Abstract
Alginates exhibit unique material properties suitable for medical and industrial applications. However, if produced by Pseudomonas aeruginosa, it is an important virulence factor in infection of cystic fibrosis patients. The alginate biosynthesis machinery is activated by c-di-GMP imparted by the inner membrane protein, MucR. Here, it was shown that MucR impairs alginate production in response to nitrate in P. aeruginosa. Subsequent site-specific mutagenesis of MucR revealed that the second MHYT sensor motif (MHYT II, amino acids 121–124) of MucR sensor domain was involved in nitrate sensing. We also showed that both c-di-GMP synthesizing and degrading active sites of MucR were important for alginate production. Although nitrate and deletion of MucR impaired alginate promoter activity and global c-di-GMP levels, alginate yields were not directly correlated with alginate promoter activity or c-di-GMP levels, suggesting that nitrate and MucR modulate alginate production at a post-translational level through a localized pool of c-di-GMP. Nitrate increased pel promoter activity in the mucR mutant while in the same mutant the psl promoter activity was independent of nitrate. Nitrate and deletion of mucR did not impact on swarming motility but impaired attachment to solid surfaces. Nitrate and deletion of mucR promoted the formation of biofilms with increased thickness, cell density, and survival. Overall, this study provided insight into the functional role of MucR with respect to nitrate-mediated regulation of alginate biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alginate is composed of β-1,4 linked β-d-mannuronic acid and α-l-guluronic acid. It is commercially harvested from brown seaweed for applications in food, biotechnology, and medicine. However, pure alginate of defined properties is usually mandatory in medical applications, and seaweed may struggle to meet this requirement due to seasonal variation. Under this circumstance, bacteria could supersede seaweed as the primary source of alginate for medical applications because alginate produced by bacteria is highly homogenous. On the other hand, during infection of the cystic fibrosis (CF) lung, the human pathogen Pseudomonas aeruginosa over-produces alginate which clogs patients’ airways and protects the pathogen against host immune responses and antibiotics, making this disease difficult to treat (Cohen and Prince 2012; Hoiby et al. 2010). Therefore, any progress on the understanding of regulatory mechanisms of alginate production in bacteria will contribute to the development of strategies to manipulate alginate production for high premium products and/or disease treatment.
In P. aeruginosa, alginate is synthesized by a multiprotein complex. The alginate precursor, GDP-mannuronic acid, is polymerized by the inner membrane (IM) proteins Alg8 and Alg44 (Oglesby et al. 2008; Remminghorst et al. 2009; Remminghorst and Rehm 2006a, b). The resulting polymer is translocated across the periplasm by a multiprotein scaffold where it is modified and then secreted from the cell (Albrecht and Schiller 2005; Baker et al. 2014; Farrell and Tipton 2012; Franklin and Ohman 2002; Gutsche et al. 2006; Hay et al. 2012; Jain et al. 2003; Jain and Ohman 1998; Rehman et al. 2013; Robles-Price et al. 2004; Tan et al. 2014).
The alginate biosynthesis machinery is activated by the ubiquitous bacterial secondary messenger, cyclic di-guanidine monophosphate (c-di-GMP), which binds to the PilZ domain of Alg44 (Amikam and Galperin 2006; Merighi et al. 2007). Another IM protein, MucR, is thought to modulate a localized pool of c-di-GMP in proximity to Alg44 to control alginate production at post-translational level (Hay et al. 2009). MucR has an IM sensor domain and two cytosolic output domains. The sensor has three MHYT motifs (named after its amino acid sequence) suggested to co-ordinate a copper ion, allowing perception of nitric oxide (NO) and other gases (Galperin et al. 2001; Hay et al. 2009). NO is released during denitrification along with other nitroactive intermediates which are linked to suppressed alginate production (Wood et al. 2007; (Zumft 1997). However, whether MucR’s sensor domain is involved in sensing these nitroactive intermediates, leading to suppressed alginate production, remains unknown.
The two cytosolic output domains of MucR, GGDEF and EAL (named after their conserved catalytic amino acid sequences), are involved in c-di-GMP synthesis [diguanylate cyclase (DGC) activity] and degradation [phosphodiesterase (PDE) activity], respectively (Hay et al. 2009; Li et al. 2013). In vitro studies indicate that although both GGDEF and EAL domains are catalytically active (Li et al. 2013), it has not yet been determined whether both activities (or active sites) are necessary for alginate production.
The majority of DGCs and PDEs characterized to date form multimeric quaternary structures—dimers or tetramers (Barends et al. 2009; De et al. 2009; Phippen et al. 2014; Rao et al. 2008; Romling et al. 2013; Sharma et al. 2014; Tarutina et al. 2006; Tchigvintsev et al. 2010; Wassmann et al. 2007). This is particularly important for DGCs, where dimerization is critical for their enzymatic activity (Romling et al. 2013). Since MucR has DGC and PDE activity, we anticipate that it should also form higher MW complexes. Furthermore, a recent study shows that a shift in oligomeric state can affect an enzyme’s catalytic activity. For instance, DcpA, another c-di-GMP modulating enzyme, functions as DGC and PDE in its dimeric form while in its monomeric form it only has PDE activity (Sharma et al. 2014). However, whether the signal perceived by MucR impacts its oligomeric state is unknown.
