhibiting the growth of cells expressing haloalkane dehalogenase inactive enzyme meanwhile cells with active enzymes survive were used in subsequent selection approaches.
In preliminary experiments, the expression of DhlA and EchA was L-rhamnose-induced, but its consumption as carbon source disturbed the selection process, especially for substrates 1 and 2. Therefore, the expression system was switched to the inducer isopropyl β-D-1-thiogalacto-pyranoside (IPTG) to determine more suitable selection conditions (Fig. 3), with CIF as new negative control.
Growth of bacteria expressing DhaA, DhlA or the haloalkane dehalogenase inactive CIF in selection media with suitable combinations of glucose and toxin concentrations. Substrates 3–7 20 mM with 0.1 % (4 and 6), 0.2 % (3 and 7), 0.4 % (5) and without glucose (1 and 2, both 15 mM). Relative growth was calculated in dependency to the growth of toxin-free cultures under same cultivation conditions
The glucose concentration was stepwise lowered from 0.4 % for substrates 3–7 to adjust the selection pressure for an optimal inhibition of haloalkane dehalogenase-inactive cells. For substrates 1, 4 and 6, a high background of inactive cells is expected, because in the toxicity tests, the relative growth, meaning the inhibition ratio depending of the culture growing in presence of toxin compared to a toxin-free growth control, was higher than 45 % (Fig. 3).
Application of the selection process with non-functional DhlA-mutants
With these preliminary results at hand, selection assays with saturation libraries of different inactive DhaA/DhlA mutants were performed. For this, first the essential residues D106/D124 (nucleophile) and H272/H289 (base) involved in the catalytic mechanism of DhaA/DhlA (respectively) were replaced by alanine substitutions to ensure that only inactive variants are used before the subsequent selection experiments.
As a proof of concept, these mutants were created (double mutant D106A/H272A and single mutants D106A or H272A for DhaA; double mutant D124A/H289A and single mutants D124A or H289A for DhlA). Subsequently, the same positions were then saturated with NNK (nucleophile), NDT (base) codons or both (dual library, ‘DUAL’). This method ensured the complete inactivation of the enzymes and a complete repression of background activity from non-mutated wild-type haloalkane dehalogenases. The initial substitution of catalytically important residues guaranteed the proper statistical analysis of the calculated reversion rates. Only variants, which were reverted to the wild-type enzyme, should be selected. The resulting library size is up to 384 possible variants per mutagenesis: The saturation mutagenesis with the degenerate NNK codon includes all 20 amino acids in 32 codons. Aspartate is only encoded by GAT, according to one codon out of 32 possible triplets (3.125 %). The degenerate NDT triplet covers 12 amino acids in 12 codons, whereas the chance for histidine (CAT) is 8.33 %. The randomization of both positions leads to 384 possible combinations of all codons with the final chance to get an aspartate/histidine pair of thus 0.26 %. To visualize the cellular behaviour during the selection process, growth curves were performed by daily measurements (Fig. 4). Especially for substrates 3, 5 and 7, the negative control could not grow under the same conditions and died out throughout the selection.
Despite choosing the conditions for the selection carefully, in the case of 1-bromobutane or 1-iodopropane, the negative control showed growth even when only the minimum concentration of glucose was used (Fig. 5).
Two additional experiments were performed to vary the selection conditions. First, the induction of the cells was performed at a higher OD600 of 0.8 to test the habituation of the cells to the media. The second concept was a 100-fold dilution of the selection media after 48 h to thin out the culture. As a result, there were no differences detectable concerning the enrichment of active haloalkane dehalogenase variants. The selection approach comprised up to 106 cells. These results could also be successfully reproduced with two additional libraries containing inactive haloalkane dehalogenase variants as a starting point. After 48 h, the relative amount of active wild-type haloalkane dehalogenase was more than 50 % in the DhlA library A124NNK and nearly 100 % in the DhlA A289NDT library as confirmed by sequencing.
Suitable dilutions of cultures transformed with genetic material from saturation mutagenesis libraries were spread on LB agar plates to achieve single colonies. These were transferred to microtiter plates and analysed with the pH assay with phenol red for haloalkane dehalogenase activity (Holloway et al. 1997). The assay process was automatized on a robotic platform to reach maximum throughput and to screen a sufficient colony number of the selected combinatorial libraries (data unpublished). The reversion rate was calculated from phenol red-positive variants (hence active haloalkane dehalogenases) compared to the total number of analyzed clones. Again, positive colonies were sequenced to confirm the detection of wild-type enzyme. The best reversion rates were 75–95 % in case of DhaA and 44–100 % for DhlA, depending on the substrate used (Table 1).
