Abstract
Insects occupy a central position in the biosphere. They are able to resist infections even though they lack an adaptive immune system. Drosophila melanogaster has been used as a potent genetic model to understand innate immunity both in invertebrates and vertebrates. Its immune system includes both humoral and cellular arms. Here, we review how the distinct immune responses are triggered upon sensing infections, with an emphasis on the mechanisms that lead to systemic humoral immune responses. As in plants, the components of the cell wall of microorganisms are detected by dedicated receptors. There is also an induction of the systemic immune response upon sensing the proteolytic activities of microbial virulence factors. The antiviral response mostly relies on sensing double-stranded RNAs generated during the viral infection cycle. This event subsequently triggers either the viral short interfering RNA pathway or a cGAS-like/STING/NF-κB signaling pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The study of invertebrate immunity can be traced back to the pioneering work of Elie Metchnikoff on the defensive role of phagocytes against blastomycetes infection in Daphnia, which ushered the study of immunity in both invertebrates and vertebrates (Metchnikoff 1884). Thus, the initial focus of studies on insect host defense against microbial infections was on cellular immunity. It was only in 1918 that Glaser published a study showing the existence of an inducible humoral immune response in caterpillars (Glaser 1918). The antimicrobial activities found in insects were however not identified in insects until the biochemical isolation of the first antimicrobial peptides (AMPs) by Hans Boman in the late 1970s (Steiner et al. 1981). One can easily figure out the advantages for the organism to induce an immune response only when needed. This however implies that evolution has selected mechanisms that allow hosts to sense the presence of microbial infections. These have been identified in the Drosophila melanogaster model over the past 20 years (Buchon et al. 2014; Ferrandon et al. 2007; Hultmark 1993; Lemaitre and Hoffmann 2007).
The study of the humoral immune system has been slow as compared to that of the mammalian one, which was highly facilitated in the latter by the ease of working with antibodies. This however led to a view of the immune system that was largely centered on adaptive immunity at the expense of innate immunity, the importance of which was only recognized rather recently (Janeway 1989; Janeway and Medzhitov, 2002). Actually, one strength of invertebrate models is that they allow the dissection of innate immunity unencumbered by the additional layer of specific, albeit slow, protection provided by acquired immunity. Furthermore, the very large majority of extant species rely solely on innate immunity for host defense against infections. AMPs were initially isolated from the Hyalophora cecropia moth pupa that is almost as large as a mouse (Hultmark et al. 1980; Steiner et al. 1981). Additional early biochemical studies led to the discovery of further AMP families, both in Lepidopterans and in large Diptera. This allowed cloning of the AMP genes and the study of their promoters, which revealed the existence of NF-κB binding sites in the promoters of AMP genes. It was however transgenic analysis in D. melanogaster that rigorously established the importance of these sites for the inducibility of the immune response in vivo (Engström et al. 1993; Meister et al. 1994). The next step was the discovery in 1996 of the major role played by the Toll pathway in the host defense against fungal infections, even though the study of NF-κB signaling relied on the induction of the systemic immune response with bacterial and not fungal challenge (Lemaitre et al. 1996). Transgenic analysis as well as the study of already available mutant lines underscore the power of the Drosophila model that benefits from more than 100 years of genetic studies as well as the development of transgenesis in the early 1980s (Rubin and Spradling 1982). Nowadays, genetic engineering allows expressing or inactivating genes at will in a cell type/tissue and time-dependent manner with numerous useful stocks being obtainable from stock centers on several continents (Bellen et al. 2010). Thus, the study of a gene can be performed at multiple scales, from that of molecules to that of the organisms or even at the population biology and evolution level since the genome of more than 100 species of Drosophila is now available, e.g., in Kim et al. (2021). This has resulted in an important body of knowledge easily classified by gene product (flybase.org) ever since the D. melanogaster genome was sequenced in 2000 (Adams et al. 2000). However, the most powerful tool of Drosophila genetics remains the ability to perform large-scale screens, a strategy that besides the identification of the genes required for Drosophila development in the early 1980s (Nüsslein-Volhard and Wieschaus 1980) allowed the molecular identification of sensors of the immune response that activate the expression of AMP genes, more than 20 years after the isolation of the first AMPs (Choe et al. 2002, 2005; Gobert et al. 2003; Gottar et al. 2002, 2006; Michel et al. 2001; Pili-Floury et al. 2004).
Here, this primer provides a global view of the mechanisms used by Drosophila to perceive the presence of infectious microorganisms and that allow suiting at least to some degree the specificity of the response toward the microbial nature of the invader (Lemaitre et al. 1997). We shall first succinctly describe the different arms of Drosophila host defense. Next, we shall focus on the sensors that detect specific microbial compounds of the cell wall as well as those that perceive the enzymatic activity of virulence factors. We shall also discuss some microbial metabolites released by microorganisms in the gut lumen and that are thought to contribute to the distinction between beneficial or harmless microbiota versus pathogenic germs. Finally, we shall briefly describe the receptors likely involved in the cellular immune response and will provide an overview of the sensing of viral infections.
Overview of the Drosophila host defense against microbial infections
To study and comprehend host defenses, the choice of a microorganism is not neutral. In plants, the deciphering of innate immunity initially rested on the study of host–pathogen interactions, which was grounded on the needs from agriculture to protect crops derived from selected cultivars. It initially led to a view in which virulence factors from pathogens, against which plants had evolved effectors to specifically neutralize them, were actually called avirulence factors (Jones and Dangl 2006). In contrast, the use of nonpathogenic or weakly pathogenic microorganisms profoundly affected our initial understanding of Drosophila innate immunity as these microorganisms are thought to be rapidly cleared by the immune response of wild-type flies. Indeed, this allowed deciphering basal innate immunity mechanisms that function against any microorganism, pathogenic or not, without the added complexity of host–pathogen relationships that are superposed onto the basal layer. The Drosophila systemic immune response relies essentially on two NF-κB pathways. Some pathogens such as Salmonella enterica, Aeromonas salmonicida, or Aspergillus fumigatus are able to neutralize NF-κB immune signaling pathways (Jones et al. 2008, 2012; Pahl et al. 1996), a phenomenon that would hamper the identification and dissection of such pathways. In addition, it provided a paradigm in which to study the action of virulence factors, for instance, by the transgenic expression of virulence factors from highly virulent pathogens such as Yersinia pestis (Harnish et al. 2021; Paquette et al. 2012).
The systemic humoral immune response against bacterial and fungal infections
Genetic analysis of existing mutations or of mutants isolated in large-scale screens identified two NF-κB pathways that together regulate the expression of most genes induced or repressed by systemic infections (De Gregorio et al. 2002a; Ferrandon et al. 2007; Ganesan et al. 2011; Lemaitre and Hoffmann 2007). The immune deficiency (IMD) pathway is mainly activated upon infectious challenges with Gram-negative bacteria and regulates the expression of multiple AMP genes such as Diptericin or Drosocin that are active essentially on this class of bacteria. It resembles to some extent the mammalian TNF-α signaling pathway. In contrast, the Toll pathway is preferentially stimulated by prokaryotic Gram-positive bacteria and eukaryotic fungi and is required for host defense against these pathogens. It regulates the strong expression of an antifungal peptide, Drosomycin, active on filamentous fungi and also of a family of secreted peptides, the Bomanins, the function of which is just beginning to be understood (Lin et al. 2020). The Toll pathway presents analogies to interleukin (IL-1) (Gay and Keith 1991) or possibly Toll-like receptor signaling, even though Toll itself belongs to a sub-family of Toll receptors that is distinct from mammalian Toll-like receptors (Leulier and Lemaitre 2008). Importantly, it does not directly detect compounds of microbial origins but is activated by the Spätzle (SPZ) cytokine, which needs to be proteolytically matured into an active ligand upon the upstream detection of infections (Levashina et al. 1999; Weber et al. 2003).
Of note, some antimicrobial peptide genes such as those encoding cecropins, attacins, or defensin appear to be jointly regulated by both pathways whereas Metchnikowin can be induced independently by either pathway. As mentioned above, the use of nonpathogenic bacteria allowed the determination of a significant degree of specificity in stimulating one or the other pathway or their effectiveness against specific classes of pathogens. In contrast, the use of highly virulent pathogens in septic injury models such as the Gram-negative bacterium Pseudomonas aeruginosa revealed that not only the IMD but also the Toll pathway was activated and required to provide some protection against this lethal pathogen (Lau et al. 2003; Limmer et al. 2011). It follows that systemic infections with Drosophila pathogens lead to the stimulation of both NF-κB pathways, which allows the formation of Relish/Dorsal/DIF homo- or heterodimer combinations that allow the two pathways to synergize (Tanji et al. 2007, 2010).