Previous studies demonstrated that loss of alginate production enhanced swarming motility and surface attachment by P. aeruginosa and increased production of the exopolysaccharides, Pel and Psl (Hay et al. 2009; Ghafoor et al. 2011). Here, it was proposed that if nitrate suppressed alginate production, then motility, attachment, and Pel and Psl production might increase. Pel and Psl are involved in cell-cell interactions which could affect biofilm characteristics (see below).
In this study it was investigated whether (1) nitrate triggers the suppression of alginate production through the sensor domain of MucR, (2) nitrate suppresses alginate production by impairing MucR’s DGC activity, (3) nitrate controls the oligomeric state of MucR, (4) nitrate-induced suppression of alginate production is caused by lowered global intracellular c-di-GMP levels and reduced alginate promoter activity, and whether (5) nitrate-induced suppression of alginate production enhances surface attachment and swarming motility as well as Psl and Pel production, biofilm thickness, compactness, and cell survival.
Variants of MucR containing inactive sensor and output domains were generated and characterized with respect to their functional role in nitrate-mediated regulation of exopolysaccharide biosynthesis with emphasis on alginate production. The effect of nitrate on MucR oligomeric state was studied using crosslinking and immunoblot experiments. A c-di-GMP-sensitive-promoter-lacZ reporter was employed to assess the effect of nitrate and MucR on intracellular c-di-GMP levels, i.e., post-translational regulation. Moreover, the impact of nitrate-mediated MucR-dependent regulation on surface attachment, swarming motility, and biofilm characteristics was studied.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. All Escherichia coli strains were grown in LB medium at 37 °C. E. coli S17-1 was used for conjugative transfer of mob site-containing pBBR1MCS-5, pEX100T, and mini-CTX-lacZ-derived plasmids. E. coli SM10 was used for conjugative transfer of the pFLP2 plasmid. P. aeruginosa strains were grown in LB or Pseudomonas Isolation medium, and on Pseudomonas Isolation Agar (PIA) (Difco) at 37 °C. For E. coli, antibiotics were used at the following concentrations (μg ml−1): ampicillin, 100; gentamycin (Gm), 10; and tetracycline, 12.5. For P. aeruginosa, the following concentrations were used (μg ml−1): carbenicillin, 300; Gm, 300; and Tet, 150.
Isolation, analysis, and manipulation of DNA
General cloning procedures were followed. Deoxynucleoside triphosphates, Taq, and Platinum Pfx polymerases were from Invitrogen. Restriction enzymes were from New England Biolabs. DNA was cloned using the pGEMt-easy kit (Promega, USA). DNA was verified by sequencing using an ABI310 automatic sequencer.
Construction of mucR mutant
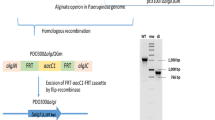
An isogenic marker free mucR mutant was generated in P. aeruginosa PDO300, through homologous recombination (Hay et al. 2009).
Construction of plasmids for expression of MucR variants
The mucR mutant was complemented with pBBR1MCS-5:mucR (Hay et al. 2009). Mutant variants of mucR were generated by inverse PCR and gene synthesis (Chiu et al. 2004) (Genscript). For each mutation, two primer pairs were designed: a pair of tailed forward/reverse primers and a pair of non-tailed-forward/reverse primers. The tailed primers are extended forms of the non-tailed primers which carry the desired mutation(s) at their 5′ ends. pBBR1MCS-5:mucR was used as template in PCR reactions containing the relevant primers (Chiu et al. 2004). PCR mixtures were digested with DpnI to remove template DNA and a hybridization step was performed (Chiu et al. 2004). Final sequence confirmed plasmids were introduced into PDO300ΔmucR. Successful transconjugants were selected on PIA Gm.
Alginate quantification and uronic acid assays
Sample preparation from solid media
Alginate yields were determined as described previously (Remminghorst and Rehm 2006b). Briefly, strains were grown overnight in LB medium. Cells were washed twice with 1 vol sterile saline and suspended to an OD of 6.0. For each plate, 1 ml of cells was suspended in 200 μl, spread onto a PIA Gm plate and incubated for 72 h at 37 °C. To examine the effect of nitrate on alginate production, the medium was supplemented with potassium nitrate to 1 % w/v.
Biomass from plates were suspended in saline and centrifuged at 7500×g at 4 °C for 30 min. Crude alginate was precipitated from the supernatant with 1 vol ice-cold isopropanol. Alginate and cells were lyophilized. Alginate samples were suspended in buffer (0.05 M Tris–HCl, 10 mM MgCl2) to a final concentration of 0.5 % w/v and incubated with DNase I (15 μg ml−1) and RNase A (15 μg ml−1) for 6 h at 37 °C with shaking at 200 rpm. After further 18 h of incubation with Pronase E (20 μg ml−1), samples were dialyzed (12 kDa cut-off) against 5 l of 0.9 % w/v NaCl for 24 h at 4 °C. Samples were precipitated with ice-cold isopropanol, freeze-dried, and subjected to uronic acid assay.
Sample preparation from liquid media
Uronic acid content of culture supernatants was analyzed (Hay et al. 2010). Briefly, strains were grown at 37 °C overnight in 20 ml of LB medium + Gm. Culture supernatants were filtered using Vivaspin 500 spin columns (10 kDa MW cut-off; GE Healthcare) by centrifugation (16,000×g for 1 h at 4 °C) and uronic acid content of filtered and non-filtered supernatant were determined.
