Abstract
This study provides a comprehensive, long-term microbiological study of a continuously operated, mesophilic, agricultural biogas plant fed with whole-crop silages of maize and rye, cattle manure and cattle slurry. The microbial community structure was accessed by high-throughput 16S rRNA gene amplicon sequencing. For the characterisation of the microbial dynamics, the community profiling method terminal restriction fragment length polymorphism (TRFLP) in combination with a cloning-sequencing approach as well as a LC-MS/MS approach for protein identification were applied. Our results revealed that the anaerobic digestion is a highly sensitive process: small variations in the process performance induce fluctuations in the microbial community composition and activity. In this context, it could be proven that certain microbial species were better adapted to changing process condition such as temperature (interspecies competition) and that there is a physiological compensation between different microorganisms so that the reactor efficiency was not adversely affected despite of structural and functional changes within the microbial community.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One central concern of an environmentally friendly, sustainable energy policy is the use of high-yield methane-containing biogas as alternative energy source (Weiland 2010; Appels et al. 2011; Lebuhn et al. 2014). During the anaerobic digestion, conversion of organic material to biogas is mediated by a complex interacting microbial consortium (Talbot et al. 2008; Ward et al. 2008; Nelson et al. 2011). The process can be disturbed when a single degradation step is out of balance. Frequent reasons are temperature increase caused by a self-induced heating effect or the overproduction of volatile fatty acids (VFA) in combination with inappropriate buffer capacity (e.g. Blume et al. 2010; Fotidis et al. 2013; Lindorfer et al. 2008). In case of large-scale biogas plants (BGPs), such imbalances can lead to dramatic economic losses (Weiland 2010; Nelson et al. 2011). Hence, a well-balanced microbial consortium providing high efficiency as well as resilience towards disturbances is commonly regarded as the most crucial for the reactor performance and the biogas production.
The biochemical metabolic pathways at different process stages of anaerobic digestion are well described, and molecular biological analysis of the process microbiology with respect to the composition and dynamics of the involved microorganisms revealed a high community complexity and broad range of metabolic interactions (e.g. Nettmann et al. 2010; Wang et al. 2010; Carballa et al. 2011; Regueiro et al. 2012; Fotidis et al. 2013). The following molecular biological methods are commonly used to study the structural and dynamic variations of biogas-producing microbial communities: cloning/sequencing (C/S) (e.g. Rademacher et al. 2012), quantitative real-time PCR (Q-PCR) (e.g. Blume et al. 2010), community profiling techniques such as the terminal restriction fragment length polymorphism (TRFLP) (e.g. Rademacher et al. 2012), the denaturing gradient gel electrophoresis (DGGE) (e.g. Liu et al. 2009) or the microarray technology (Franke-Whittle et al. 2009; Goberna et al. 2010) and also the high-throughput metagenome or amplicon sequencing (Schlüter et al. 2008; Krause et al. 2008; Jaenicke et al. 2011; Zakrzewski et al. 2012). These analyses mainly base on the investigation of the 16S rRNA gene as this gene gives the most reliable information about the phylogenetic relation of organisms and is therefore useful for a description of microbial communities (Lane et al. 1985; Talbot et al. 2008; Tringe and Hugenholtz 2008; Kim et al. 2011). In addition to these DNA-based methods, proteomics is increasingly applied to characterise a microbial community (Benndorf and Reichl 2014; Vanwonterghem et al. 2014; Heyer et al. 2015). Recently, metaproteomic analyses were successfully applied to investigate functional microbial communities of anaerobic digesters (Hanreich et al. 2013; Heyer et al. 2013; Kohrs et al. 2014). Ideally, a comprehensive investigation combining genomic and proteomic approaches in correlation with physicochemical process parameters can show how community structure and function are determined in its respective habitat.
Recent genomic studies of lab- and full-scale anaerobic digesters revealed that the composition of the microbial community is strongly affected by process conditions, e.g. temperature, substrate composition or pH value (e.g. Carballa et al. 2011; Nettmann et al. 2010; Pycke et al. 2011; Wang et al. 2010). Unfortunately, no signatures of a “healthy”, well-balanced, efficiently working microbial consortium for the production of biogas were described so far. Furthermore, one of the main questions which is still under discussion is the optimal degree of community complexity, a less complex community of highly specialised workhorses or a more complex community with functional redundant and often less efficient species. Accordingly, there is an urgent need to better understand the levels of microbial community organisation and the functionality within BGPs. Identification of key organisms (biomarkers) related to specific process conditions could serve as basis for microbiological monitoring, control and management.
In this study, we characterised the microbial community of an agricultural biogas plant using a polyphasic, complementary approach of different molecular biological methods, namely 16S rRNA gene amplicon sequencing, TRFLP fingerprinting in combination with a cloning/sequencing approach and metaproteomic analysis. The aim of this study was to investigate the microbial community resistance against process fluctuations as an important property for ecosystem stability. We assumed that even small variations, e.g. in the temperature regime or a combined effect of temperature, pH value and the ammonium nitrogen content (NH4 +-N), will cause structural and functional variations within the microbial community. We further supposed that these variations will compensate the changing process condition without influencing the overall biogas production process.
Materials and methods
Reactor performance and sampling
The analysed agricultural, mesophilic (43 ± 2 °C) BGP is operating since 2007 and consists of two parallel-operated main fermenters (MF) and one residue storage tank. Each continuously stirred tank reactor (CSTR) has a volume of 1500 m3. The BGP was continuously fed with 35.1 × 103 ± 2.4 × 103 kgFM solid and liquid feedstock mixture (energy crops and agricultural waste) per day, whereby only 21 % of the fresh mass (FM) was volatile solids (VS) and therefore available for biodegradation. The BGP was fed with whole-crop silages of maize (46 % fresh mass in 2011 resp. 54 % FM in 2012) and rye (14 % FM in 2011 resp. 9 % FM in 2012) as well as cattle manure (ca. 15 % FM) and cattle slurry (ca. 22 % FM). The BGP was operated with an organic loading rate (OLR) of 1.7 ± 0.2 kgVS m−3 day−1 and a hydraulic retention time (HRT) of 129 ± 9 days. The average biogas yield was nearly constant with 0.7 ± 0.07 m3 kgVS −1 and a methane content of 51.4 ± 2.1 %. As the biogas plant consists of two MF, the partial biogas volume fluxes were added up as total biogas volume flux. The biogas yield was determined by the daily total biogas volume flux (measured before entering the combined heat and power unit (CHP) unit), the daily average biogas composition and the daily VS supply.