In addition to the Toll and IMD pathways, the c-Jun N-terminal kinase (JNK), Map/Erk kinase kinase 1 (MEKK1), and Janus kinase-signal transducers and activators of transcription (JAK-STAT) pathways are also activated during infection but appear to play apparently ancillary roles and likely mediate homeostatic responses to stresses occurring during infection rather than directly fighting off pathogens (Agaisse et al. 2003; Brun et al. 2006; Ekengren et al. 2001; West and Silverman 2018).
Melanization and prophenoloxidase activation
An important host defense found in arthropods is melanization (Soderhall and Cerenius 1998). Besides its role in pigmentation, it is known to play important roles in sealing off wounds in complement of coagulation as well as host defense against infections. Melanization involves initially tyrosine metabolites, notably dihydrophenylalanine (DOPA) and dopamine that become oxidized into corresponding quinones by the action of phenol oxidases (POs), which then assemble into polymers that form eumelanin (Sugumaran and Barek 2016). This set of reactions also produces cytotoxic molecules that are likely active on microorganisms although the exact mode of action remains to be determined (Nappi et al. 1995). It is an open possibility that the production of cytotoxic molecules may be at least partially uncoupled from the blackening reaction at the wounding site (Dudzic et al. 2019). As active PO is toxic for the fly, it is found in the form of inactive pro-forms called pro-PO (PPO) that needs to be matured by proteolytic cleavage by partially characterized immune protease cascades. Thus, similar to the activation of the Toll pathway, PO activation results from the wound or microorganism-triggered proteolytic cascades. Melanization has been reported to be required in the host defense against fungal and some Gram-positive bacterial infections (Binggeli et al. 2014; Dudzic et al. 2015, 2019; Neyen et al. 2015; Wang et al. in preparation). Three genes encode PPOs, and they appear to have somewhat overlapping functions, although PPO1 and the Sp7 protease have been reported to be specifically required to fight off Staphylococcus aureus (Dudzic et al. 2019). In contrast, we have found PPO2, and not Sp7, to be involved in the host defense against fungal infections and at least one intestinal infection with a Gram-negative bacterium (Wang et al. in preparation; Chen et al. in preparation).
Of note, coagulation also participates in host defense against bacterial infections by trapping pathogens in a clot (Loof et al. 2011; Theopold et al. 2004). The biochemistry of activation of coagulation by clip-domain-containing proteases downstream of sensors of cell wall components has been especially well-studied in the horseshoe crab (Muta and Iwanaga 1996). These proteolytic cascades are evocative of those at work in Drosophila for Toll pathway activation, including the structure of coagulogen, which is related to that of Spätzle (Bergner et al. 1997).
Local immune responses at barrier epithelia
Insects have an exoskeleton made up of a chitino-proteinaceous cuticle that provides a strong physical protection against wounds and infections. It also covers the tracheal respiratory system, the foregut, and hindgut, although the composition of the latter may be adapted to its physiological functions such as water, ions, and amino acid resorption. The Drosophila cuticle is shed at each larval molt and replaced by an initially soft cuticle prior to its tanning. Even though the cuticle constitutes a strong barrier to infections, entomopathogenic fungi have evolved strategies that allow them to enzymatically degrade the cuticle at specialized structures known as appressoria developed by the germinating fungus. The fungus then penetrates into the hemocoel by exerting a strong osmosis-driven physical pressure through microscopic holes.
The development of fluorescent reporter transgenes for the expression of AMP genes has revealed expression patterns that can either be constitutive or inducible (Ferrandon et al. 1998). An example of constitutive expression is that of Drosomycin in the spermathecae, organs that store the sperm from males after insemination. A partial, “spontaneous” expression of Drosomycin-GFP was observed in larval tracheae in culture vials. It was possible to induce its widespread expression throughout the tracheal system by dipping the larvae in a concentrated solution of the Gram-negative bacterium Erwinia carotovora carotovora (Ecc15) now called Pectobacterium carotovorum. Interestingly, the inducible responses were found to be regulated by the IMD and not the Toll pathway, even though Drosomycin is essentially controlled by the Toll pathway during the systemic humoral immune response (Davis and Engstrom 2012; Ferrandon 2013; Onfelt Tingvall et al. 2001; Tzou et al. 2000).
Finally, the Toll and IMD pathways are known to be activated in the nervous system but also under circumstances that are not directly related to immunity, e.g., trauma or sleep homeostasis (Blum et al. 2021; Cao et al. 2013). As in mammals, the blood–brain barrier is likely to form an efficient barrier against the dissemination of infections. Nevertheless, some pathogens able to cross this barrier have been recently identified and these novel models will likely allow a better understanding of immune responses in the nervous system, which are poorly understood at present (Benmimoun et al. 2020). Interestingly, a link between the nervous system and host defense against systemic infections in insect larvae had already been established in the 1930s by Serguei Metalnikov at the Institut Pasteur.
The cellular immune response
The cellular immune response in adult Drosophila mostly relies on phagocytosis (Melcarne et al. 2019a; Stuart and Ezekowitz 2008). A limited number of hemocytes is found in adults and correspond to mostly sessile cells (Lanot et al. 2001). Until recently, adult hemocytes were thought to be constituted mostly of macrophage-like plasmatocytes. However, some 10% of adult hemocytes express PPO1 and might therefore play a key role in melanization defenses (Boulet et al. 2021). The link between melanization and hemocytes needs to be more thoroughly investigated in adult flies. In larvae, two major cell types exist, plasmatocytes and crystal cells, the latter containing PPO2 crystals, which rupture upon infection through a JNK-mediated process and release their content in the hemolymph (Bidla et al. 2007; Binggeli et al. 2014). A third cell type, the lamellocyte, is inducible by parasitoid wasp egg deposition within the hemocoel of the second instar larva and together with plasmatocytes encapsulate the egg prior to PPO2-PPO3-dependent melanization (Dudzic et al. 2015). The melanized capsule isolates and may ultimately kill the wasp egg. We shall focus below on the putative phagocytic receptors as well as potential opsonins.
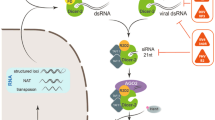
The Drosophila antiviral defenses
Whereas in mammals the interferon response is a primary line of defense against viral infections in most cell types, it has not been selected as the main antiviral defense in all protostome phyla (Schneider and Imler 2021). Indeed, while some evidence for an interferon-like response in mollusks has recently been provided, insects do not encode major components of this pathway in their genomes, even though one Drosophila antiviral effector, Vago, has been proposed to function in an analogous manner by activating the JAK-STAT pathway in Culex mosquito cells (Deddouche et al. 2008; Lafont et al. 2020; Paradkar et al. 2012; Qiao et al. 2021). Instead, insect antiviral immunity appears to rely to a large extent on RNA interference (RNAi), as is the case for plants and for some mammalian cell types such as embryonic stem cells. This defense involves Dicer-2 (DCR-2) and Argonaute2 (AGO2). In brief, DCR-2 processes long viral dsRNA such as those produced during viral replication into small RNAs known as vsiRNAs. These siRNAs are then loaded onto the AGO2-containing RNA-induced silencing complex (RISC), and one strand acts as a specificity determinant for the cleavage of viral RNAs. These responses take place in infected cells. Recently, a systemic aspect has been revealed that relies on the production of virus-derived complementary DNAs (vDNA) that in turn produce secreted secondary siRNAs released in exosome-like vesicles. These vDNAs appear to be involved in priming the immune response against secondary infections and also in transgenerational antiviral immunity (Goic et al. 2013; Mondotte et al. 2018, 2020; Poirier et al. 2018; Tassetto et al. 2017). Besides the siRNA pathway, virus-inducible gene expression programs regulated by several signaling pathways have been reported. A first one is the single JAK-STAT pathway of Drosophila, which plays a role in host defense against Dicistroviridae RNA viruses such as the Drosophila C virus or cricket paralysis virus (CrPV). It is also required to control the DNA virus Invertebrate iridescent virus (IIV)-6. JAK-STAT pathway mutants are more susceptible to these viral infections (Dostert et al. 2005; West and Silverman, 2018). This pathway appears to be more involved in the resilience to cellular damages occurring during viral infections than to a direct antiviral activity since the viral burden does not appear to be strongly affected when the JAK-STAT pathway is affected. Alternatively, viruses might release effectors suppressing JAK-STAT-inducible antiviral compounds or the JAK-STAT pathway itself, as appears to be the case for the recently described Kallithea DNA virus with respect to Toll signaling (Palmer et al. 2019). This however does not seem to be the case for the RNA viruses that induce the expression of the vir-1 JAK-STAT pathway target gene (Dostert et al. 2005).