Uronic acid assay
Uronic acid content of samples isolated from solid and liquid media was determined (Blumenkrantz and Asboe-Hansen 1973; Remminghorst and Rehm 2006a).
Crosslinking of MucR
Strains were grown on PIA media. Biomass from three plates were harvested and washed six times. Wash buffer A (150 mM NaCl) and wash buffer B (150 mM NaCl and 50 mM KNO3) were used to wash cells grown in the absence and presence of nitrate, respectively. Cells were incubated for 30 min at 37 °C in 5 ml of crosslinking buffer [10 mM HEPES, 150 mM NaCl, pH 7.4, 20 % v/v of DMSO, 5 mM disuccinimidyl glutarate (DSG) crosslinker (Thermo Scientific Pierce)]. For each treatment, a negative control was included; these cells were suspended in crosslinking buffer minus DSG. Reactions were stopped by 20 mM Tris–HCl (pH 7.4).
Cells were washed twice with buffer (10 mM HEPES, 150 mM NaCl, pH 7.7), lysed by sonication, and IM fractions were prepared and probed for MucR.
Immunoblot analysis of envelope fractions
Cells were treated on ice for 1 h with lysozyme buffer (10 mM HEPES, 150 mM NaCl, pH 7.4, ×1 conc. of Roche EDTA free protease inhibitor) and lysed by sonication (on ice, 12 cycles—15 s of sonication and 15 s cool down). Unlysed cells and debris were removed by centrifugation (15,000×g for 30 min at 4 °C). The clear cell lysate was centrifuged (100,000×g for 90 min at 4 °C), sedimenting the envelope fraction which was suspended in 300 μl 10 mM HEPES, 150 mM NaCl, pH 7.4 containing 0.7 % w/v N-lauroyl-sarcosine and subject to gentle agitation at 25 °C for 1 h to solubilize the IM fraction. The sample was centrifuged (100,000×g for 90 min at 4 °C) and the pellet discarded. The remaining supernatant containing the IM fraction was aliquoted and stored at −80 °C.
Protein concentrations were determined by Bradford Assay (Bio-Rad Protein Assay Kit; Bio-Rad Laboratories, Inc.). Proteins were run on SDS-PAGE (3.9 % stacking and 8 % separating gel) and transferred to a nitrocellulose membrane using iBlot® Dry Blotting system (Invitrogen). Membranes were washed thrice with TBST (Tris, NaCl, Tween 20 0.1 % v/v) and blocked at 25 °C with 5 % BSA in TBST for 1 h. Membranes were washed thrice and probed at 4 °C overnight with rabbit polyclonal anti-MucR antibody (Genescript) in TBST containing 1 % w/v BSA at 1:10,000. After washing, membranes were probed with goat-anti-rabbit antibody conjugated to horse radish peroxidase for 1 h at room temperature (1:10,000 in TBST containing 1 % w/v BSA). After washing, substrate was added and incubated for 5 min, and image developed.
Measuring intracellular c-di-GMP levels using a c-di-GMP sensitive lacZ reporter
A c-di-GMP sensitive lacZ reporter plasmid was constructed as previously described (Baraquet et al. 2012). Integration of this plasmid, mini-CTX-lacZ-pelAwt, was confirmed by colony PCR, and the vector backbone was removed (Hoang et al. 2000). This construct was used to determine intracellular c-di-GMP levels through beta-galactosidase assay.
Measurement of algD, pelA, and pslA promoter activities
Promoter activities of algD, pelA, and pslA were determined using previously described promoter-specific lacZ fusion reporter plasmids: miniCTXPalglacZ, pTZ110:pelA, and pTZ110:pslA (Ghafoor et al. 2011; Hay et al. 2012; Overhage et al. 2005), through beta-galactosidase reporter assays.
Beta-galactosidase reporter assays
Beta-galactosidase (LacZ) activity was measured according to Miller (Miller 1972; Zhang and Bremer 1995). Briefly, strains were grown on PIA medium for 72 h at 37 °C. Cells were taken, washed twice with saline, and adjusted to an O.D of 0.7. LacZ activity was measured in 96-well microtiter plates. Five microliters of sample was added to 20 μl of buffer A [100 mM Na2HPO4, 20 mM KCl, 2 mM MgSO4, 0.8 mg ml−1 of (CTAB) hexadecyltrimethylammonium bromide, 0.4 mg ml−1 of sodium deoxycholate, and 5.4 μl ml−1 of β-mercaptoethanol]. To this mixture, 150 μl of buffer B [60 mM Na2HPO4, 40 mM NaH2PO4, 2.7 μl ml−1 of β-mercaptoethanol, and 1 mg ml−1 of o-nitrophenyl-β-d-galactoside (ONPG) as substrate] was added and incubated for 30 to 90 min at 30 °C. Buffer B minus ONPG was used as a blank for each sample. Reactions were stopped by adding 175 μl of buffer D: 1 M sodium carbonate (Na2CO3). Absorbance at 405 nm was measured and LacZ activities were calculated in Miller Units (Zhang and Bremer 1995).