Samples were taken from the first MF according to VDLUFA (2011) to be representative of the fermenter contents at its current state (supplementary “Text S1”). Sampling was carried out in two campaigns: in July 2011 and over a time period of 14 weeks from February to May 2012.
The process data (e. g. feeding, process temperature, biogas yield and methane content) were recorded daily and provided by the biogas plant operator.
The following chemical analyses were conducted with weekly sampled feedstock and fermenter material according to VDLUFA (1997): total solids (TS); volatile solids (VS); total Kjeldahl nitrogen (TKN); total ammonium nitrogen (NH4 +-N); soluble volatile fatty acids (VFA) in terms of acetate, propionate, iso- and n-butyrate, iso- and n-valerate and capronate; pH; total volatile fatty acids/total alkalinity (TVFA/TA; ratio between TVFA and TA); and conductivity. For a better estimation of the biomass conversion process, crude protein (XP); crude fat (XL); neutral detergent fibre (NDF); acid detergent fibre (ADF) and acid detergent lignin content (ADL) were additionally determined. Calculation of specific process parameters can be found in supplementary material “Text S2”.
Molecular biological and proteomic analyses were performed monthly (at calendar week (CW) 30 in 2011 as well as CW 06, 10, 14 and 19 in 2012), whereby the digestate samples were stored at −80 °C until further preparation.
Extraction of total microbial DNA
Total microbial DNA was extracted using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories Inc., USA) according to manufacturer’s instructions. The mechanical cell disruption was performed using the FastPrep® instrument (MP Biomedical, USA). For each crude sample, DNA from three subsamples was extracted. The extracted DNA was then used as template for the TRFLP analyses as well as for the construction of 16S rRNA gene sequence libraries and the 16S rRNA gene amplicon datasets to characterise the diversity and dynamics of the bacterial and archaeal communities.
Analysis of the microbial community structure by high-throughput 16S rRNA gene amplicon sequencing
Taxonomic characterisation of the microbial community was done for the sample CW 19, 2012, applying high-throughput 16S rRNA gene amplicon sequencing as described previously (Caporaso et al. 2012). Briefly, to amplify the hypervariable region V4 (as recommended by Klindworth et al. 2013) of the 16S rRNA gene, the bacterial/archaeal primers 515F (5′ - GTGCCAGCMGCCGCGGTAA - 3′) and 806R (5′ - GGACTACHVGGGTWTCTAAT - 3′) (Caporaso et al. 2012), complemented with Multiplex Identifier (MID) tags as well as Illumina-specific sequencing adaptor sequences, were used. Fragments of the expected length of approximately 300 bp were amplified by PCR in three technical replicates. Purification of PCR products was done with AMPureXP® magnetic beads (Beckman Coulter Inc.). Further qualitative and quantitative analyses of the generated amplicons were performed using the Agilent 2100 Bioanalyzer system (Agilent Inc.). Constructed 16S rRNA gene amplicon libraries were pooled in equimolar amounts and diluted for the sequencing procedure to 10 nM.
Obtained amplicon libraries were sequenced on Illumina MiSeq system by applying the paired-end protocol. All sequences obtained in this study have been deposited to the European Molecular Biology Laboratory (EMBL) and are available under the project accession number PRJEB8240. Raw sequences were processed using the UPARSE pipeline (Edgar 2013) based on Usearch 7.0 (Edgar 2010) with default settings. Processed operational taxonomic units (OTUs) were taxonomically classified by means of the RDP classifier 2.7 in 16S modus (Wang et al. 2007) using the release version 11.1 of the RDP database (http://rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp). Only assignments featuring a confidence value of at least 0.8 (phylum rank) were considered. In the next step, raw sequences were mapped on the OTUs to get quantitative assignments. Finally, the CopyRighter software (Angly et al. 2014) was applied for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. The taxonomic profile was visualised using Krona tool (Ondov et al. 2011).
Microbial community dynamics analysed by TRFLP fingerprinting and 16S rRNA gene sequence libraries
The TRFLP analyses were in general carried out following the protocols proposed by Rademacher et al. (2012) and Klang et al. (2015). Briefly, the bacterial and archaeal 16S rRNA genes were amplified (two replicates per crude DNA extract) using the primer pairs 27F/926MRr (Bacteria) and Ar109f/Ar912r (Archaea) whereby the forward primer were fluorescently labelled. After purification of the PCR products, 150–200 ng were digested with MspI and Hin6I in case of the bacterial assay or with AluI for the archaeal assay. The digestion fragments were separated using a GenomeLab™ GeXP Genetic Analysis System (Beckman Coulter, Krefeld, Germany). The obtained data were pre-analysed using the GeXP analysis software (version 10.2) whereby only profiles were considered for further analyses whose internal standard had a standard deviation of 0.39 nucleotides (nt) or less (Rademacher et al. 2012). A detailed analysis was then performed using the software package BioNumerics 7.1 (Applied Maths, Belgium) according to Klang et al. (2015). The identification of “true” terminal restriction fragments (TRFs) by distinguishing background and baseline “noise” or false positives (bleed through peaks) from signals of correctly fluorescent-labelled fragments as well as the alignment (band matching) of TRFs with a threshold of 0.1 based on the evaluation of the peak height. Finally, TRFs were visualised by their relative distribution within the profile pattern.
For the identification of the detected TRFs, a 16S rRNA gene sequence library was constructed from sample CW 19, 2012, whereby the obtained sequences were processed using the software package BioNumerics 7.1 (Applied Maths, Belgium) following the protocol by Klang et al. (2015). After a quality check of the sequences, OTUs were defined at 97 % (Bacteria) resp. 99 % (Archaea) sequence similarity required for the identification at the species level (Kim et al. 2011). All new sequences obtained in this study have been deposited to the European Molecular Biology Laboratory (EMBL) and are available under accession numbers HG530255-HG530303 (Bacteria) and HG530304-HG530309 (Archaea). The defined OTUs were cut virtually using the restriction digest tool of BioNumerics 7.1 to assign the detected TRFs of the reactor samples. Finally, OTUs were phylogenetically identified using the RDP Naïve Bayesian rRNA Classifier Version 2.6 (Wang et al. 2007).