The NF-κB signaling pathways have also been reported to be involved in host defense against viral infections, as supported by the existence of viral suppressors (Palmer et al. 2019). Interestingly, the Toll pathway appears to be effective against several RNA viruses in an oral but not in a systemic infection model, even though it gets activated in the fat body in both types of infection. Of note, this defense does not involve the Dorsal-related immunity factor (DIF), which mediates the systemic antimicrobial response in adults, but its neighboring gene Dorsal, which mediates the developmental role of the Toll pathway. Dorsal migrates to the nucleus only in infected fat body cells and not in other tissues. However, blocking the Toll pathway by RNAi in the fat body or other tissues failed to lead to an enhanced susceptibility to the ingested virus (Ferreira et al. 2014). It may indirectly interfere with a step of the viral infection process occurring only upon intestinal infection, e.g., translocation of the virus particles across the digestive barrier.
As regards the IMD pathway, it may be involved in the regulation of apoptosis, an effective antiviral defense, in complex manners (Schneider and Imler 2021). The finding that several viruses have incorporated suppressors of the IMD pathway suggests that it may exert a selective pressure on viruses. Recently, it has been found that a noncanonical IMD pathway involving Drosophila melanogaster STING, IKKß, and the NF-κB transcription factor Relish is required for host defense (Goto et al. 2020) and bears some resemblance to the antiviral response mediated by STING in mammals. How exactly it gets activated will be described further below.
Behavioral immunity
Insects anticipate to some extent the exposure to potential pathogens and adapt their behavior accordingly. Repulsive volatile compounds such as geosmin can be detected through specific odorant receptors, whereas others such as commercial lipopolysaccharide (LPS) (notoriously known to be heavily contaminated by other cell wall compounds such as peptidoglycan) are perceived through gustatory receptors (Soldano et al. 2016; Stensmyr et al. 2012). Interestingly, the sensing of infections in the gut leads to the production of the Unpaired3 IL-6-like cytokine that metabolically reprograms the ensheathing glia and thereby affects their metabolic coupling to neurons. Thus, signaling from the gut leads to an enhanced avoidance of Ecc15-laced food (Cai et al. 2021). This aspect of innate immunity has been recently reviewed (Montanari and Royet 2021).
Sensing of Gram-negative bacterial infections: detection of DAP type peptidoglycan
The Gram-negative cell wall is a composite structure that comprises the cytoplasmic membrane, a gelatinous periplasm that includes a thin but rigid layer of peptidoglycan (PGN) and a lipid bilayer that forms the outer membrane (OM). The inner part of the OM is made of phospholipids and lipoproteins that connect to the PGN layer whereas the outer layer is made up of LPS and also integrates proteins such as porins as well as surface proteins. LPS is also known as endotoxin and triggers the mammalian inflammatory response at very low concentrations. Even though LPS is exquisitely detected in Limulus species, chelicerate arthropods, the cell wall compound sensed by the Drosophila innate immune system during bacterial infections is PGN, be it of Gram-positive or Gram-negative origin (Kaneko et al. 2004; Leulier et al. 2003). PGN is a complex polymer made of long glycan chains of alternating ß-(1,4) linked N-acetylglucosamine and N-acetyl muramic acid monomers cross-linked by peptide chains of three to five amino-acids that form a mesh-like layer. In Gram-negative bacteria, the third amino-acid of the linker peptide is meso-diaminopimelic acid (DAP), whereas in many Gram-positive bacteria, a lysine (LYS) is found at the same position. An amidated DAP is found in Gram-positive bacilli. Tracheal cytotoxin (TCT) is a glycan dimer with an anhydro form of muramic acid coupled to a DAP-containing peptide linker found at the end of PGN chains. TCT can be released during bacterial division and growth when PGN gets remodeled. Both DAP-PGN and TCT are detected by the Drosophila immune system and trigger the IMD pathway whereas LYS-PGN is a better inducer of the Toll pathway than DAP-PGN. PGNs are essentially sensed, or enzymatically inactivated, by members of the peptidoglycan recognition protein (PGRP) family (Fig. 1) (Ferrandon et al. 2007; Royet and Dziarski 2007).
The Drosophila genome encodes 13 PGRP family members that share a common 160 amino acid PGRP domain with similarity to N-acetylmuramoyl-L-alanine amidases such as the T7 bacteriophage lysozyme (Werner et al. 2000). Only a few members of the family (PGRP-SCs and PGRP-LB) have maintained a catalytically active enzymatic site that allow them to cleave the lactyl-amide bond between the glycan chain and the first amino acid of the stem peptide of PGN (Bischoff et al. 2006; Charroux et al. 2018; Costechareyre et al. 2016; Garver et al. 2006; Gendrin et al. 2009; Guo et al. 2014; Mellroth et al. 2003; Paredes et al. 2011; Zaidman-Remy et al. 2006, 2011). Most of the other family members are involved in sensing PGN (Choe et al. 2002; Gottar et al. 2002; Iatsenko et al. 2016; Kaneko et al. 2006; Michel et al. 2001; Ramet et al. 2002; Takehana et al. 2002, 2004). PGRPs can either be short proteins noted PGRP-S that are secreted or long proteins (PGRP-L). Most PGRP-Ls include a transmembrane domain. For instance, PGRP-LC is a type II transmembrane domain receptor whereas full length PGRP-LE is cytosolic. The complexity of the family is enhanced by the existence of multiple splice isoforms that have been shown to have important functional properties as regards PGRP-LC and PGRP-LB.
The crystal structures of several PGRPs are available and revealed the major mechanism of binding to PGN and how the DAP- and LYS-PGN are being discriminated by distinct family members. The structures of PGRP-LB, PGRP-SA and human PGRP-Iα revealed that PGN is bound to the sensor through a large hydrophilic L-shaped cleft (Chang et al. 2004; Kim et al. 2003; Reiser et al. 2004). The DAP-type PGN sensors PGRP-LC and PGRP-LE complexed to TCT have been crystallized and their structures solved (Chang et al. 2006; Lim et al. 2006). These studies identified specific amino-acids including an arginine buried at the bottom of the docking groove that is required for forming a bidentate salt bridge with the carboxylate end of meso-DAP. These residues are not conserved in PGRPs that bind preferentially to LYS-PGN. These studies also revealed the head-to tail infinite polymerization of PGRP-LE bound to TCT, since the binding of the first PGRP-LE monomer induces the formation of a second binding site in which the disaccharide moiety is present at the dimer interface and not the peptide stem that is bound at the first binding site (Lim et al. 2006). In the case of PGRP-LC, the PGRP-LCx isoform complexes TCT whereas PGRP-LCa binds only to the TCT-PGRP-LCx complex (Fig. 1), as it is unable to directly bind to PGN because its binding groove is interrupted by two helical insertions (Chang et al. 2005). In contrast, modeling studies suggest that PGRP-LCx alone is able to bind to polymeric PGN by forming arrays in which molecules binds to a PGN disaccharide every five units of the chain (Chang et al. 2006).
Sensors of bacteria activating the IMD pathway. The IMD pathway is activated after recognition of Gram-negative bacteria, through the DAP-type peptidoglycans (PGNs) present in their cell wall and that may be released during bacterial growth and division or upon attack by AntiMicrobial Peptides (AMPs). DAP-type PGNs are sensed, or enzymatically inactivated, by members of the PeptidoGlycan Recognition Protein (PGRP) family. Polymeric DAP-type PGNs can be detected by PGRP-LCx whereas the monomeric DAP-type PGNs tracheal cytotoxin (TCT) is sensed by a PGRP-LCx-PGRP-LCa dimer. PGRP-LE functions as an intracellular sensor of TCT in barrier epithelia such as the gut or the Malpighian tubules and also in octopaminergic neurons. The cryptic RIP Homotypic Interaction Motif (cRHIM) domain found in PGRP-LCx, -LCa and -LE allows their oligomerization that seeds the formation of amyloid fibers, that the IMD adapter elongates through its own cRHIM domain. The amyloid fibers triggers downstream IMD pathway signaling that ultimately activates the Relish transcription factor. This process can be negatively regulated by the induction of the PIRK repressor. PGRP-SCs and PGRP-LB prevent detrimental IMD pathway activation by degrading polymeric or monomeric PGNs into metabolites unable to trigger the IMD pathway, a process that is counteracted by the binding of PGRP-SD to PGN. PGRP-LF may prevent the formation of PGRP-LC multimers and hence negatively regulate the initiation of IMD pathway signaling. PGRP-LA may enhance IMD pathway activation solely in barrier epithelia (not shown)
As mentioned above, only the PGRP-LCx isoform is able to bind to PGN whereas the PGRP-LCa is required to sense TCT both in vitro and in vivo. It was possible to test the role of each PGRP-LC isoform in vivo by complementation experiments of a null PGRP-LC mutant (Neyen et al. 2012). The PGRP-LCx alone was sufficient to confer resistance against Ecc15 and also immune-responsiveness to this bacterium whereas the LCa and LCy other isoforms were not active, alone or in combination. Indeed, like PGRP-LCa, PGRP-LCy is structurally unable to bind to PGN but in contrast to the former is not required to sense TCT. More recently, PGRP-LC isoforms with an alternative first coding exons have been identified, which encode alternative cytoplasmic tails for LCx, LCy, and LCa. These rPGRP-LC alternative forms are more induced by an immune challenge than the other isoforms. They play a negative role and target the receptors for endosomal degradation via the ESCRT machinery in a negative regulatory loop. Thus, the function of these isoforms is to down-regulate and terminate IMD pathway signaling upon sensing PGN released by killed bacteria but not upon TCT challenge. This system may allow the organism to adapt to the level of the threat presented by live but not killed bacteria (Neyen et al. 2016).