Solid surface attachment assay
Attachment to a solid surface was assessed according to Merrit et al. (2005). Briefly, relevant strains were grown to saturation in PI medium. Cell cultures were supplemented with either KNO3 (0.1 vol × 10 stock to a final concentration of 1 % w/v) or 0.1 vol. water (negative control). One hundred microliters was transferred to seven wells of a sterile 96-well microtiter plate and incubated at 37 °C for 90 min. Unbound cells were removed by inverting and washing the plate. Bound cells were stained with 125 μl of 0.1 % crystal violet and incubated at 25 °C for 15 min. Unbound crystal violet was removed by two washes as above and air dried. Bound crystal violet was solubilized with 200 μl of dimethyl sulfoxide and attachment efficiency was inferred from absorbance at 595 nm.
Swarming motility assay
Swarming motility was assessed as described previously (Tremblay et al. 2007). Briefly, plates consisted of modified M9 medium ± 1 % w/v of KNO3. Autoclaved medium was poured in petri dishes and dried under laminar flow for 1 h. Swarm plates were inoculated with 5 μl of stationary-phase bacterial culture and incubated at 30 °C for 16 h.
Continuous-culture flow cell biofilms
P. aeruginosa strains were grown in continuous-culture flow cells (channel dimensions of 4 mm × 40 mm × 1.5 mm) at 37 °C (Hay et al. 2009). Channels were inoculated with 0.5 ml of early-stationary-phase cultures and incubated without flow for 4 h at 25 °C. Then a flow rate of 0.3 ml min−1 (Reynolds number of 5) was maintained for 20 h. Cells were stained using the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Inc., Eugene, OR) and visualized via confocal laser scanning microscopy (Leica SP5 DM6000B). Biofilm characteristics were calculated using Imaris software (Ghafoor et al. 2011). Briefly, the average height and compactness were determined as previously described (Ghafoor et al. 2011). Also, a ratio of dead to live cells was reached by dividing the total red with the total green fluorescence (Ghafoor et al. 2011).
Results
Suppression of alginate production by nitrate
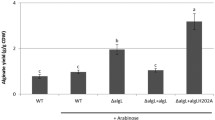
Previously, it was reported that inactivation of mucR in P. aeruginosa PDO300 severely reduced alginate production (Hay et al. 2009). In the present study, we showed that expressing mucR in trans in the mucR mutant restored alginate production (Fig. 1a). An empty vector also partially restored alginate production in the mutant, a known response due to antibiotic stress (Hay et al. 2009; Remminghorst and Rehm 2006a, b).
Alginate yield (g g−1 CDW, mean ± SD) of strains grown in the absence (white) and presence (black) of nitrate. a Nitrate suppresses alginate production through MucR. PDO300 = PDO300(pBBR1MCS-5); dR = PDO300ΔmucR(pBBR1MCS-5), and dR + R = PDO300ΔmucR(pBBR1MCS-5:mucR). b The sensor domain of MucR is involved in nitrate sensing. MHYTI, MHYTII, and MHYTII = PDO300ΔmucR expressing variants of MucR (from the plasmid pBBR1MCS-5) containing individually inactivated MHYT motifs (MHYT to MAYT mutation). Roman numeral indicates which MHYT motif was mutated. c The GGDEF and EAL domains of MucR are important for alginate production. GGDEF and EAL = PDO300ΔmucR expressing variants of MucR with inactivated GGDEF and EAL domains (GGDEF to GGAAF and EAL to AAL mutations), respectively. In panels (a), (b), and (c), treatments with different letters are significantly different (post hoc Tukey HSD test, n = 3, p < 0.05). g g−1 CDW = grams of alginate per gram of cellular dry weight
Supplementing the growth medium of PDO300(pBBR1MCS-5) with nitrate (potassium nitrate, 1 % w/v) abolished alginate production (Fig. 1a). However, nitrate did not affect alginate production in the mucR mutant harboring an empty vector (Fig. 1a). In contrast, nitrate significantly reduced alginate yield by the mutant expressing mucR in trans, PDO300ΔmucR(pBBR1MCS-5:mucR) (Fig. 1a). Subsequent immunoblotting shows that MucR was detected at full length in the IM fractions of PDO300(pBBR1MCS-5) and PDO300ΔmucR(pBBR1MCS-5:mucR) (Fig. S1).
Analysis of uronic acid (UA) content of non-filtered and filtered culture supernatants indicated that nitrate suppressed alginate production by impairing polymerization (Table S1). UA of the non-filtered fraction (TOTAL UA) represented the total alginate produced, both high molecular weight and low molecular weight/degraded polymer. In contrast, the UA content of the filtered fraction represented only the low molecular weight/degraded polymer (smaller than 10 kDa) which was referred to as free uronic acid (FUA) content. In the absence of nitrate, PDO300 produced 627 ± 27.6 and 106 ± 17.3 μg ml−1 of UA in the total UA and FUA fractions, respectively (Table S1). Thus, roughly a sixth of the alginate was low molecular weight/degraded polymer, a finding consistent with previous studies (Hay et al. 2010). In contrast, when grown in the presence of nitrate, PDO300 yielded around 100 μg ml−1 of UA in both fractions, indicating that nitrate impaired polymerization (Table S1). Interestingly, inactivation of mucR also led to background levels of UA, independent of nitrate (Table S1), suggesting that MucR is critical for polymerization during growth in liquid medium.