Monitoring of the metabolic activity by analysis of the microbial metaproteome
For proteome analysis, protein extraction, quantification, separation of 100 μg proteins on SDS-PAGE, as well as tryptic digestion and peptide separation on a liquid chromatography (LC) system, were carried out according to Heyer et al. (2013). LC system was coupled online to a Velos Pro/OrbiTrap Elite hybrid mass spectrometer (MS, Thermo Fisher Scientific). For MS acquisition, a data-dependent MS/MS method was chosen. Precursor scanning was performed in the orbital trap at a resolution of 30,000 and a m/z range of 350-2,000 in positive ion mode. Subsequently, fragment scan was preceded in the linear ion trap with “normal” parameter settings (mass range, scan rate) for the top 20 most intense precursors selected for collision-induced dissociation. Acquired data files were loaded with Proteome Discoverer 1.4 (Thermo Fisher Scientific, Bremen, Germany) without any post processing and directly exported as Mascot Generic Format (MGF) files with default parameter settings. For database search, MGFs were uploaded to ProteinScape (version 3.0.0.337, Bruker Daltonik, Bremen, Germany) and automatically submitted to MASCOT search against UniProt database (version 2013/02/20) using the following parameters: trypsin, one missed cleavage, monoisotopic mass, carbamidomethyl (C) as fixed and oxidation (M) as variable modifications, ±0.03 Da precursor tolerance, ±0.5 Da MS/MS tolerance, 1 13C and +2/+3 peptide charge. Mascot DAT files were exported to MetaProteomeAnalyzer software (Muth et al. 2015) including the MascotDatfile library (Hensens et al. 2007) for parsing results. Protein hits were accepted with an overall dataset false discovery rate of minor 5 %. For taxonomic classification of the microbial community, redundantly assigned peptide-to-protein matches have to be avoided. The assigned protein taxonomies were inherited to its peptides. Afterwards, all proteins sharing at least one peptide were fused to so-called metaproteins (MPs). For taxonomic classification of MP, the common ancestor of all unique peptides in the phylogenetic tree was determined and finally assigned to MP. Only protein and MP hits with a spectral count (SpC) of at least 2 were considered as verified. The metaproteomic data are available on request from the corresponding author and can be examined in more details using the MetaProteomeAnalyzer Viewer application provided (see https://code.google.com/p/meta-proteome-analyzer/ for download and supporting material).
Statistical analysis
Various ecological indices were applied which mainly were based on the microbial resource management concept (Verstraete et al. 2007; Marzorati et al. 2008; Read et al. 2011). The richness (R) was determined as the total number of detected TRFs. The Jaccard index was computed as it measures the similarity between samples and hence can be used as an index for dynamic community changes (Marzorati et al. 2008; Pycke et al. 2011). It is defined as the intersection of two samples derived by the sum of TRFs in samples A and B minus the intersection. From an ecological point, this means the higher the Jaccard index, the more similar the compared samples. Additionally, we defined the Lorenz curve and the derived Gini coefficient for each sample which is related to information about the community organisation (Verstraete et al. 2007; Marzorati et al. 2008; Wittebolle et al. 2009). The Lorenz curves were used to illustrate the community disparity compared to the perfect equality line. Based on the Lorenz curves, the Gini coefficient was calculated as the normalised area between a given Lorenz curve and the perfect evenness line. The higher the Gini coefficient, the more uneven is the community.
In order to identify representative (indicator) species or more precisely TRFs of particular environmental conditions (e. g. different temperature regimes), the indicator species analysis (ISA; Dufrêne and Legendre 1997) was performed with the software package of PC Ord Version 6 (McCune and Mefford 2011). This analysis considers the proportional abundance of a TRF within a sample relative to the abundance in all samples and the relative frequency within the sample. As a result, ISA produces indicator values (IV ranging from 0 to 100, absent to exclusively present) for each TRF in defined groups of a given environment.
Additionally, we calculated correlation coefficients among and between the operational and biological parameters using a linear regression. Tests for the statistical significance were conducted to evaluate if the correlations were considerable or not. Statistical significance was established at the p < 0.05 level.
Results
Biogas plant performance
The MF performance of the analysed agricultural BGP in July 2011 and over a 3-month time period from February to May 2012 is shown in Fig. 1 and Table S1. Constant operation of the BGP (e.g. in terms of the feeding and temperature) over the entire sampling period resulted in constant process values and hence a stable reactor performance shown by the biogas yield. The average biogas yield was nearly constant with 0.7 ± 0.07 m3 kgVS −1 and a methane content of 51.4 ± 2.1 %. This biogas/methane yield was the result of the anaerobic co-digestion of whole-crop maize and rye silage combined with cattle manure and cattle slurry. In regard to the process efficiency, overall 76.2 ± 2.8 % of the total VS were degraded and thus representing a well-operating agricultural plant. As expected, the highest biomass conversion was detected for easy degradable carbohydrates (e. g. sugar) and for cellulose with more than 80 %, followed by hemicellulose and raw fat with around 70 % and raw proteins with 60 % (data not shown). Based on the potential maximum biogas yield of the supplied feedstocks (maize and rye silage, 0.9 m3 kgVS −1; cattle slurry, 0.6 m3 kgVS −1; Mähnert and Linke 2009), the large-scale overall biomass conversion and the resulting biogas yield demonstrate high performance and efficiency of biogas technology.
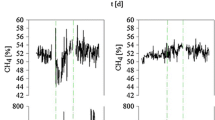
Feedstock supply given as fresh mass input (a), process temperature, ammonium nitrogen (NH4 +-N), free ammonia nitrogen (NH3-N) content and biogas yield (b), as well as volatile fatty acids (VFA) and pH value (c), during the sampling campaign in July 2011 and over a time period of 14 weeks from February to May 2012. Sampling points which were chosen for the biological analyses are marked in red. All values which are shown in the graphs are additionally given in Table S1. Missing values (gaps) in (b) for the NH4 +-N and NH3-N content are caused by laboratory problems. CW calendar weeks, AAE acetic acid equivalents, AS acetic acid, PS propionic acid, nBA n-butyric acid, iBA iso-butyric acid, nVA n-valeric acid, iVA iso-valeric acid, CA capronic acid
Additionally, a stable pH of 7.9 ± 0.1 and a constant time course of TVFA/TA ratio of 0.1 further indicated a well-functioning biogas reactor. The ammonium nitrogen (NH4 +-N) and the free ammonia nitrogen (NH3-N) contents showed average values of 2.5 ± 0.2 and 0.3 ± 0.08 g L−1, resp. In contrast to most other process parameters observed, the total VFA content showed significant changes and ranged between 0.07 and 2.17 g L−1 AAE (acetic acid equivalents) (Fig. 1, Table S1).
Microbial community structure based on high-throughput 16S rRNA gene amplicon sequencing
To determine the taxonomic profile of the biogas community, high-throughput sequencing of the hypervariable V4 region of the 16S rRNA gene was performed on the Illumina MiSeq system by applying the paired-end protocol.