PGRP-LE belongs to the PGRP-L family yet does not appear to have a signal peptide, even though a shorter form consisting essentially of the sole PGRP domain has been detected in the hemolymph. Its overexpression has been reported to induce melanization, presumably by triggering the proteolytic cascades in the hemolymph that activate PPOs (Takehana et al. 2002). Functionally, PGRP-LE is dispensable for triggering the IMD pathway in the systemic immune response induced by Ecc15 but not Escherichia coli. It is however required for full activation of the systemic immune response induced by the ingestion of Ecc15 bacteria in a sensitized PGRP-LB genetic background (see also below) (Neyen et al. 2016). Further studies have shown that PGRP-LE functions as an intracellular sensor of TCT in barrier epithelia such as the gut or the Malpighian tubules and also in the brain (Bosco-Drayon et al. 2012; Charroux et al. 2018; Kaneko et al. 2006; Kurz et al. 2017).
TCT can be sensed intracellularly by cytosolic PGRP-LE, which raises the question as to how TCT produced by extracellular bacteria reaches the cytoplasm. While it had been shown that the Drosophila SLC15 transporter family Yin is able to enhance the import of muramyl dipeptide and its subsequent sensing by NOD2 when ectopically expressed in mammalian cells, further studies in Drosophila demonstrated that this transporter does not play a significant role in transporting TCT intracellularly for PGRP-LE sensing (Capo et al. 2017; Charriere et al. 2010). In contrast, the SLC46 family CG8046 transporter is required for such a role, at least in Malpighian tubules (Paik et al. 2017). Another possibility for TCT delivery to the cytosol might involve the fusion of TCT-containing outer membrane vesicles secreted by bacteria, a phenomenon not investigated in Drosophila so far.
The intracellular part of PGRP-LC isoforms (rPGRP-LCs excepted) contain a cryptic RIP Homotypic Interaction Motif (cRHIM) domain, which is also found in PGRP-LE, the IMD adapter, and PIRK, a negative regulator of IMD pathway signaling. RHIM domains in mammalian RIP kinase (RIPK) proteins form functional amyloids during necroptosis (Grootjans et al. 2017; Liu et al. 2017). The PGRP-LE, PGRP-LC, and IMD proteins form amyloid fibers in vitro and also in Drosophila S2* cells when overexpressed (Kleino et al. 2017). The disruption of these amyloid fibers led to an impaired IMD immune response in S2 cells and in vivo in flies as well. It is thought that PGRP-LC and PGRP-LE when bound to their ligands seed the formation of amyloid fibers, whereas the much more abundant IMD adapter would then elongate the fibers (Kleino et al. 2017). This process can be negatively regulated by the induction of the PIRK repressor that poisons the formation of novel fibrils and might possibly disassemble existing fibrils. This later possibility remains however to be demonstrated.
Together with sensors of viral infections, PGRP-LE is one of the few known Drosophila cytosolic receptors that trigger innate immune responses. Interestingly, it has also been shown to target intracellular Listeria monocytogenes, a DAP-PGN containing Gram-positive bacterium, for xenophagy (Yano et al. 2008).
Modulation of PGRP-LC signaling by other PGRP family members
The sensing of bacteria with a DAP-PGN cell wall relies on sensing PGN glycan chains or derived metabolites. Several of the PGRP family members enhance or diminish the interactions of PGRP-LC and PGRP-LE with their cognate elicitors. While PGRP-SD had originally been proposed to function in Toll pathway signaling to enhance the detection of Gram-positive bacteria such as S. aureus (Bischoff et al. 2004; Wang et al. 2008), this finding was not confirmed upon studying a CRISPR-Cas9 null mutant that revealed instead a positive role in sensing Gram-negative bacteria, in keeping with its three-dimensional structure that revealed features consistent with PGRP-SD binding to DAP- and not LYS-PGN (Iatsenko et al. 2016; Leone et al. 2008). PGRP-SD is induced through IMD pathway activation and its phenotype is not visible at the very early stages of IMD signaling. It may protect PGN from binding to PGRPs that have a negative impact on IMD pathway signaling (Iatsenko et al. 2016). One is PGRP-LF, which contains two PGRP extracellular domains but a very short cytoplasmic tail unable to initiate downstream canonical IMD signaling (Maillet et al. 2008). While PGRP-LF had originally been proposed to compete with PGRP-LC for DAP-PGN binding, the structural analysis of its two PGRP domains revealed that these domains are unexpectedly unable to bind PGN even though the key amino-acids for binding to DAP-PGN are present in its sequence. However, the PGRP-LF ectodomain is able to bind to PGRP-LCx with an affinity similar to that measured for the PGRP-LCa ectodomain binding to PGRP-LCx in the absence of TCT. Both interactions are increased seven-fold when TCT is added. Thus, by binding to PGRP-LCx, PGRP-LF may prevent the formation of signaling PGRP-LC multimers and hence negatively regulate the initiation of IMD pathway signaling (Basbous et al. 2011). Indeed, the IMD pathway is constitutively active in the absence of PGRP-LF.
PGRP-LA, PGRP-LC, and PGRP-LF form a cluster of genes on the third chromosome. The sequence of PGRP-LA indicates it is unable to bind to PGN. PGRP-LA is poorly expressed in the fat body but is enriched in barrier epithelia. Accordingly, a PGRP-LA mutant did not display any phenotype with respect to the systemic immune response triggered by Ecc15; yet, the ectopic overexpression of its D splice isoform strongly activated the IMD pathway, likely by seeding amyloid fibers through its cRHIM domain. Interestingly, the PGRP-LA loss-of-function mutant displayed a decreased induction of the IMD pathway in larval tracheae and in the adult midgut. Taken together, these results suggest that PGRP-LA promotes IMD pathway activation in local epithelial immune responses (Gendrin et al. 2013).
Two of the three Drosophila nonaspannin proteins, which contain nine transmembrane domains have been shown to interact with PGRP-LC (Perrin et al. 2015). TM9SF4 appears to be required for the trafficking or maintenance of PGRP-LC to the surface of S2 cells. Its downregulation, however, did not impair IMD pathway signaling in cultured cells and in flies. It is an open question whether the sensitivity of TM9SF4 mutants to bacterial infections is caused by defects in the cellular immune response since PGRP-LC appears to play at best an ancillary role in this process (Bergeret et al. 2008; Perrin et al. 2015).
PGRP-SBs, PGRP-SCs, and PGRP-LB have been shown to be functional amidases (Bischoff et al. 2006; Charroux et al. 2018; Costechareyre et al. 2016; Garver et al. 2006; Gendrin et al. 2009; Guo et al. 2014; Mellroth et al. 2003; Paredes et al. 2011; Zaidman-Remy et al. 2006, 2011). Some of them play major roles in preventing detrimental IMD pathway activation by degrading polymeric or monomeric PGN into metabolites unable to trigger the IMD pathway. PGRP-SBs have been shown to be secreted in the hemocoel and to be active against DAP-type PGN. However, the deletion of both PGRP-SB1 and PGRP-SB2 together or in combination with other PGRP amidases did not reveal any immune phenotype (Paredes et al. 2011; Zaidman-Remy et al. 2011). Both PGRP-SC2 and PGRP-LB are strongly expressed in the gut (Costechareyre et al. 2016). Yet, only the latter plays a major role in degrading PGN in the gut lumen and in preventing further systemic activation of the IMD pathway in the fat body, even though PGRP-SC2 mutants display enhanced aging of the gut through increased dysbiosis of the microbiota and concurrent hyperproliferation of intestinal stem cells (Costechareyre et al. 2016; Guo et al. 2014). In the framework of the systemic immune response, it appears that PGRP-SC2 is the major player to degrade immuno-stimulatory PGN. While PGRP-SC1 has been described as being required for phagocytosis and Toll pathway activation independently of its amidase activity, it does not appear to play any regulatory role in the IMD pathway (Costechareyre et al. 2016; Garver et al. 2006; Paredes et al. 2011).