Involvement of the MucR sensor domain in nitrate sensing
To examine if the proposed MHYT sensor domain of MucR had a role in nitrate sensing, variants of MucR containing single, double, and triple histidine to alanine substitutions at their MHYT motifs (H59A, H122A, and H189A)—numbered I, II, and III from the N terminus—were generated and introduced separately into the mucR mutant. We showed that replacing histidine of MHYT I or III by alanine, respectively, did not impair nitrate-induced suppression of alginate biosynthesis (Fig. 1b). However, replacing the histidine of the second MHYT motif (MHYT II) by alanine abolished nitrate-induced suppression of alginate production (Fig. 1b). These MucR variants were detected at full length in IM fractions by immunoblot (Fig. S1). Subsequent mutation of any two or all three MHYT motifs also interfered with nitrate sensing (Fig. S2).
Importance of MucR’s GGDEF and EAL domains for alginate production
To determine the role of MucR’s GGDEF and EAL domains in nitrate-dependent regulation of alginate production, two variants of MucR were generated, one with a disabled GGDEF domain (GGDEF to GGAAF mutation) and the other with an inactivated EAL domain (EAL to AAL mutation). Introducing either variant into the mucR deletion mutant led to alginate yields comparable to the mucR mutant harboring the empty vector (Fig. 1c), indicating that both DGC and PDE active sites of MucR are required for alginate production. Both variants of MucR were detected at full length by immunoblot in IM fractions (Fig. S1).
Non-impact of nitrate on the oligomeric state of MucR
Previous studies have shown that c-di-GMP modulating enzymes form higher oligomeric states—dimers and tetramers (Romling et al. 2013). To examine if MucR also formed higher MW complexes, we performed crosslinking and immunoblot experiments using PDO300(pBBR1MCS-5). Cells were treated with or without a crosslinking reagent (DSG) and IM fractions were probed with anti-MucR antibodies. In untreated cells, MucR was detected as a monomer (∼75 kDa). However, in DSG-treated cells, MucR was detected at a substantially higher MW (∼300 kDa) (Fig. S3). This shift in MW was independent of nitrate (Fig. S3).
Modulation of c-di-GMP levels and alginate promoter activity by nitrate through MucR
To assess whether nitrate-induced suppression of alginate production was associated with depleted global intracellular c-di-GMP levels and reduced alginate promoter (PalgD) activity, we measured c-di-GMP levels and PalgD activity using previously described c-di-GMP-sensitive- and PalgD-lacZ fusion reporters (Hay et al. 2012; Baraquet et al. 2012). Nitrate reduced c-di-GMP levels by 15 % and PalgD activity by 50 % in PDO300. Similarly, deletion of mucR reduced c-di-GMP levels by 30 % and PalgD activity by 50 %. Furthermore, inactivation of the sensor and output domains of MucR also reduced PalgD activity by 50 % (Fig. S4). However, nitrate did not further reduce c-di-GMP or PalgD activity in the mucR mutant (Fig. 2). We next examined if over-producing a highly active DGC, WspR, in PDO300 could increase PalgD activity. Our results showed that when PDO300 was grown in the absence and presence of nitrate, over-production of WspR increased PalgD by 20 and 40 %, respectively (Fig. S5). Overall, these results suggested that PalgD activity and c-di-GMP levels are linked.
The effect of MucR and nitrate on global intracellular c-di-GMP levels and alginate promoter activity. a C-di-GMP levels (Miller Units, mean ± SD) of strains grown in the absence/presence (white/black) of nitrate. C-di-GMP was measured using a c-di-GMP sensitive promoter-lacZ reporter. PDO300 and dR = PDO300 and PDO300ΔmucR strains containing c-di-GMP-lacZ reporter and empty pBBR1MCS-5 vector, respectively. b Alginate promoter activities (Miller Units, mean ± SD) of strains grown in the absence/presence (white/black) of nitrate. PalgD activity was measured using chromosomally integrated miniCTX-PalgD-lacZ reporter. PDO300 and dR = PDO300 and PDO300ΔmucR strains containing miniCTX-PalgD-lacZ reporter and empty pBBR1MCS-5 vector, respectively. In panels (a) and (b), n = 6 and 3, respectively. Treatments with different letters are significantly different (post hoc Tukey HSD Test, p < 0.05)
Effect of nitrate and MucR on surface attachment, swarming motility, pel and psl promoter activity as well as biofilm characteristics
Previously, alginate-negative strains were shown to be enhanced in surface attachment and swarming motility (Hay et al. 2009). Hence, we predicted that nitrate-induced suppression of alginate production could enhance these phenotypes. Here, we showed that nitrate impaired surface attachment by PDO300. However, it did not affect attachment by mucR mutant which was already impaired compared to PDO300 (Fig. 3). We also showed that neither nitrate nor deletion of mucR affected swarming motility (Fig. S6).
Earlier studies suggested that alginate negative mutants produced more Psl and Pel (Ghafoor et al. 2011; Hay et al. 2009). Given that nitrate suppressed alginate production in PDO300 (Fig. 1a), we anticipated that nitrate would also boost Psl and Pel production. However, using plasmid-borne PpslA- and PpelA-lacZ reporters, we show that in PDO300, nitrate did not affect pslA or pelA promoter activities (Fig. 4). Similarly, nitrate did not affect pslA promoter activity in the mucR mutant, which was approximately 40 % higher than PDO300. In contrast, nitrate increased pelA activity in the mucR the mutant by a similar amount.