Discussion
While using the selection assay, we were able to screen successfully for haloalkane dehalogenase activity in saturation mutagenesis libraries. Reverted and thus active mutants were selected from three different mutant libraries representing increasing complexity. After incubation under continuous selection, reversion rates up to 100 % were achieved with the fastest enrichment within 24 h. Only active variants and in certain cases mutants with an integrated stop codon (data not shown) were found, but no other inactive variants of DhaA and DhlA. The toxicity of different substrates was investigated to gain an effective growth inhibition with an adjustable selection pressure. Even if substrates 1, 4 and 6 promoted the growth of the negative control, an accumulation of haloalkane dehalogenase active cells was found in the saturation libraries. The decreased toxicity of those substrates cannot be explained exactly, because there was not found any specific trend in their toxic alkylation mechanisms. For the approaches without glucose addition, substrate 2 shows more promising results for the selection of DhaA cultures. More generally, the growth results of bacteria expressing the haloalkane dehalogenases used seem to be specific for one or the other enzyme (Fig. 3) and are in agreement to their catalytic activities for the respective substrates (Koudelakova et al. 2013; Koudelakova et al. 2011). Therefore, the selection setup requires fine-tuning for the proper selection of DhaA (substrates 2, 4 and 6) or DhlA (substrates 1, 3, 5 and 7). Lower toxin concentrations used in the toxicity tests were not sufficient for the bacterial selection, and higher toxin amounts even can inhibit cells expressing functional haloalkane dehalogenases.
Regarding the growth curves of the cultures with the respective substrates, the results were quite diverse due to the substrate specificity of the haloalkane dehalogenases and the overall selection pressure mediated by the limiting growth substrate and the applied toxin concentration. The negative control CIF showed also growth for substrates 1 and 2 during the selection process. The release of glycerol from the cells containing active haloalkane dehalogenase will presumably promote the overall proliferation resulting in a time frame too small for sufficient selection. It has to be taken into account that dying cells, bearing expressed protein derived from preinduced cultures, liberate a multitude of potential metabolites.
However, cells expressing active haloalkane dehalogenases reached higher optical densities at any time point for all substrates, enhancing the feasibility of their successful accumulation. Moreover, the strength of the selection was focused on increasing the content of cells with active haloalkane dehalogenase enzyme in the combinatorial saturation libraries (Table 1). Their growth behaviour was quite similar compared to the pure wild-type selection cultures, but also reached slightly higher cell densities (Fig. 4). This is possible due to the faster enrichment and growth of the respective wild-type cells, favouring the consumption of glucose. The majority of cells containing both saturation libraries (DUAL) will die due to the toxin and set free additional substrates, which promotes a stronger proliferation. Only well-defined selection pressure leads to the consumption of the carbon source exclusively by the minority of the bacteria expressing the desired active haloalkane dehalogenases. These cultures reached the stationary phase (the maximum possible cell growth depending on the added substrates) the earliest after 2 days, but at the latest after 5-day selection. For the proper selection of active wild-type haloalkane dehalogenases, a minimum of 1-day cultivation was necessary to reach a 346- to 384-fold enrichment, corresponding to a reversion of 90–100 % wild-type in the combinatorial saturation libraries: When the assay was set up, only one combination of 384 codons provides the essential aspartate/histidine pair, which is necessary for the catalytic activity detected in the phenol red assay. A reversion rate of 90 % resulted from an achieved distribution of 346 variants out of the 384, which are now wild-type enzyme. The enzyme activities could be thus rescued from a starting distribution of 0.26 % for all substrates. Additionally, single saturation libraries were selected, and a high rate of activity retention was found. Those rates were higher for NDT than NNK saturation because of the higher number of possible codons and in consequence the lower chance to achieve a wild-type reversion.
Even in the approaches with 1-bromobutane and 1-iodopropane, the wild-type activity could be successfully rescued. With the chance of selecting false negatives, because of growth of the negative control (without haloalkane dehalogenase activity), a selection is difficult with the latter substrates. Increasing the substrate concentrations or decreasing the glucose amount also led to the segregation of haloalkane dehalogenase-expressing cells. Nevertheless, with the substrates 4 and 6, we were able to enrich the original activity in the culture, and thus, they could be suitable for examining other dehalogenases (Fetzner and Lingens 1994; Janssen et al. 2001). Random samples of positive clones in the phenol red assay were sequenced and clearly identified as reverted wild-type haloalkane dehalogenases.