The constitutive activation of the IMD pathway in the gut resulting from the inactivation of some of its negative regulators is detrimental to the life span of flies (Guo et al. 2014; Paredes et al. 2011; Ryu et al. 2008). In the framework of the systemic immune response to infection, one might expect that any detrimental role of enhanced IMD pathway activation would lead to a shortened survival to infection, as has indeed been observed with the injection of Pseudomonas entomophila (Costechareyre et al. 2016). Unexpectedly, mutants for PGRP-LB and PIRK/RUDRA lead respectively to no change and increased survival to an Ecc15 challenge, in keeping with an increased expression of AMPs to which Ecc15 is at least partially sensitive (Aggarwal et al. 2008; Zaidman-Remi et al. 2006). Thus, the exact level of IMD pathway activation results in differing effects according to the infectious pathogen.
Impact of bacterial infections on the local and systemic immune responses
The digestive system represents one of the major and most extensive interface with the environment. Indeed, flies feed on a microbe-rich diet such as decaying fruits and the gut barrier is not as tight as the exoskeleton cuticle. Digestion occurs in the midgut, a section of the digestive tract not protected by cuticle but by a semi-permeable chitinoproteinacous membrane known as the peritrophic matrix that prevents the direct contact of most bacteria with the underlying intestinal epithelium (Lemaitre and Miguel-Aliaga 2013). The gut lumen and a digestive tract diverticulum, the crop, are colonized by the microbiota, which strongly influences the biology of the fly at multiple levels (Broderick and Lemaitre 2012; Pais et al. 2018). It is therefore essential to be able to fight off pathogenic bacteria without unnecessarily clearing off potentially beneficial microbiota. The microbiota is not as diverse as in humans and is made of some 20–30 species with a handful of prevailing ones such as Lactobacillus plantarum or Acetobacter pomorum. Very few species are actually able to truly colonize the digestive tract. PGN derived from the microbiota or ingested bacteria are sensed by enterocytes through PGRP-LE and PGRP-LC, which are distributed in distinct sub-regions of the midgut with PGRP-LC being more prevalent in the anterior part and PGRP-LE in the posterior regions (Bosco-Drayon et al. 2012). As for the systemic immune response, PGRP-LC detects extracellular PGN, whereas PGRP-LE binds to intracellular TCT. PGRP-LE plays a major role in setting up the tolerance level of the microbiota by regulating the expression of IMD pathway negative regulators (Charroux et al. 2018). Interestingly, the level of TCT to which PGRP-LE is exposed is modulated by intracellular forms of PGRP-LB, thereby allowing a fine control of IMD pathway activation within enterocytes independently of systemic IMD pathway activation (Charroux et al. 2018). The extracellular form of PGRP-LB secreted in the lumen limits the passage of PGN through the gut by an unknown mechanism that shields it from the action of the intracellular forms of PGRP-LB in enterocytes. When an intestinal infection occurs, PGN levels increase yet PGRP-LB manages to limit the passage of PGN to the hemocoel. As a result, the induction of the systemic immune response is limited. PGN originating from the gut also reaches octopaminergic neurons where it is sensed by PGRP-LE and activates the IMD pathway to inhibit egg laying (Kurz et al. 2017). As in enterocytes, an intracellular form of PGRP-LB modulates what appears to be a form of behavioral immunity.
Interorgan communications between the gut and the fat body may prime the organism for a potential systemic infection. This is documented in larvae: the ingestion of Ecc15 induces the activation of the systemic immune response, especially that of the IMD pathway (Basset et al. 2000; Foley and O'Farrell, 2003; Shia et al. 2009; Wu et al. 2012; Yang et al. 2019). In contrast, there is hardly any activation of the systemic innate immune response when Ecc15 bacteria are ingested by adult flies (Basset et al. 2000), unless the flies are mutant for PGRP-LB or PGRP-SC2 (Neyen et al. 2012; Zaidman-Remy et al. 2006). Reactive oxygen species generated by the dual oxidase (DUOX) enzyme in enterocytes or by the nitric oxide (NO) synthase (NOS) appear to be required for gut to fat body communication in larvae (Foley and O'Farrell, 2003; Wu et al. 2012). Of note, the work on NO signaling has been performed using chemical inhibitors or NO donors. An ultimate confirmation awaits reproducing these data with a null NOS mutant (Yakubovich et al. 2010). Several studies have reported an involvement of hemocytes in relaying the signal from gut to fat body in larvae (Basset et al. 2000; Charroux and Royet 2009; Shia et al. 2009; Yang et al. 2019). In one study, IMD pathway activation is required in enterocytes and leads to the production by hemocytes of aldose reductase by an unknown mechanism. This leads to an increased concentration in the hemolymph of polyols such as sorbitol or galactitol obtained from the catalysis of glucose or galactose by aldose reductase. These polyols in turn activate the metalloproteinase 2 (MMP2) that would cleave off the extracellular domain of PGRP-LC on the surface of fat body cells and thereby initiate IMD pathway activation (Yang et al. 2019). Of note, the ectopic transgenic expression of mmp2 had earlier been shown to lead to the cleavage of the extracellular part of the PGRP-LC receptor and to the activation of the IMD pathway. The significance of this finding was difficult to assess since IMD pathway activation was normal in mmp2 mutant larvae after septic injury (Schmidt et al. 2011). Thus, the Yang et al. study provided a physiological context in which to interpret this observation. It will be interesting to determine whether PGN fragments are also involved in the activation of the systemic immune response in larvae, for instance, by testing whether it is enhanced in PGRP-SC mutants, since the major PGN degradation function of PGRP-SC takes place in the hemolymph (Costechareyre et al. 2016). Conversely, it remains to be shown that the polyol pathway is also active in adults.
Sensing of Gram-positive bacterial and fungal infections extracellularly triggers the Toll pathway
The Toll pathway is required in the host defense against two strikingly distinct categories of pathogens, prokaryotic Gram-positive bacteria and eukaryotic fungi. In contrast to the IMD pathway where the activation of signaling takes place at the level of the cytoplasmic membrane or intracellularly, the sensing of infections that trigger the Toll pathway takes place extracellularly in the hemocoel (Fig. 2). As noted earlier, Toll does not directly recognize microbial compounds but binds to the Spätzle cytokine, which gets activated by proteolytic cleavage by SPE (Jang et al. 2006). SPE gets activated by two proteolytic cascades, one that involves the upper protease ModSP and the other the Persephone (PSH) protease (Buchon et al. 2009; El Chamy et al. 2008; Gottar et al. 2006). ModSP is activated by sensors that detect LYS-PGN or ß-(1–3)-glucans, whereas PSH acts as a bait for the proteases secreted by bacterial or fungal pathogens.