We next examined if mucR and nitrate affected biofilm characteristics, including the average thickness, cellular density, and ratio of live to dead cells. In general, nitrate-containing medium led to thicker, more compact biofilms with fewer dead cells while deletion of mucR had a similar effect (Table 2).
Discussion
In the present study, nitrate was identified as a signal perceived by MucR, leading to suppressed alginate production (Fig. 1a). We also showed that inactivation of the second MHYT motif (aa 121–124) blocked nitrate-induced suppression of alginate production (Fig. 1b), suggesting that this motif is critical in nitrate perception. Nitrate is utilized by Pseudomonas spp. as alternative electron acceptor and respective denitrification intermediates had been linked to reduced alginate production (Vollack and Zumft 2001; Wood et al. 2007; Worlitzsch et al. 2002; Zumft 1997). One of these intermediates, NO, binds strongly with transition metals such as the copper ion proposed to reside in MucR’s sensor domain. NO has also been shown to induce biofilm dispersal dependent of MucR (Li et al. 2013). Our results are in accordance with these previous findings and suggest that MucR senses nitrate through denitrification intermediates such as nitric oxide. However, the possibility that MucR directly senses nitrate and/or changes in redox potential (due to denitrification) cannot be ruled out.
Inactivation of either GGDEF or EAL domain of MucR impaired alginate production (Fig. 1c), indicating that both c-di-GMP synthesizing and degrading activities of MucR are required for alginate production. This is consistent with results of a previous study showing that MucR’s DGC activity is enhanced by its EAL domain (Li et al. 2013). Furthermore, because MucR functions as a DGC and PDE during biofilm and planktonic growth modes, respectively (Li et al. 2013), we proposed that the EAL domain of MucR could be bi-functional, serving a regulatory role during biofilm mode—to enhance MucR’s DGC activity, while taking on a PDE role during planktonic mode. Taken together, these results demonstrated for the first time that both GGDEF and EAL active sites of MucR are required for alginate production.
C-di-GMP modulating enzymes often form dimers and tetramers (Barends et al. 2009; De et al. 2009; Phippen et al. 2014; Rao et al. 2008; Romling et al. 2013; Sharma et al. 2014; Tarutina et al. 2006; Tchigvintsev et al. 2010; Wassmann et al. 2007). Dimerization is usually essential for DGC activity (Romling et al. 2013). Since MucR is an active DGC during biofilm growth, we investigated whether it would also form oligomeric states. Our crosslinking and immunoblot experiments suggested that MucR forms higher molecular weight complexes, independent of nitrate (Fig. S3). This is consistent with previous studies alluding to MucR forming higher oligomeric states (Hay et al. 2009). However, our data showed that nitrate did not affect MucR’s oligomeric state. Hence, dimerization and DGC activity might not be part of the molecular mechanism of nitrate-mediated MucR-dependent regulation.
Previously, MucR was proposed to modulate alginate production at a post-translational level through a localized pool of c-di-GMP (Hay et al. 2009). In the current study, we tested whether nitrate-induced suppression of alginate production was caused by lowered global intracellular c-di-GMP levels and reduced alginate promoter activity. While c-di-GMP levels were correlated quite well with alginate promoter activity (Fig. 2 and Fig. S5), alginate yield was not directly associated with c-di-GMP levels or alginate promoter activity (Figs. 1 and 2 and Fig. S4), indicating that further post-translational regulatory mechanisms are involved in alginate production.
Interestingly, neither nitrate nor inactivation of MucR could abolish PalgD activity or c-di-GMP levels (Fig. 2 and Fig. S4), suggesting that other factors contribute to these phenotypes. For instance, PalgD activity is also driven by transcription factors AlgU, AmrZ, AlgR, AlgP, and AlgB, which are highly active in PDO300 (Damron and Goldberg 2012; Hay et al. 2014). Furthermore, P. aeruginosa has over 30 different enzymes which modulate c-di-GMP levels (Kulasakara et al. 2006).
Previous studies have shown that alginate negative mutants displayed enhanced surface attachment, swarming motility, and production of Psl and Pel exopolysaccharides (Ghafoor et al. 2011; Hay et al. 2009). Hence, we predicted that nitrate-induced suppression of alginate production might enhance these phenotypes. However, our results showed that while nitrate impaired attachment in PDO300 (Fig. 3), it had no effect on swarming motility (Fig. S6). Furthermore, nitrate did not elevate pslA or pelA promoter activities in PDO300 (Fig. 4). However, pelA promoter activity was enhanced by nitrate in the mucR mutant (Fig. 4) while deletion of mucR increased pslA promoter activity (Fig. 4). This differential regulation may contribute to the already highly complex regulatory network controlling and fine tuning the production of exopolysaccharides and other matrix components during biofilm development (Ghafoor et al. 2011).