The development and identification of new biocatalysts with activities against haloalkane pollutants are possible with this approach (Gribble 2010). The adaption of this method can also be useful for the selection of enantioselective dehalogenases in an approach with enantiopure compounds. The selection of metagenome libraries with this concept may also lead to the discovery of new dehalogenases. These results envisage that a similar selection assay for other hydrolases (such as epoxide hydrolases using glycidol derivatives as substrate (Archer 1997; Swaving and deBont 1998)) may work as well (Acevedo-Rocha et al. 2014). In this selection assay on the one hand, the formation of a growth substrate from a toxic compound offers the possibility to avoid the use and construction of deficient strains with the risk of reversion by mutation (Roth et al. 2006; Steele and Jinks-Robertson 1992). On the other hand, the toxicity of readily available compounds led to the selective enrichment of a specific dehalogenase depending on the individually preferred substrate and enables the possibility of the selection for enzymes with precisely defined characteristics, e.g., the adaption of additional factors like cosolvents (Stepankova et al. 2013), which are easily adaptable to our concept.
References
Acevedo-Rocha CG, Agudo R, Reetz MT (2014) Directed evolution of stereoselective enzymes based on genetic selection as opposed to screening systems. J Biotechnol 191:3–10
Altenbuchner J (1988) A new E.coli cloning vector containing a melanin marker for insertion screening. Nucleic Acids Res 16(17):8710
Archer IVJ (1997) Epoxide hydrolases as asymmetric catalysts. Tetrahedron 53(46):15617–15662
Arnold FH (1998) Design by directed evolution. Acc Chem Res 31(3):125–131
Arnold FH, Volkov AA (1999) Directed evolution of biocatalysts. Curr Opin Chem Biol 3(1):54–59
Bahl CD, Morisseau C, Bomberger JM, Stanton BA, Hammock BD, O’Toole GA, Madden DR (2010) Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor. J Bacteriol 192(7):1785–1795
Bornscheuer UT (2013) Protein engineering as a tool for the development of novel bioproduction systems. Adv Biochem Eng Biotechnol 137:25–40
Bornscheuer UT, Pohl M (2001) Improved biocatalysts by directed evolution and rational protein design. Curr Opin Chem Biol 5(2):137–143
Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K (2012) Engineering the third wave of biocatalysis. Nature 485(7397):185–194
Bosma T, Damborsky J, Stucki G, Janssen DB (2002) Biodegradation of 1,2,3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene. Appl Environ Microbiol 68(7):3582–3587
Böttcher D, Schmidt M, Bornscheuer UT (2008) Alteration of substrate specificity and stereoselectivity of lipases and esterases protein engineering handbook. Wiley-VCH Verlag GmbH & Co. KGaA, pp 753–775
Chaloupkova R, Sykorova J, Prokop Z, Jesenska A, Monincovaa M, Pavlova M, Tsuda M, Nagata Y, Damborsky J (2003) Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J Biol Chem 278(52):52622–52628
Chen K, Arnold FH (1993) Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc Natl Acad Sci 90(12):5618–5622
Chovancova E, Kosinski J, Bujnicki JM, Damborsky J (2007) Phylogenetic analysis of haloalkane dehalogenases. Proteins 67(2):305–316
Christians FC, Loeb LA (1996) Novel human DNA alkyltransferases obtained by random substitution and genetic selection in bacteria. Proc Natl Acad Sci 93(12):6124–6128
Dvorak P, Bidmanova S, Damborsky J, Prokop Z (2014) Immobilized synthetic pathway for biodegradation of toxic recalcitrant pollutant 1,2,3-trichloropropane. Environ Sci Technol 48(12):6859–6866
Fernández-Álvaro E, Snajdrova R, Jochens H, Davids T, Böttcher D, Bornscheuer UT (2011) A combination of in vivo selection and cell sorting for the identification of enantioselective biocatalysts. Angew Chem 50:8584–8587
Fetzner S, Lingens F (1994) Bacterial dehalogenases: biochemistry, genetics, and biotechnological applications. Microbiol Rev 58(4):641–685
Gribble GW (2010) Naturally occurring organohalogen compounds: a comprehensive update. Springer, Wien
Holloway P, Trevors JT, Lee H (1997) A colorimetric assay for detecting haloalkane dehalogenase activity. J Microbiol Methods 32(1):31–36
Janssen DB (2004) Evolving haloalkane dehalogenases. Curr Opin Chem Biol 8(2):150–159
Janssen DB, Pries F, Vanderploeg J, Kazemier B, Terpstra P, Witholt B (1989) Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhla gene. J Bacteriol 171(12):6791–6799
Janssen DB, Oppentocht JE, Poelarends GJ (2001) Microbial dehalogenation. Curr Opin Biotechnol 12(3):254–258