Sequencing of the 16S rRNA gene amplicons on average resulted in 91,871 ± 12,551 raw reads (supplementary material Table S2). After processing (quality control, chimaera detection, taxonomic classification and 16S rRNA gene copy number correction), approximately 80,000 sequences per replicate were obtained whereby 99.9 % of the 16S rRNA gene sequences were assigned to the domains Bacteria and Archaea. Figure 2 illustrates that Bacteria with 99 % of all sequences dominated within the biogas reactor, whereas 1 % of the sequences were classified as Archaea. Overall, 42 % of the identified sequence reads could be assigned to ten different phyla. The predominant phyla were Firmicutes with 27 ± 1.5 % and Bacteroidetes with 13 ± 1.0 % at the bacterial level and Euryarchaeota with 1 % at the archaeal level. The taxonomic profiling of the microbial community revealed in total 15 different classes, 8 different orders and 21 different families. Only 30 % of the sequence reads could be assigned at the order rank with Clostridiales as the most abundant one (15 to 18 %), followed by Bacteroidales with 11 to 12 % and Methanosarcinales with 0.6 to 0.8 %. At the family rank, the 16S rDNA amplicon library uncovered the dominance of Porphyromonadaceae with 10 % (order Bacteroidales), Clostridiales incertae sedis XI with 5 to 6 % and Ruminococcaceae with 4 to 6 % (both order Clostridiales). The archaeal community was predominated by the genus Methanosarcina (order Methanosarcinales) with 0.6 to 0.8 %.
Taxonomic profiling of the biogas plant microbial community of CW 19, 2012, based on 16S rRNA gene amplicon sequences. Data from three replicates were combined in order to describe the taxonomic profile of the analysed anaerobic digester. 16S rRNA gene sequences were classified using the RDP classifier. For the taxonomic profile, only assignments with a confidence level of 0.8 were considered. The profile was visualised using Krona tools (Ondov et al. 2011)
Microbial community dynamic profiling by TRFLP analyses
For time-dependent profiling of the microbial community, the fingerprinting method TRFLP targeting either the bacterial or the archaeal 16S rRNA gene was used (Fig. 3). In case of the bacterial community, a total of 27 different TRFs were found most prevalent whereby 21 TRFs were identified by the constructed 16S rRNA gene sequence library (see supplementary material Table S3).
Structural composition of the bacterial (a) and archaeal (b) communities involved in the biomethanation process of the analysed BGP. The relative abundance of the detected terminal restriction fragments (TRFs) as a function of the percental fluorescence intensity of each individual TRF in relation to the total fluorescence intensity is shown. Numbers symbolise the dominant TRFs in base pairs (bp)
With 19 to 29 %, TRF-150 bp was the most abundant bacterial TRF in all samples. In the 16S rRNA gene sequence library, five different OTUs were found that showed a virtual TRF of 150 bp with an overall abundance of 25 % confirming the quantification by TRFLP analysis. These OTUs were clearly related to the phylum Firmicutes, presumably to the class Clostridia. Within this group, OTU-B039 (representing 17 % of the sequences) was further related to the order Clostridiales.
Four further bacterial TRFs were determined as predominant: TRF-178 bp (9 to 11 %) and TRF-181 bp (6 to 10 %) were also assigned to the phylum Firmicutes while TRF-86 bp (6 to 9 %) and TRF-93 bp (3 to 8 %) were related to the family Porphyromonadaceae (order Bacteroidales, phylum Bacteroidetes). In the latter case, the corresponding OTUs of TRF-86 bp and TRF-93 bp covered around 15 % of the entire 16S rRNA gene sequence database, which is in accordance with the TRFLP results.
With eight different TRFs in total, the archaeal community was less diverse compared to the bacterial one. With up to 63 %, the TRF-627 bp dominated all samples. TRF-470 bp showed a high abundance in CW 30, 2011 (45 %), whereas it reached only 3 to 5 % within the other samples. Three further archaeal TRFs (TRF-84 bp, TRF-174 bp and TRF-318 bp) were found in all five samples, each with an average of 5 %. Generally, almost all detected TRFs were assigned to the order Methanosarcinales (see supplementary material Table S3). The constructed archaeal 16S rDNA sequence library showed a clear dominance of one OTU with 87 % which had a theoretical TRF of 627 bp. This OTU was assigned to Methanosarcina barkeri (99 % sequence identity to NR_074253). Furthermore, TRF-470 bp was related to be a relative of Methanosarcina acetivorans (90 % sequence identity to NR_044724), while TRF-340 bp (detected only in CW 30, 2011, with 2 % and CW 19, 2012, with 4 %) was identified to be a relative of Methanobacterium formicicium (95 % sequence identity to NR_025028) from the order Methanobacteriales.
In conclusion, the bacterial community organisation was more even (Gini coefficients between 0.34 and 0.43) than the archaeal one (Gini coefficients between 0.45 and 0.61) as shown in Table 1. Furthermore, Jaccard indices were calculated in order to characterise the dynamics of the microbial community during the biogas production process. The estimated Jaccard indices showed that the microbial communities of the first sample (CW 30, 2011) and the last sample (CW 19, 2012) had a similarity value of 65 %. Both communities differ with up to 60 % from the microbial communities of CW 06, 2012, CW 10, 2012, and CW 14, 2012, whereby the last three showed similarity values of 68 to 80 % indicating the period with the most stable community structure (Table 1).
Functional profiling by metaproteomic analysis
Both bacterial (mean of 27.2 ± 14) and archaeal MPs (mean of 24.6 ± 7) were identified and can be investigated in detail using the provided MPA viewer. Archaeal MP hits were assigned to Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway maps in order to evaluate the abundance of enzymes involved in acetoclastic (KEGG module M00357) and hydrogenotrophic methanogenesis (KEGG module M00567). For acetoclastic methanogenesis, acetyl-CoA decarbonylase/synthase complex (ACDS) and its respective subunits (alpha, beta, gamma, delta and epsilon) were detected in all samples (Fig. 4, Table S4). Methylenetetrahydromethanopterin dehydrogenase has been detected as well, but with minor abundance, indicating a weak activity of methanogens that use the hydrogenotrophic pathway of methane formation. Proteins which are involved in both pathways showed the highest abundance ranging from 55 to 70 % (Fig. 4): tetrahydromethanopterin S-methyltransferase (subunit A, H and E), V-type H+-transporting ATPase subunit B and methyl-coenzyme M reductase (subunits alpha, beta and gamma). The latter is the key enzyme in the final step of methanogenesis. In addition, trimethylamine methyltransferase MttB has also been detected in all samples. This protein is involved in the formation of methane using tertiary amines (KEGG module M00563). Additionally, methanol-5-hydroxybenzimidazolylcobamide co-methyltransferase was identified in two of five samples (Fig. 4) indicating partial activity of methanogens using methanol (KEGG module M00356) as intermediate.