The role of MucR and nitrate in transcriptional regulation of Psl and Pel polysaccharide biosynthesis operons. Promoter activities (Miller Units, mean ± SD) of a pslA and b pelA, measured in PDO300 and PDO300ΔmucR (dR) using plasmid borne promoter-specific lacZ fusion reporters. White/black = grown in the absence/presence of nitrate. Treatments with different letters are significantly different (post hoc Tukey HSD test, n = 3, p < 0.05)
To assess whether nitrate-induced suppression of alginate production would coincide with increased biofilm thickness, compactness, and cell survival, biofilms grown in flow cell chambers were analyzed by CLSM. Our results showed that nitrate enhanced these biofilm characteristics (Table 2). Interestingly, nitrate also enhanced these characteristics in the mucR mutant, which also formed more compact biofilms with enhanced survival (Table 2). These results are consistent with the fact that nitrate is utilized as an alternative terminal electron acceptor under hypoxic conditions. Furthermore, these phenotypes were linked to elevated production of Pel and Psl polymers which are involved in surface attachment, intercellular interaction, micro-colony formation, and biofilm development (Colvin et al. 2012; Ghafoor et al. 2011; Ma et al. 2009; Vasseur et al. 2005; Yang et al. 2011; Zhao et al. 2013). A recent study has shown that another protein, NbdA—which also has an MHYT-GGDEF-EAL domain structure but only has PDE activity—plays a more dominant role (than MucR) in NO-induced biofilm dispersal (Li et al. 2013). However, whether NbdA also contributes to enhanced biofilm thickness, compactness, and cell survival is yet to be determined.
Overall, the current study provides new insight into the role of MucR in nitrate-dependent suppression of alginate production. On the basis of our results and findings from previous studies, we proposed a model for MucR-mediated, nitrate-dependent suppression of alginate production (Fig. 5a). We also showed that MucR and nitrate differentially regulate alginate, Pel and Psl exopolysaccharide production, and surface attachment as well as biofilm characteristics. The role of MucR and nitrate in regulation of these other phenotypes is summarized in Fig. 5b and c. Nitrate enhanced PpelA activity in the absence of mucR; nitrate did not modulate PpslA activity while MucR impaired PpslA activity, and nitrate impaired PalgD activity through MucR (Fig. 5b). Figure 5c shows that nitrate enhanced attachment and biofilm characteristics (average height, compactness, and survival) while deletion of mucR has a similar effect. It also shows that neither nitrate nor deletion of mucR affected swarming motility.
Model for nitrate-MucR-dependent regulation of various phenotypes. a Alginate production. MucR imparts a localized pool of c-di-GMP, driving alginate production predominately at a post-translational level. When nitrate is present, it is denitrified, releasing nitric oxide, causing nitroactive stress, and shifts in redox potential which are sensed by MucR’s MHYT domain. This interferes with MucR’s DGC activity, leading to suppressed alginate production. The EAL domain of MucR enhances DGC activity. Both GGDEF and EAL active sites are required for alginate production. Potential signals perceived by MucR are represented in rectangular boxes (i.e., nitrate, NO, nitroactive stress, or redox potential). b Regulation of pelA, pslA, and algD promoter activities. Nitrate enhances PpelA activity in the absence of mucR; nitrate does not modulate PpslA activity while MucR negatively modulates PpslA activity, and nitrate impairs PalgD activity through MucR. c Regulation of swarming motility, surface attachment, and biofilm characteristics (i.e., thickness, compactness, and cell survival). Neither nitrate nor MucR affect swarming motility. MucR enhances attachment while nitrate impairs it. Nitrate enhances biofilm characteristics while deletion of mucR has a similar effect
References
Albrecht MT, Schiller NL (2005) Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol 187:3869–3872
Amikam D, Galperin MY (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6
Baker P, Ricer T, Moynihan PJ, Kitova EN, Walvoort MTC, Little DJ, Whitney JC, Dawson K, Weadge JT, Robinson H, Ohman DE, Codee JDC, Klassen JS, Clarke AJ, Howell PL (2014) P. aeruginosa SGNH hydrolase-like proteins AlgJ and AlgX have similar topology but separate and distinct roles in alginate acetylation. Plos Pathog 10(8), e1004334. doi:10.1371/journal.ppat.1004334
Baraquet C, Murakami K, Parsek MR, Harwood CS (2012) The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218
Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RL, Gomelsky M, Schlichting I (2009) Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018
Blumenkrantz N, Asboe-Hansen G (1973) New method for quantitative determination of uronic acids. Analyt Biochem 54:484–489
Chiu J, March PE, Lee R, Tillett D (2004) Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res 32:e174. doi:10.1093/nar/gnh172
Cohen TS, Prince A (2012) Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat Med 18:509–519
Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR (2012) The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 14:1913–1928
Damron FH, Goldberg JB (2012) Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol 84:595–607
De N, Navarro MVAS, Raghavan RV, Sondermann H (2009) Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J Mol Biol 393:619–633
Farrell EK, Tipton PA (2012) Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa. Biochemistry 51:10259–10266
Franklin MJ, Ohman DE (2002) Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J Bacteriol 184:3000–3007
Galperin MY, Gaidenko TA, Mulkidjanian AY, Nakano M, Price CW (2001) MHYT, a new integral membrane sensor domain. FEMS Microbiol Lett 205:17–23
Ghafoor A, Hay ID, Rehm BHA (2011) Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 77:5238–5246
Gutsche J, Remminghorst U, Rehm BHA (2006) Biochemical analysis of alginate biosynthesis protein AlgX from Pseudomonas aeruginosa: purification of an AlgX-MucD (AlgY) protein complex. Biochemie 88:245–251
Hay ID, Remminghorst U, Rehm BHA (2009) MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl Environ Microbiol 75:1110–1120