Sensors of microorganisms activating the Toll pathway. Lys-type peptidoglycans (LYS-PGNs) found at the surface of Gram-positive bacteria are sensed by a complex of GNBP1 and PGRP-SA, the former processing LYS-PGN for detection by PGRP-SA. ß-glucans found at the surface of fungi can be detected by GNBP3. GNBP1 and GNBP3 can activate ModSP through their enzymatically-inactive ß-glucanase domain, that in turn activates Grass. Next, Grass is thought to activate both PSH and Hayan, which function redundantly. The sensing of microbial proteases through Persephone (PSH) bait domain leads to the formation of a cleaved pre-activated inactive PSH that gets further processed by the circulating 26-29-p cathepsin protease to a mature PSH. PSH can also be directly activated by subtilisin (not shown). Active PSH and Hayan can both activate SPE, that processes Spätzle to activate the Toll intracellular signaling pathway. The Necrotic serpin avoids the spontaneous self-activation of PSH. In addition to the activation of Toll intracellular signaling, Hayan activates the Sp7 protease as well as the two prophenoloxidases PPO1 and PPO2. These extracellular enzymes are involved in microbial killing and activation of melanization, which may be separate processes
Genetic screens allowed to identify PGRP-SA and GNBP1 as being required in host defense against several Gram-positive bacterial strains and GNBP3 as being involved in fighting off fungal infections (Gobert et al. 2003; Gottar et al. 2006; Michel et al. 2001). While the X-ray structure of PGRP-SA was established early on, that of the N-terminal domain of GNBP3 was determined later on (Mishima et al. 2009; Takahasi et al. 2009), in keeping with the slower identification of GNBP3 that required screens that also tested fungal elicitors of the immune response. The structure of PGRP PGN-binding domains has been discussed above. As for PGRPs, the first GNBPs have been isolated biochemically in larger insects such as the silkworm Bombyx mori through their ability to bind to microbial cell wall compounds, namely ß-glucans (Ochiai and Ashida 1988; Yoshida et al. 1986, 1996). A first member of the silkworm GNBP family was purified based on its ability to bind to the cell wall of Gram-negative bacteria, hence the name (Lee et al. 1996). Like other members of the family, GNBP1 contains a N-terminal ß-(1–3)-glucan recognition domain (ßGRP) as well as a C-terminal domain with homology to ß-(1–3) and ß (1–3, 1–4) glucanases, the catalytic site of which has not been conserved. However, even though it has been reported to bind to LPS in vitro, genetic analysis unambiguously ascribed a role for GNBP1 in sensing Gram-positive and not Gram-negative bacterial infections (Gobert et al. 2003; Kim et al. 2000; Pili-Floury et al. 2004). The PGRP-SA receptor had been identified earlier as also being required for the activation of the Toll pathway by Gram-positive bacteria with LYS-PGN (Michel et al. 2001). The overexpression of both PGRP-SA and GNBP1 is sufficient to ectopically activate the Toll pathway in the absence of infection (Gobert et al. 2003). Mechanistically, GNBP1 appears to have a muramidase activity that hydrolyzes PGN from Micrococcus luteus or Enterococcus faecalis, but not S. aureus. This muramidase activity is enhanced when GNBP1 is bound to PGRP-SA. Interestingly, the produced polymeric muropeptides can trigger the Toll pathway in a PGRP-SA-dependent but GNBP1-independent manner. Thus, GNBP1 processes some LYS-PGN to present it to PGRP-SA (Filipe et al. 2005; Wang et al. 2006). Of note, in the beetle Tenebrio molitor, it has been proposed that lysozyme processes PGN and thereby provides a substrate for clustering PGRP-SA that then would recruit GNBP1 and initiate downstream signaling (Park et al. 2007).
In contrast, GNBP3 is required for sensing the fungal cell wall (Gottar et al. 2006; Mishima et al. 2009). The N-terminal ß-GRP domain has been shown to bind to relatively long ß-glucan chains with an optimal length ranging between 20 and 40 glucan units. The structure of the immunoglobulin-fold ßGRP domain has been solved, and two distinct models have been proposed to account for its binding to ß-glucans (Mishima et al. 2009; Takahasi et al. 2009). One feature possibly accounting for the ability of the ßGRP domain to bind to long and not short polysaccharide chains has been proposed to rely on a loop that covers the putative glucan binding site that would be displaced only by long glucan chains and then stabilized by two hydrophobic residues present in the lid, which is conserved only in ß-glucan-binding members of the GNBP family but not in GNBP1 (Mishima et al. 2009).
It has been proposed that the function of the ß-glucanase-like domain of GNBPs mediates the activation of the downstream proteolytic cascade, namely the modular serine protease ModSP that functions just downstream of both GNBP1 and GNBP3 (Buchon et al. 2009; Roh et al. 2009). Of note, three genes in the Drosophila genome encode the ßGRP domain only and would therefore not be expected to be directly involved in triggering the proteolytic cascades that activate the Toll pathway or melanization. One of the corresponding proteins, GNBP-like3, is one of the most strongly induced protein by a fungal immune challenge (Levy et al. 2004). It has been reported to be acting as an AMP against Gram-positive bacteria (Barajas-Azpeleta et al. 2018), findings that are not congruent with our analysis of null mutants that reveals an action against a pathogenic yeast (Huang, Quintin et al. in preparation). Finally, two cryptic GNBP members are found in the D. melanogaster genome but are affected by mutations found also in wild fly populations retrieved from diverse locations worldwide and also in closely related Drosophila species (Quintin 2009).
Sensing the enzymatic activity of microbial virulence factors elicits the systemic humoral immune response
The activation of the Drosophila systemic humoral immune response described so far relies on receptors that sense cell wall components. This view corresponds to the Pathogen (or microbial)-associated molecular pattern–Triggered Immunity (PTI) theorized in plants (Jones and Dangl, 2006) and in animals (Gaidt et al. 2021). It has been proposed that pathogens are selected for their expression of virulence factors, some of which might target PTI components. In response, the host has to develop countermeasures that target these virulence factors, a form of immunity known as effector-triggered immunity (ETI). This would usher cycles of selection for other virulence factors and their host antidotes. While evidence for such cycles remains scarce in Drosophila, the sensing of microbial proteases through the Persephone (PSH) protease is evocative of ETI (El Chamy et al. 2008; Gottar et al. 2006). An alternative view is that pathogens may mask the cell wall components that are detected by the innate immunity basal sensing system (Box 1). The detection of the enzymatic activity of secreted virulence factors would allow the host to still perceive the presence of an infection within the hemocoel. A precursor study suggested that some microbial proteases are required for Dorsal nuclear uptake in cultured fat bodies. This activity was however absent when hemolymph was not added to the cultured fat bodies (Bettencourt et al. 2004). The PSH Clip-domain-containing protease is present in hemolymph and contains a bait domain able to be cleaved by various microbial proteases. This leads to the formation of an inactive PSH that gets further processed by a circulating cathepsin protease known as 26–29-p (Issa et al. 2018). Clip domain proteases are secreted as zymogens that get activated by proteolytic cleavage of the Clip prodomain by the protease upstream in the cascade at a site just upstream of the catalytic domain (Veillard et al. 2016). This site contains residues detected usually by enzymes with a trypsin-like specificity. PSH contains an unusual histidine residue instead of the commonly found arginine or lysine, which can be cleaved only by subtilisin secreted by Bacillus subtilis or by the 26–29-p cathepsin, only once the bait domain has been processed. The bait domain is evocative of that found in mammalian α2-macroglobulin, a general suicide inhibitor of all classes of proteases. Formally, the situation is akin to that found in plants in which targets of virulence factors are monitored by guards that detect pathogen-induced modifications of the target: the bait region is a target that allows the 26–29-p guard to trigger the Toll pathway by activating the PSH protease that will activate the downstream SPE protease. Of note, for the PTI part of the pathway elicited by the sensing of cell wall compounds, PSH and another protease known as Hayan appear to function redundantly upstream of SPE (Dudzic et al. 2019). A dedicated serine protease inhibitor, the necrotic serpin (NEC), avoids the spontaneous self-activation of PSH. nec mutants are short-lived and display a constitutive activation of the Toll pathway as well as disseminated melanization (Levashina et al. 1999; Ligoxygakis et al. 2002).
As regards the activation of the IMD pathway by bacterial proteases, some evidence has been presented in S2 cells that the E. coli cell wall–associated protease OMP is required for the cleavage of PGRP-LC after exposure to live bacteria (Schmidt et al. 2011). This finding awaits however further in vivo validation. The IMD pathway can also be activated by a bacterial virulence factor, this time intracellularly. The cytotoxic necrotizing factor 1 (CNF1) produced by some pathogenic E. coli strains catalytically targets the Rho GTPase family member Rac2 for a post-transcriptional modification, the deamidation of a specific glutamine residue that transforms it into a glutamic acid. This transformation abolishes the GTPase activity of Rac2 and locks it into a GTP-bound locked active state. It then activates the IMD pathway independently of its action on the cytoskeleton by directly binding to IMD itself. The activation of the IMD pathway occurs downstream of PGRP receptors and is protective against the pathogen. Interestingly, a similar scheme of NF-κB signaling activation at the level of the IMD-homologs Rip1 and Rip2 occurs also in mammals (Boyer et al. 2011, 2012).
Besides PTI and ETI, the innate immune response has been proposed to respond to so-called danger signals. In this respect, it is interesting to note that the Toll pathway is triggered through PSH in larvae mutant for the caspase Dronc, suggesting that it gets activated when apoptosis is prevented, presumably through the release in circulation of some unidentified host protease during the ensuing necrosis. The beneficial aspects of the immune response in this particular setting remain unclear (Ming et al. 2014) but may become relevant in the course of an infection when cells get damaged. As regards the IMD pathway, it does get activated in a PGRP-independent manner in DNaseII mutants (Mukae et al. 2002). DNaseII is required to digest DNA from apoptotic bodies in the lysosomes of macrophages. How intracellular DNA is sensed in Drosophila remains unknown at present. As described further below, the recently identified Drosophila cGAS/STING pathway does not appear to be activated by DNA but by double-stranded RNA (dsRNA) and thus is not a prime candidate. One component involved in this evolutionary conserved response is Eyes absent, which is found in a complex with IKKß (Liu et al. 2012).