Jesenska A, Pavlova M, Strouhal M, Chaloupkova R, Tesinska I, Monincova M, Prokop Z, Bartos M, Pavlik I, Rychlik I, Mobius P, Nagata Y, Damborsky J (2005) Cloning, biochemical properties, and distribution of mycobacterial haloalkane dehalogenases. Appl Environ Microbiol 71(11):6736–6745
Jochens H, Hesseler M, Stiba K, Padhi SK, Kazlauskas RJ, Bornscheuer UT (2011) Protein engineering of alpha/beta-hydrolase fold enzymes. ChemBioChem 12(10):1508–1517
Kawasaki H, Kuriyama H, Tonomura K (1995) Use of haloacetate dehalogenase genes as selection markers for Escherichia coli and Pseudomonas vectors. Biodegradation 6(3):213–216
Kazlauskas R (2009) Engineering enantioselectivity in enzyme-catalyzed reactions. In: Lutz S, Bornscheuer UT (eds) Protein engineering handbook. Wiley-VCH, Weinheim, pp 15–47
Keuning S, Janssen DB, Witholt B (1985) Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol 163(2):635–639
Koudelakova T, Chovancova E, Brezovsky J, Monincova M, Fortova A, Jarkovsky J, Damborsky J (2011) Substrate specificity of haloalkane dehalogenases. Biochem J 435(2):345–354
Koudelakova T, Bidmanova S, Dvorak P, Pavelka A, Chaloupkova R, Prokop Z, Damborsky J (2013) Haloalkane dehalogenases: biotechnological applications. Biotechnol J 8(1):32–45
Kulakova AN, Larkin MJ, Kulakov LA (1997) The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB 13064. Microbiology 143(Pt 1):109–115
MacBeath G, Kast P, Hilvert D (1998) Redesigning enzyme topology by directed evolution. Science 279(5358):1958–1961
Miyazaki K (2003) Creating random mutagenesis libraries by megaprimer PCR of whole plasmid (MEGAWHOP). Methods Mol Biol 231:23–28
Nardini M, Ridder IS, Rozeboom HJ, Kalk KH, Rink R, Janssen DB, Dijkstra BW (1999) The X-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1 - An enzyme to detoxify harmful epoxides. J Biol Chem 274(21):14579–14586
Newman J, Peat TS, Richard R, Kan L, Swanson PE, Affholter JA, Holmes IH, Schindler JF, Unkefer CJ, Terwilliger TC (1999) Haloalkane dehalogenases: structure of a Rhodococcus enzyme. Biochemistry 38(49):16105–16114
Pavlova M, Klvana M, Prokop Z, Chaloupkova R, Banas P, Otyepka M, Wade RC, Tsuda M, Nagata Y, Damborsky J (2009) Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat Chem Biol 5(10):727–733
Prokop Z, Sato Y, Brezovsky J, Mozga T, Chaloupkova R, Koudelakova T, Jerabek P, Stepankova V, Natsume R, van Leeuwen JG, Janssen DB, Florian J, Nagata Y, Senda T, Damborsky J (2010) Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering. Angew Chem Int Ed Engl 49(35):6111–6115
Reetz MT, Hobenreich H, Soni P, Fernandez L (2008) A genetic selection system for evolving enantioselectivity of enzymes. Chem Commun 43:5502–5504
Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI (2006) Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol 60:477–501
Schmidt-Dannert C, Arnold FH (1999) Directed evolution of industrial enzymes. Trends Biotechnol 17(4):135–136
Steele DF, Jinks-Robertson S (1992) An examination of adaptive reversion in Saccharomyces cerevisiae. Genetics 132(1):9–21
Stepankova V, Bidmanova S, Koudelakova T, Prokop Z, Chaloupkova R, Damborsky J (2013) Strategies for stabilization of enzymes in organic solvents. ACS Catal 3(12):2823–2836
Swaving J, de Bont JAM (1998) Microbial transformation of epoxides. Enzym Microb Technol 22(1):19–26
Tang LX, van Merode AEJ, Spelberg JHL, Fraaije MW, Janssen DB (2003) Steady-state kinetics and tryptophan fluorescence properties of halohydrin dehalogenase from Agrobacterium radiobacter. Roles of W139 and W249 in the active site and halide-induced conformational change. Biochemistry 42(47):14057–14065
van Leeuwen JG, Wijma HJ, Floor RJ, van der Laan JM, Janssen DB (2012) Directed evolution strategies for enantiocomplementary haloalkane dehalogenases: from chemical waste to enantiopure building blocks. ChemBioChem 13(1):137–148
Yano T, Oue S, Kagamiyama H (1998) Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc Natl Acad Sci 95(10):5511–5515
Acknowledgments
We are grateful to Jiří Damborský (Department of Experimental Biology, Masaryk University, Brno, Czech Republic) for providing the gene encoding the haloalkane dehalogenase DhaA from Rhodococcus rhodochrous.
Conflict of interests
The authors declare that there is no financial or any other conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fibinger, M.P.C., Davids, T., Böttcher, D. et al. A selection assay for haloalkane dehalogenase activity based on toxic substrates. Appl Microbiol Biotechnol 99, 8955–8962 (2015). https://doi.org/10.1007/s00253-015-6686-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6686-y