Hay ID, Rehman ZU, Rehm BHA (2010) Membrane topology of outer membrane protein AlgE, which is required for alginate production in Pseudomonas aeruginosa. Appl Environ Microbiol 76:1806–1812
Hay ID, Schmidt O, Filitcheva J, Rehm BHA (2012) Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl Microbiol Biotechnol 93:215–227
Hay ID, Wang Y, Moradali MF, Rehman ZU, Rehm BHA (2014) Genetics and regulation of bacterial alginate production. Environ Microbiol 16:2997–3011
Hoang TT, Kutchma AJ, Becher A, Schweizer HP (2000) Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72
Hoiby N, Ciofu O, Bjarnsholt T (2010) Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674
Jain S, Ohman DE (1998) Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol 180:634–641
Jain S, Franklin MJ, Ertesvag H, Valla S, Ohman DE (2003) The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol Microbiol 47:1123–1133
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd, Peterson, KM (1995) Four new derivatives of the broadhost-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S (2006) Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844
Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N (2013) NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol 195:3531–3542
Ma LM, Conover M, Lu HP, Parsek MR, Bayles K, Wozniak DJ (2009) Assembly and development of the Pseudomonas aeruginosa biofilm matrix. Plos Pathog 5(3), e1000354. doi:10.1371/journal.ppat.1000354
Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JIA, Jensen P, Johnsen AH, Givskov M, Ohman DE, Molin S, Hoiby N, Kharazmi A (1999) Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology-Sgm 145:1349–1357
Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S (2007) The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol 65:876–895
Merritt JH, Kadouri DE, O’Toole GA (2005) Growing and analyzing static biofilms, current protocols in microbiology, vol 1. Wiley, Hoboken
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor
Oglesby LL, Jain S, Ohman DE (2008) Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154:1605–1615
Overhage J, Schemionek M, Webb JS, Rehm BHA (2005) Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl Environ Microbiol 71:4407–4413
Phippen CW, Mikolajek H, Schlaefli HG, Keevil CW, Webb JS, Tews I (2014) Formation and dimerization of the phosphodiesterase active site of the Pseudomonas aeruginosa MorA, a bi-functional c-di-GMP regulator. FEBS Lett 588:4631–4636
Rao F, Yang Y, Qi Y, Liang ZX (2008) Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol 190:3622–3631
Rehman ZU, Wang YJ, Moradali MF, Hay ID, Rehm BHA (2013) Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol 79:3264–3272
Remminghorst U, Rehm BHA (2006a) Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett 580:3883–3888
Remminghorst U, Rehm BHA (2006b) In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl Environ Microbiol 72:298–305
Remminghorst U, Hay ID, Rehm BHA (2009) Molecular characterization of Alg8, a putative glycosyltransferase, involved in alginate polymerisation. J Biotechnol 140:176–183
Robles-Price A, Wong TY, Sletta H, Valla S, Schiller NL (2004) AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol 186:7369–7377
Romling U, Galperin MY, Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol R 77:1–52
Sharma IM, Prakash S, Dhanaraman T, Chatterji D (2014) Characterization of a dual-active enzyme, DcpA, involved in cyclic diguanosine monophosphate turnover in Mycobacterium smegmatis. Microbiology 160:2304–2318
Simon R, Priefer U, Pu¨hler A (1983) A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gramnegative bacteria. Bio/Technology 1:784–791
Tan JQ, Rouse SL, Li DF, Pye VE, Vogeley L, Brinth AR, El Arnaout T, Whitney JC, Howell PL, Sansom MSP, Caffrey M (2014) A conformational landscape for alginate secretion across the outer membrane of Pseudomonas aeruginosa. Acta Crystallogr D 70:2054–2068
Tarutina M, Ryjenkov DA, Gomelsky M (2006) An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem 281:34751–34758
Tchigvintsev A, Xu XH, Singer A, Chang C, Brown G, Proudfoott M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF (2010) Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J Mol Biol 402:524–538
Tremblay J, Richardson AP, Lepine F, Deziel E (2007) Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ Microbiol 9:2622–2630
Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A (2005) The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151:985–997
Vollack KU, Zumft WG (2001) Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J Bacteriol 183:2516–2526
Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T (2007) Structure of BeF3-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15:915–927
Wood SR, Firoved AM, Ornatowski W, Mai T, Deretic V, Timmins GS (2007) Nitrosative stress inhibits production of the virulence factor alginate in mucoid Pseudomonas aeruginosa. Free Rad Res 41:208–215
Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Doring G (2002) Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325
Yang L, Hu YF, Liu Y, Zhang JD, Ulstrup J, Molin S (2011) Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ Microbiol 13:1705–1717
Zhang XG, Bremer H (1995) Control of the Escherichia coli Rrnb P1 promoter strength by ppgpp. J Biol Chem 270:11181–11189
Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GCL (2013) Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497:388–391
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol R 61:533–616
Acknowledgments
I. D. Hay and Y. Wang were funded by Massey University Doctoral Scholarships. Z. U. Rehman was funded by the Higher Education Commission of Pakistan. The work was funded by Massey University. The authors would like to acknowledge the Massey University Genome Service for sequencing new plasmid constructs, and the Manawatu Microscopy and Imaging Centre for assistance in biofilm analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 382 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Hay, I.D., Rehman, Z.U. et al. Membrane-anchored MucR mediates nitrate-dependent regulation of alginate production in Pseudomonas aeruginosa . Appl Microbiol Biotechnol 99, 7253–7265 (2015). https://doi.org/10.1007/s00253-015-6591-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6591-4