Relative distribution of the detected archaeal metaproteins of the analysed BGP. The relative abundance of enzymes involved in the acetoclastic pathway (KEGG module M00357, green) or the hydrogenotrophic pathway (KEGG module M00567, orange), proteins which are involved in both pathways (KEGG module M00357 and M00567, dark grey) as well as the relative abundance of trimethylamine methyltransferase MttB (KEGG module M00563, light grey) and methanol-corrinoid protein co-methyltransferase (KEGG module M00356, black) is shown
Spectral counts (SpC) recorded for MPs of the acetoclastic methanogenesis clearly showed a higher abundance for samples CW 30, 2011 (33 %), as well as from CW 06, 2012, to CW 14, 2012, with around 20 % compared to SpC for the hydrogenotrophic proteins with 3 to 4 % (Fig. 4) corresponding to an acetoclastic to hydrogenotrophic ratio of 10 for CW 30, 2011, resp. of 5 for CW 06, 2012, to CW 14, 2012. Most methane was formed from acetate. For the sampling point CW 19, 2012, an evident increase in SpC for the hydrogenotrophic-related enzyme methylenetetrahydromethanopterin dehydrogenase was recorded resulting in a decrease of the acetoclastic to hydrogenotrophic ratio to a factor of 2.
Taxonomic assignments of MP using the common ancestor strategy allowed the classification at different taxonomic levels. Most of the identified archaeal MPs, especially enzymes from the ACDS complex, were assigned to the archaeal genus Methanosarcina. Additionally, for the MPs methyl-coenzyme M reductase and methanol-5-hydroxybenzimidazolylcobamide co-methyltransferase, the evaluation revealed a highly specific assignment on species level to M. barkeri. The hydrogenotrophic-related enzyme methylenetetrahydromethanopterin dehydrogenase was assigned at the order level to Methanomicrobiales. V-typeH+-transporting ATPase subunit B and tetrahydromethanopterin S-methyltransferase were matched at the class level to Methanomicrobia.
Correlation of the system microbiology with prevalent process parameters
It appears that the system microbiology, especially the community organisation, was correlated with the process temperature as the bacterial and archaeal communities were more uneven in samples at higher process temperature (45 °C in the first and last samples, Fig. 5). Additionally, the ISA revealed that species with a generally lower abundance (the specialists) were mainly affected. Four bacterial TRFs (TRF-99 bp, TRF-167 bp, TRF-296 bp and TRF-490 bp; related to the order Clostridiales) and one archaeal TRF (TRF-340 bp, related to the genus Methanobacterium) were exclusively (indicator value (IV) > 80, p = 0.0002) found in the samples of CW 30, 2011, and CW 19, 2012 (Table S5). This indicates a competitive advantage of these species over other community members at higher temperature regimes.
A combination of higher temperature, higher NH4 +-N concentration but lower pH and subsequently lower NH3-N concentration led to a high abundance of the TRF-470 bp which is related to the mixotrophic archaeal family Methanosarcinaceae. This suggests that the TRF-470 bp-related organism(s) probably was/were responsible for maintaining the acetoclastic pathway of methane production in CW 30, 2011, with an IV of 75 (p = 0.0008) for the prevalent NH3-N content and with an IV of 80 (p = 0.0004) for the amount of acetoclastic proteins (Table S6). In contrast to that, hydrogenotrophic methanogenesis (especially the orange TRF-340 bp) became more abundant when the NH3-N content exceeded 400 mg L−1 (Table S6).
Discussion
Conventional procedural-chemical parameters are still the best source for monitoring the process performance of anaerobic digesters and are routinely used in large-scale BGPs for controlling (Drosg 2013). Biogas formation itself, however, is driven by the physiological activity of the present microbial community. Consequently, actual microbiological research efforts focus on correlating process parameters and process microbiology (structure and activity) in order to optimise the environmental conditions for the growth, fitness and activity of microorganisms and hence to improve the biogas production. In this context, the presented study investigated new insights into the microbial community behaviour by combining genomic and proteomic approaches.
General biogas plant performance
Our results generally showed that the overall biogas production process was stable during the trial period as we found no statistical significant correlations between the process parameters feedstock, OLR, HRT, process temperature, VFA, NH4 +-N or NH3-N content and the biogas resp. the methane yield (data not shown).
However, our results also revealed that the anaerobic digestion is a complex, sensitive and hence dynamic process. All prevalent procedural-chemical parameters are subjected to small temporal variations as shown in Fig. 1. It is well known that the temperature is one of the most important factors driving the anaerobic digestion process as it affects the microbial activity and hence the hydrolysis, acido-/acetogenesis and methanogenesis rates (Donoso-Bravo et al. 2009). According to Weiland (2010), the analysed BGP was operated at a temperature regime between the optimal values for meso-resp. thermophilic conditions: While the anaerobic digestion was performed at 45 °C in July 2011, from February 2012 to May 2012, a successive increase in the process temperature from 41 to 45 °C occurred (Fig. 1b, Table S1). These temperature changes can have several reasons: (i) a heating effect due to an increase in the ambient temperature in combination with a decrease of heat emission into the environment or (ii) the fermenter showed a self-induced heating effect as reported by Lindorfer et al. (2008). Independent from the reason, the temperature increase of the fermenter can lead to several process imbalances such as an increase in the NH3-N content as it was found, for example, in CW 19, 2012. But the NH3-N content is not only a function of changing temperature conditions but also a function of the NH4 +-N content and the pH value (Fig. 1b, Table S1). As it is generally accepted that NH3-N is an important regulator for methane formation (Chen et al. 2008), the observed acid accumulation (from CW 06, 2012, to CW 08, 2012, and from CW 11, 2012, to CW 13, 2012; Fig. 1c, Table S1) can be interpreted as a failing degradation of the produced fatty acids causing a slight decrease in the biogas yield (Fig. 1b, Table S1).
As the anaerobic digestion is mediated by a complex interacting microbial consortium, the aim of this study was an ecological interpretation of the behaviour of the process microbiology under particular environmental condition, and further to evaluate potential process key players. The fact that the mentioned variations in the prevalent procedural-chemical process parameters had no significant negative effect on the overall biogas production led to the assumption that the microbial community was either resistant, resilient and/or functional redundant and therefore enabled to compensate the sudden stress situation (Allison and Martiny 2008).