The other major humoral response is melanization. It is closely intertwined with the extracellular part of the Toll pathway down to the step just prior to SPE activation (Dudzic et al. 2019; Matskevich et al. 2010). For instance, the overexpression of GNBP3 or GNBP1/PGRP-SA can lead to PPO cleavage even in spätzle mutants (Matskevich et al. 2010). Thus, the GNBP1/PGRP-SA and GNBP3 sensors of microbial cell wall compounds trigger the ModSP/Grass proteolytic cascade, which leads to SPE activation through the redundant proteases PSH and Hayan that appear to have been evolutionarily duplicated rather recently in the Melanogaster group of Drosophila flies; they also likely act redundantly in parallel for the activation of PPOs. Interestingly, the bait region of PSH is also found in Hayan albeit slightly truncated, suggesting that Hayan may also be activated by microbial proteases as is the case for PSH (Dudzic et al. 2019). The finding that GNBP3 can be found in biochemical complexes together with PPOs further suggests that the melanization enzymes might be brought in close proximity to invading fungi, except if this association is disrupted as observed when the flies are infected by the entomopathogenic fungus Beauveria bassiana through the cuticle (Matskevich et al. 2010).
Many septic injury models produce by definition a wound, which is not neutral for the host as it triggers several responses that lead to wound healing and the maintenance of homeostasis (Box 2). The response to wound such as the production of ROS or AMPs may also tell prospective pathogens that they have entered an unfriendly territory and coax them to express virulence programs that may not be used upon intestinal infections and subsequent escape from the digestive tract (Limmer et al. 2011; Nehme et al. 2007).
In summary, the detection of infections that leads to the activation of humoral responses depends both on binding to specific microbial cell wall components and the detection of the enzymatic activity of virulence factors that are secreted either extracellularly such as proteases or intracellularly such as CNF1. It is not established at present that the activation of the immune responses triggered in Dronc mutants are relevant to microbial infections, whereas EYA is not required for IMD pathway activation induced by a microbial challenge (Liu et al. 2012).
Influence of metabolites released by the microbiota or pathogens in the gut on Drosophila biology.
The gut barrier represents a special frontier between the environment and the internal milieu of the host. The ingested food is rich in microorganisms, some of which are potentially beneficial or detrimental. It is thus important to adjust immune responses so that the microbiota is tolerated under normal conditions but that a strong response against ingested pathogens can be appropriately triggered when needed. In the current paradigm, intestinal host defenses rely on the production of microbe-killing ROS and the production of AMPs by enterocytes. We shall not focus on the different impacts of the microbiota on the gut and more generally on host physiology as these have been recently reviewed (Ferguson and Foley 2021; Lesperance and Broderick 2020). Here, we discuss how specific microbial metabolites modulate immune responses and the homeostasis of the intestinal epithelium (Ferrandon, 2013; Kim and Lee 2014).
A major constituent of the microbiota found in laboratory-raised Drosophila is Acetobacter pomorum. It has been shown to release acetate that is required for its beneficial action on growth of larvae raised on nutrient-poor food (Shin et al. 2011). In the adult gut, the short-chain fatty acid acetate acts on lipid homeostasis in enterocytes of the anterior midgut through the IMD pathway (Kamareddine et al. 2018). Unexpectedly, the critical cell type in which the IMD pathway is activated is not the enterocyte but entero-endocrine cells able to express the endocrine peptide tachykinin. Secreted tachykinin acts on neighboring enterocytes and represses triglyceride synthesis (Song et al. 2014). Acetate promotes IMD pathway signaling in enteroendocrine cells and favors the expression of AMPs albeit weakly and of tachykinin. Acetate does not directly activate the IMD pathway: it is transported inside enteroendocrine cells by the monocarboxylic acid transporter Tarag and increases the acetyl-CoA pool. It has been proposed that it would influence the activity of the acetyltransferase Tip60 complex that in turn would increase the expression of PGRP-LC indirectly through acetylation of the histone H2Aν. This acetylated histone would then promote the transcription of ecdysone-regulated genes including PGRP-LC (Jugder et al. 2021). It remains unclear whether the increased levels of PGRP-LC are sufficient on their own to promote tachykinin transcription through the IMD pathway or whether the enhanced PGRP-LC levels would allow it to react in a more sensitive manner to PGN released by the microbiota. Of note, IMD pathway activation paradoxically promotes Vibrio cholerae infection partially by wasting host metabolic reserves. The susceptibility to infection was decreased when components of the acetate response pathway were silenced by RNA interference in enteroendocrine cells.
The phenomenon described above may not occur under all conditions as the constitution of the microbiota is influenced by the composition and pH of the food. In some laboratories, Acetobacter species are not major components of the microbiota or are absent and Lactobacillus species predominate. A major metabolite produced by Lactobacillus is lactate, which is transported within enterocytes by monocarboxylic acid transporters. It produces NADH by lactate dehydrogenase and triggers the production of ROS through the Nox1 NADPH oxidase (Iatsenko et al. 2018; Jones et al. 2013). This leads to an enhanced proliferation of intestinal stem cells. It also triggers the Nrf2-dependent antioxidant response and leads to an enhanced protection against oxidative injury (Jones et al. 2015). However, in dysbiosis conditions in which the number of Lactobacillus plantarum is increasing such as when IMD signaling is impaired in PGRP-SD mutants, the production of ROS becomes detrimental and leads to a shortened lifespan through premature dysplasia of the intestinal epithelium (Fast et al. 2018; Iatsenko et al. 2018).
ROS have been proposed to represent a major host defense against intestinal infections in Drosophila based on multiple lines of evidence. The most important one is provided by knocking down DUOX expression, a NADPH oxidase with two domains, an intracellular NADPH domain, and an extracellular peroxidase domain thought to generate HOCl (Ha et al. 2005a). The silencing of Duox by RNAi silencing using a ubiquitous driver or a caudal-GAL4 driver expressed only in the posterior midgut leads to an enhanced susceptibility to ingested Ecc15, Salmonella typhimurium, E. coli, and unexpectedly Saccharomyces cerevisiae, an excellent source of food for flies (Ha et al. 2005a, 2009a). Proteins extracted from bacteria retrieved from wild-type infected guts are carbonylated, whereas lipids are peroxidized. A dye developed to detect HOCl strongly stains the anterior midgut but surprisingly not the posterior part of the midgut (given the silencing experiment with the caudal driver) of flies that have ingested Ecc15 (Lee et al. 2013). The heme-containing peroxidase known as Immune-regulated catalase (IRC) is thought to limit the self-damages inflicted by DUOX. Indeed, IRC mutants succumb to the ingestion of killed Ecc15, presumably from the ROS generated by DUOX (Ha et al. 2005b). The DUOX-dependent intestinal defense may be prevalent and only ROS-resistant microorganisms seem to be susceptible to IMD pathway intestinal defenses that appears to act as a second line of defense (Ryu et al. 2006).
The question that arises is how this defense system is elicited by pathogens and not by commensals. Biochemical purification studies from Ecc15 culture supernatant suggest that the bacterial metabolite uracil is secreted by pathobionts and not by commensal microorganisms (Lee et al. 2013). In keeping with these results, a URA− Ecc15 strain unable to secrete uracil no longer leads to a positive R19S staining and persist longer in the gut. However, the identification of a putative uracil receptor able to activate DUOX within enterocytes remains elusive to this date. Uracil would also trigger the hedgehog (HH) pathway, again in undefined manner. This receptor is thought to be endocytosed upon uracil binding together with the HH-dependent Cad99C to form a GαQ-PLCß signaling endosome that leads through the release of intracellular calcium stores to the biochemical activation of DUOX enzymatic activity (Lee et al. 2015). In conjunction with the upper part of the IMD pathway (PGRP-LC and IMD but not Relish) that diverges onto the p38 pathway, the GαQ-Plcß axis also leads to the induction of the Duox gene at the transcriptional level through the ATF2 transcription factor (Ha et al. 2009b). Interestingly, PLCß appears to be required for the repression of the p38 pathway when only commensal bacteria are present in the gut lumen.
In conclusion, metabolites released by gut bacteria modulate or trigger immune responses that are independent of classical AMP expression regulation with important impact on the homeostasis of the gut epithelium.