Community structure and its metabolic potential
Based on the 16S rRNA gene amplicon database, the biogas-producing microbial community was dominated by Bacteria with 99 % whereby the Archaea were only represented with 1 %. These findings have previously been reported, e. g. by Liu et al. (2009); Carballa et al. (2011); Regueiro et al. (2012) and Sundberg et al. (2013), and are in agreement with the fact that Bacteria are involved in the first three steps of biomass transformation with a high variety of substrates whereby Archaea are restricted to a very narrow substrate spectrum in terms of acetate, methyl-group containing compounds as well as CO2 and H2.
According to Bergey’s Manual of Systematic Bacteriology (Krieg et al. 2010; Vos et al. 2011), all detected bacterial community members are typically involved in the anaerobic degradation of the supplied feedstock as they are described to have cellulolytic, saccharolytic, glycolytic, lipolytic, proteolytic and/or acido-/acetogenic capacities. Regarding the biomass transformation cascade, the 16S rRNA gene amplicon database revealed, for example, species from the genera Ruminococcus, Acetivibrio (phylum Firmicutes, family Ruminococcaceae) and Petrimonas (phylum Bacteroidetes, family Porphyromonadaceae) which are reported to be involved in the anaerobic digestion of various carbohydrates including monosaccharide and disaccharides, hemicellulose, cellulose and/or pectin. The proteolytic genera Sedimentibacter (phylum Firmicutes, family XI incertae sedis) and Proteiniphilum (phylum Bacteroidetes, family Porphyromonadaceae) were also recorded. Additionally, the 16S rRNA gene amplicon database further recorded the occurrence of acetogenic species from the family Syntrophomonadaceae (phylum Firmicutes) which oxidise saturated fatty acids such as butyrate in syntrophic association with hydrogenotrophic methanogens, for example, of the genera Methanospirillum from the order Methanomicrobiales (Schink and Stams 2013; Schmidt et al. 2013). In conclusion, the occurring bacterial species are enabled to perform a successive and complementary biomass conversion with a functional redundancy among diverse phylogenetic groups.
Concerning the archaeal community and hence the major pathway of methane formation in full-scale anaerobic digesters, previous studies reported a predominance of the hydrogenotrophic methanogenesis mediated by species from the orders Methanobacteriales and Methanomicrobiales (e. g. Nettmann et al. 2010; Regueiro et al. 2012; Fotidis et al. 2013). In contrast, both genomic and metaproteomic analyses of the here presented study clearly showed a dominance of acetoclastic methanogenesis derived from the activity of species from the genus Methanosarcina (family Methanosarcinaceae). Among the methanogenic Archaea, the family Methanosarcinaceae includes the most versatile methanogens (De Vrieze et al. 2012). They can use acetate, methanol, methylated amines, carbon monooxide and CO2/H2 as substrates (Liu and Whitman 2008). This high metabolic diversity is reflected by a high number of genes encoding for methanogenesis proteins resulting in a relative large genome sizes as it was shown for M. barkeri strain Fusario with 4.8 Mb or Methanosarcina acetivorans strain C2A with 5.8 Mb (Galagan et al. 2002; Maeder et al. 2006). The presence of proteins involved in methanogenesis from acetate, methanol and methylamines in the recorded metaproteomic data confirms that this extensive genetic potential is used, and Methanosarcina species succeed by following a generalist strategy as they switch between substrates and cope with variable substrate concentrations (Galagan et al. 2002; Maeder et al. 2006).
Community dynamics and adaptation to changing environmental conditions
Our results indicate that the analysed BGP consisted of a well-established microbial community which is able to compensate variations in the surrounding environment. According to the microbial resource management (MRM) approach (Verstraete et al. 2007; Marzorati et al. 2008; Read et al. 2011) and as shown by the study of Carballa et al. (2011), the medium community organisation values found in this study (Gini coefficient between 0.34 and 0.43 for the bacterial and 0.45 and 0.61 for the archaeal community; Table 1) indicate a well-balanced community composition. It encompassed not only generalists that are mostly defined by their predominant occurrence but also specialists which are able to compensate environmental fluctuations and hence are responsible for maintaining the ecosystem functionality. For example, the process parameters of the BGP, in particular the VFA concentration, showed fluctuations between CW 06, 2012, and CW 14, 2012 (Fig. 1c), whereby the biogas yield (Fig. 1b) as well as the community structure (Fig. 3) and activity (Fig. 4) were not significantly affected. Up to 80 % similarity of the community structure (Table 1) and a constant metaproteome represented by an acetoclastic to hydrogenotrophic ratio of 5 (Fig. 4) are indicative for the ability of the microbial community to compensate these environmental changes without any significant negative effects on the process. These findings prove the assumption that the occurring microbial community is not only resistant but also resilient against sudden stress situations as the degree to which the microbial community remains unchanged in face of a disturbance was quite low (Allison and Martiny 2008).
Both, community and functional data, revealed that the process temperature was one major factor shaping the microbial community. At the first and last sampling point with a process temperature of 45 °C, the bacterial and archaeal community organisation was more uneven (Fig. 5). Not surprisingly, the ISA for these sampling points clearly identified the exclusive occurrence of bacterial species from the order Clostridiales indicating the well-known competitive advantage over species from the phylum Bacteroidetes at higher temperature (Rademacher et al. 2012). However, as the process temperature of the analysed BGP has never reached commonly defined thermophilic conditions (50–60 °C), species of the phylum Bacteroidetes were constantly represented with at least 20 to 30 % of the entire community. Additionally, higher temperature also supported the occurrence of hydrogenotrophic methanogens symbolised by the detected TRF-340bp, a relative of Methanobacterium formicicum. However, the activity of the TRF-related organism(s) was obviously suppressed in CW 30, 2011, as we found a tenfold higher abundance of MPs of the acetoclastic pathway in CW 30, 2011, compared to those of the hydrogenotrophic pathway (Fig. 4). In contrast, hydrogenotrophic MPs were enriched in CW 19, 2012, and the acetoclastic to hydrogenotrophic ratio decreased to a factor of 2. Probably the higher concentration of free ammonia nitrogen (NH3-N, 440 mg L−1) in CW 19, 2012, caused an inhibition of the mixotrophic archaeal family Methanosarcinaceae (especially a close relative of Methanosarcina acetivorans represented by TRF-470 bp) and opened a niche for hydrogenotrophic methanogens from the genus Methanobacterium. To conclude, these findings distinctly showed that both microbial or more precisely the archaeal communities (from CW 30, 2011, and CW 19, 2012) were functional similar as they carried out a functional process at a similar rate, regardless of the differences in the composition (Allison and Martiny 2008).
Towards biomarkers for the biomethanation process
The here presented complementary application of 16S rRNA gene amplicon sequencing (entire community structure), community fingerprinting by TRFLP (community dynamics) and metaproteomics (community activity) together with a monitoring of procedural-chemical process parameters and the biogas yield resulted in one of the most comprehensive descriptions of full-scale agricultural BGPs. The occurring community of the analysed BGP seems to be resistant or resilient as it had a high adaptability potential to changing process conditions. These community characteristics could be ecological related to interspecies competition as well as functional redundancy and similarity among diverse phylogenetic groups. This allows fluctuation in the community structure and functionality without negative effects for the overall BGP performance and efficiency. Finally, this study impressively showed that using the strength of different methods in a polyphasic approach can compensate limitations of each applied method. For example, the here presented results revealed that different species from the mixotrophic archaeal family Methanosarcinaceae mediated either the acetoclastic (Methanosarcina acetivorans represented by TRF-470 bp) or the hydrogenotrophic pathway (Methanosarcina barkeri symbolised by TRF-627 bp) which paves the way towards the development of process-relevant molecular biomarkers.
The challenge at this point is to strengthen investigations on the characterisation on the system microbiology during disturbed resp. varying process phases. Such efforts would provide the possibility to determine process-relevant microorganisms with both a positive and a negative impact on the anaerobic biomass conversion and the related biogas yield. This may prospectively allow us to draw up recommendations for “adjustment screws” to set the process conditions better suiting the requirements of the microbial community.
References
Allison SD, Martiny JBH (2008) Resistance, resilience, and redundancy in microbial communities. PNAS 105:11512–11519
Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW (2014) CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2:11–23
Appels L, Lauwers J, Degrève J, Helsen L, Lievens B, Willems K, Van Impe J, Dewil R (2011) Anaerobic digestion in global bio-energy production: potential and research challenges. Renew Sust Energ Rev 15:4295–4301
Benndorf D, Reichl U (2014) Proteomics in environmental and technical microbiology. Eng Life Sci 14:27–46
Blume F, Bergmann I, Nettmann E, Schelle H, Rehde G, Mundt K, Klocke M (2010) Methanogenic population dynamics during semi-continuous biogas fermentation and acidification by overloading. J Appl Microbiol 109:441–450
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Carballa M, Smit M, Etchebehere C, Boon N, Verstrate W (2011) Correlation between molecular and operational parameters in continuous lab-scale anaerobic digesters. Appl Microbiol Biotechnol 89:303–314
Chen Y, Cheng JJ, Creamer KS (2008) Inhibition of anaerobic digestion process: a review. Bioresour Technol 99:4044–4064
De Vrieze J, Hennebel T, Boon N, Verstraete W (2012) Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour Technol 112:1–9
Donoso-Bravo A, Retamal C, Carballa M, Ruiz-Filippi G, Chamy R (2009) Influence of temperature on the hydrolysis, acidogenesis and methanogenesis in mesophilic anaerobic digestion: parameter identification and modeling application. Water Sci Technol 60:9–17
Drosg B (2013) Process monitoring in biogas plants. IEA Bioenergy Task 37 – Energy from Biogas
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345–366
Edgar RC (2010) Search and clustering of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Fotidis IA, Karakashev D, Angelidaki I (2013) The dominant acetate degradation pathway/methanogenic composition in full-scale anaerobic digesters operating under different ammonia levels. Int J Environ Sci Technol. doi:10.1007/s13762-013-0407-9
Franke-Whittle IH, Goberna M, Pfister V, Insam H (2009) Design and development of the ANAEROCHIP microarray for investigation of methanogenic communities in biogas reactors. J Microbiol Methods 79:279–288
Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HG, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway de Macario E, Ferry JG, Jarrell KF, Jing H, Macario AJL, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B (2002) The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Res 12:532–542
Goberna M, Gademaier M, Garcia C, Wett B, Insam H (2010) Adaptation of methanogenic communities to the cofermentation of cattle excreta and olive mill wastes at 37 °C and 55 °C. Appl Environ Microbiol 76:6564–6571
Hanreich A, Schimpf U, Zakrzewski M, Schlüter A, Benndorf D, Heyer R, Rapp E, Pühler A, Reichl U, Klocke M (2013) Metagenome and metaproteome analyses of mesophilic biogas-producing anaerobic batch fermenters indicate concerted plant carbohydrate degradation. Syst Appl Microbiol 36:330–338
Hensens K, Martens L, Vandekerckhove J, Gevaert K (2007) MascotDatfile: An open-source library to fully parse and analyse MASCOT MS/MS search results. Proteomics 7:364–366
Heyer R, Kohrs F, Benndorf D, Rapp E, Kausmann R, Heiermann M, Klocke M, Reichl U (2013) Metaproteome analysis of the microbial communities in agricultural biogas plants. New Biotechnol 30:614–622
Heyer R, Kohrs F, Reichl U, Benndorf D (2015) Metaproteomics of complex microbial communities in biogas plants. Microb Biotechnol. doi:10.1111/1751-7915-12276
Jaenicke S, Ander C, Bekel T, Bisdorf R, Dröge M, Gartemann KH, Jünemann S, Kaiser O, Krause L, Tille F, Zakrzewski M, Pühler A, Schlüter A, Goesmann A (2011) Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing. PLoS One 6(1), e14519
Kim M, Morrison M, Yu Z (2011) Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods 84:81–87
Klang J, Theuerl S, Szewzyk U, Huth M, Tölle R, Klocke M (2015) Dynamic variation of the microbial community structure during the long-time mono-fermentation of maize and sugar beet silage. Microb Biotechnol. doi:10.1111/1751-7915.12263
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41: doi:10.1093/nar/gks808
Kohrs F, Heyer R, Magnussen A, Benndorf D, Muth T, Behne A, Rapp E, Kausmann R, Heiermann M, Klocke M, Reichl U (2014) Sample prefractionation with liquid isoelectric focusing enables in depth microbial metaproteome analysis of mesophilic and thermophilic biogas plants. Anaerobe 29:59–67
Krause L, Diaz NN, Edwards RA, Gartemann KH, Krömeke H, Neuweger H, Pühler A, Runte KJ, Schlüter A, Stoye J, Szczepanowski R, Tauch A, Goesmann A (2008) Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J Biotechnol 136(1–2):91–101
Krieg NR, Ludwig W, Whitman WB, Hedlund BP, Paster BJ, Staley JT, Ward N, Brown D, Parte A (2010) Bergey’s manual of systematic bacteriology, volume 4: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. Springer, pp. 25–102
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82:6955–6959
Lebuhn M, Munk B, Effenberger M (2014) Agricultural biogas production in Germany—from practice to microbiology basics. Energy Sustain Soc 4:1–21
Lindorfer H, Waltenberger R, Köllner K, Braun R, Kirchmayr R (2008) New data on temperature optimum and temperature changes in energy crop digesters. Bioresour Technol 99:7011–7019
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic Archaea. Ann N Y Acad Sci 1125:171–189
Liu FH, Wang SB, Zhang JS, Zhang J, Yan X, Zhou HK, Zhao GP, Zhou ZH (2009) The structure of the bacterial and archaeal community in a biogas digester as revealed by denaturing gradient gel electrophoresis and 16S rDNA sequencing analysis. J Appl Microbiol 106:952–966
Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR (2006) The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J Bacteriol 188:7922–7931
Mähnert P, Linke B (2009) Kinetic study of biogas production from energy crops and animal waste slurry: effect of organic loading rate and reactor size. Environ Technol 30:93–99
Marzorati M, Wittebolle L, Boon N, Daffonchio D, Verstraete W (2008) How to get more out of molecular fingerprints: practical tools for microbial ecology. Environ Microbiol 10:1571–1581
McCune B, Mefford M J (2011) PC-ORD. Multivariate analysis of ecological data. Version 6.08 MjM Software, Gleneden Beach, Oregon, U.S.A.
Muth T, Behne A, Heyer R, Kohrs F, Benndorf D, Hoffmann M, Lehtevä M, Reuchl U, Martens L, Rapp E (2015) The MetaProteomeAnalyzer: a powerful open-source software suite for metaproteomic data analysis and interpretation. J Proteome Res 14:1557–1565
Nelson MC, Morrison M, Zhongtang Y (2011) A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol 102:3730–3739
Nettmann E, Bergmann I, Pramschüfer S, Mundt K, Plogsties V, Herrmann C, Klocke M (2010) Polyphasic analyses of methanogenic archaeal communities in agricultural biogas plants. Appl Environ Microbiol 76: 2540–2548 + Erratum Appl Environ Microbiol 77: 394.
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a web browser. BMC Bioinforma 12:385–393
Pycke BFG, Etchebehere C, van de Caveye P, Negroni A, Verstraete W, Boon N (2011) A time-course analysis of four full-scale anaerobic digesters in relation to the dynamics of change of their microbial communities. Water Sci Technol 63:769–775
Rademacher A, Nolte C, Schönberg M, Klocke M (2012) Temperature increases from 55 to 75 °C in a two-phase biogas reactor result in fundamental alterations within the bacterial and archaeal community structure. Appl Microbiol Biotechnol 96:565–576
Read S, Marzorati M, Guimarães BCM, Boon N (2011) Microbial resource management revisited: successful parameters and new concepts. Appl Microbiol Biotechnol 90:861–871
Regueiro L, Veiga P, Figueroa M, Alonso-Gutierrez J, Stams AJM, Lema JM, Carballa M (2012) Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol Res 167:581–589
Schink B, Stams AJM (2013) Syntrophism among prokaryotes. In: Rosenberg E, DeLong EF, Lory S, Stakebrandt E, Thompson F (eds) The prokaryotes, 4th edn. Springer, Berlin, Heidelberg, pp 471–493
Schlüter A, Bekel T, Diaz NN, Dondrup M, Eichenlaub R, Gartemann KH, Krahn I, Krause L, Krömeke H, Kruse O, Mussgnug JH, Neuweger H, Niehaus K, Pühler A, Runte KJ, Szczepanowski R, Tauch A, Tilker A, Viehöver P, Goesmann A (2008) The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J Biotechnol 136(1–2):77–90
Schmidt A, Müller N, Schink B, Schleheck D (2013) A proteomic view at the biochemistry of syntrophic butyrate oxidation in Syntrophomonas wolfei. PLoS ONE 8, e56905. doi:10.1371/journal.pone.0056905
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626
Talbot G, Topp E, Palin MF, Massé DI (2008) Evaluation of molecular methods used for establishing the interactions and functions of microorganisms in anaerobic reactors. Water Res 42:513–537
Tringe SG, Hugenholtz P (2008) A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 11:442–446
Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW (2014) Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol 27:55–64
VDLUFA - Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten: Methodenbuch – Band III (1997) Die chemische Untersuchung von Futtermitteln (The chemical analysis of feedstuffs). VDLUFA-Verlag, Speyer
VDLUFA - Verband Deutscher Landwirtschaftlicher Untersuchungs- und Forschungsanstalten: Methodenbuch – Band VII (2011) Umweltanalytik (environmental analysis). VDLUFA-Verlag, Speyer
Verstraete W, Wittebolle L, Heylen K, Vanparys B, de Vos P, van de Wiele T, Boon N (2007) Microbial resource management: the road to go for environmental biotechnology. Eng Life Sci 7:117–126
Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (2011) Bergey’s manual of systematic bacteriology volume 3: The Firmicutes. Spriner
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang H, Tolvanen K, Lehtomäki A, Puhakka J, Rintala J (2010) Microbial community structure in anaerobic co-digestion of grass silage and cow manure in a laboratory continuously stirred tank reactor. Biodegradation 21:135–146
Ward AJ, Hobbs PJ, Holliman PJ, Jones DL (2008) Optimisation of the anaerobic digestion of agricultural resources. Bioresource Technol 99:7928–7940
Weiland P (2010) Biogas production: current state and perspectives. Appl Microbiol Biotechnol 85:849–860
Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, de Vos P, Verstraete W, Boon N (2009) Initial community evenness favours functionality under selective stress. Nature 458:623–626
Zakrzewski M, Goesmann A, Jaenicke S, Jünemann S, Eikmeyer F, Szczepanowski R, Al-Soud WA, Sørensen S, Pühler A, Schlüter A (2012) Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. J Biotechnol 158:248–58
Acknowledgments
S. Theuerl, F. Kohrs, I. Maus and R. Kausmann were supported by the German Federal Ministry of Food, Agriculture and Consumer Protection (BMELV), grant numbers 22010711, 22028711 and 22028811 (joint research project Biogas-Biocoenosis) and 22027707 (joint research project BiogasEnzyme).
The authors would like to thank Kerstin Mundt for her excellent technical support in the laboratory.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 163 kb)
Rights and permissions
About this article
Cite this article
Theuerl, S., Kohrs, F., Benndorf, D. et al. Community shifts in a well-operating agricultural biogas plant: how process variations are handled by the microbiome. Appl Microbiol Biotechnol 99, 7791–7803 (2015). https://doi.org/10.1007/s00253-015-6627-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6627-9