Role of hemocytes in the host defense: phagocytosis and links with the systemic immune response
Much emphasis has been initially placed on the characterization of the systemic immune response due to the predominant role of AMPs in host defense. As regards Gram-negative bacteria, most AMP families regulated by the IMD pathway are active on them (Diptericins, Drosocins, Attacins, and Cecropins) (Bulet and Stocklin, 2005). Thus, the ancillary role of phagocytosis can be revealed only when the IMD pathway is inactivated at least to some degree (Elrod-Erickson et al. 2000; Nehme et al. 2011). Alternatively, a predominant role for phagocytosis has been revealed in intestinal infection models with Gram-negative bacteria that are able to escape from the gut compartment into the hemocoel such as Serratia marcescens or Pseudomonas aeruginosa (Haller et al. 2018; Limmer et al. 2011; Nehme et al. 2007). Hemocytes prevent the proliferation of bacteria that have crossed the intestinal barrier, at least initially in the case of P. aeruginosa. Interestingly, phagocytosis is powerless when these bacteria are injected in the hemocoel in a systemic infection model.
As regards Gram-positive bacteria, the function of phagocytosis in host defense against these pathogens was more easily established (Charroux and Royet 2009; Defaye et al. 2009; Nehme et al. 2011). This may reflect the relative paucity of bona fide AMPs active against Gram-positive bacteria identified so far. Indeed, Defensin is active on Gram-positive bacteria in vitro, but unexpectedly is no longer expressed in IMD pathway mutants, which resist like wild-type flies to Gram-positive bacterial infections (Rutschmann et al. 2000). A locus of ten Bomanin genes has been identified, the deletion of which phenocopies Toll pathway mutants (Clemmons et al. 2015). However, no secreted Bomanin active against Gram-positive bacteria have been identified to date. Some Gram-positive bacteria such as Staphylococcus aureus are not sensitive to the action of the core Toll pathway but are limited by phagocytosis and melanization (Defaye et al. 2009; Dudzic et al. 2019; Nehme et al. 2011). This latter defense is activated through GNBP1 and PGRP-SA (Matskevich et al. 2010). When only a few bacteria are introduced in the hemocoel, then melanization, and not phagocytosis, becomes the relevant defense (Dudzic et al. 2019).
As regards fungi, the cellular immune response may partially mediate the host defense against yeast forms of pathogens. For instance, phagocytosis acts only in a second line after the Toll pathway against injected C. glabrata (Quintin et al. 2013). We have recently identified a role for phagocytosis against injected conidia of Metarhizium robertsii, which form yeast-like hyphal bodies and not when the fungus invades the fly through the cuticle and forms hyphae (Wang et al. in preparation).
Mechanistically, invading microorganisms need to be sensed by dedicated receptors present on the surface of macrophage-like plasmatocytes present as sessile or circulating hemocytes in the hemocoel (Fig. 3). As in mammals, opsonization appears to be also involved in insects: opsonins are soluble molecules that bind to various microbes and promote their engulfment by acting as adapters that provide a conserved molecular interface to phagocytes. These molecules are briefly presented below.
Drosophila phagocytosis receptors and opsonins. Eater, NimC1 and Draper are three members of the Nimrod family, that are characterized by a subtype of epidermal growth factor (EGF) repeat called Nimrod (NIM) repeat. Eater and Draper can detect bacteria, although NimC1 can recognize yeast zymosan particles. Croquemort and Sr-C1 are two members of the scavenger receptor family that sense bacteria. The integrin made by αPS3 and βυ was shown to be involved in the phagocytosis of S. aureus. Some thioester-containing proteins (TEPs) and some members of the Nimrod B-type subfamily (NimBs) are soluble molecules that bind to various microbes and promote their engulfment by phagocytes (opsonins). NimBs can bind bacteria and TEPs had been shown to recognize both bacteria and yeasts. (Adapted from Melcarne et al. 2019a)
Nimrods
Nimrods are a group of 12 Drosophila phagocytic receptors characterized by a subtype of epidermal growth factor (EGF) repeat called Nimrod (NIM) repeat (Kurucz et al. 2007; Somogyi et al. 2008). Ten genes encoding these receptors are clustered on the second chromosome and two (eater and draper) are clustered on the third chromosome. Two subgroups of the Nimrod receptor family members containing a transmembrane domain can be described: the C-type (NimC1-NimC4 and Eater) and the Draper-type (NimA and Draper) receptors. In contrast, the NimB family members are secreted and circulate in the hemolymph.
The members of the Nimrod C-type subfamily contain multiple NIM domains, whereas the Draper type is characterized by only one such repeat. NimC4, also called Six-Microns-Under (SIMU), is involved in apoptotic cell phagocytosis upstream of Draper through phosphatidylserine recognition (Kurant et al. 2008). Eater and NimC1 mediate the engulfment of bacteria (Bretscher et al. 2015; Kocks et al. 2005; Kurucz et al. 2007; Melcarne et al. 2019b). Both receptors carry a cytosolic tail with predicted phosphorylation sites.
eater is a target of the transcription factor Serpent, a major transcription factor expressed in most hemocytes. eater is expressed in S2 cells and plasmatocytes from both larvae and adult flies. eater knockdown and deficiencies uncovering the eater gene region and neighboring genes allowed to show its important role in Gram-negative and Gram-positive bacteria phagocytosis (Kocks et al. 2005; Nehme et al. 2011). A soluble Fc-tagged receptor variant of Eater directly binds to Gram-positive bacteria and Cecropin A-pretreated Gram-negative bacteria, although the specific ligand remains unknown (Chung and Kocks 2011). The role of eater in the phagocytosis of Gram-positive, but not Gram-negative bacteria, was confirmed in an eater null knock-out mutant (Bretscher et al. 2015). Nevertheless, the host defense against the ingested Gram-negative bacteria Serratia marcescens or Pseudomonas aeruginosa remains impaired also in the KO mutant, possibly because the cell surface properties of bacteria that have crossed the gut barrier are altered (DF and coworkers, unpublished) (Limmer et al. 2011; Nehme et al. 2007). This mutant also led to the identification of a role for Eater in the adhesion of hemocytes to tissues, at the expense of its role in phagocytosis (Bretscher et al. 2015).
NimC1 was initially identified as a target of the plasmatocyte-specific P1 antibody. Knockdown experiments showed that NimC1 is required for S. aureus uptake (Kurucz et al. 2007). NimC1 binds to bacteria in vitro. In contrast to Eater, a role for NimC1 in the phagocytosis of latex beads and yeast zymosan particles, but not bacteria, was established using a NimC1 null KO mutant (Melcarne et al. 2019b). The role of NimC1 in phagocytosis of latex beads was confirmed in an in vivo knockdown experiment (Hao et al. 2018). The contribution of NimC1 to bacterial engulfment is somewhat overshadowed by that of Eater. The two receptors are also complementary to some extent as NimC1; eater double mutants are deficient for the phagocytosis of all types of bacteria tested (Melcarne et al. 2019b).
Draper carries only one NIM domain, several EGF repeats, and one Emilin (EMI) domain. Draper is expressed on plasmatocytes, glial cells, and epithelial cells. Its expression is regulated by the glial transcription factor glial cell missing (Gcm) (Freeman et al. 2003). Draper had initially been shown to recognize apoptotic cells and this function in evolutionarily conserved by its homologues CED-1 in Caenorhabditis elegans or MEGF10 in humans and JEDI in mouse (Hamon et al. 2006; Mangahas and Zhou 2005). More recently, Draper was shown to mediate S. aureus and E. coli phagocytosis (Cuttell et al. 2008; Hashimoto et al. 2009; Shiratsuchi et al. 2012). The Draper ligand on the surface of S. aureus is lipoteichoic acid (Hashimoto et al. 2009).
Scavenger receptors
Scavenger receptors had first been described in mammalian macrophages, where they recognize modified low-density lipoproteins (mLDL) (Brown and Goldstein 1983). These receptors can recognize polyanionic ligands on microbes and dying cells in many species.
Twelve class B scavenger receptors are found in Drosophila. They belong to the mammalian CD36 family, initially discovered for its role in apoptotic cell clearance (Savill et al. 1992). Many of the class B receptors are expressed in the gut where their function is not characterized. One of them, Croquemort (CRQ), is specifically expressed in plasmatocytes and is a receptor for apoptotic cells (Franc et al. 1996, 1999) and for S. aureus (Stuart et al. 2005). Several recent studies revealed a contribution of CRQ in phagosome maturation rather than in particle recognition (Guillou et al. 2016; Han et al. 2014; Meehan et al. 2016). Silencing of crq expression specifically in plasmatocytes allowed to show that it may be involved in lipid uptake as for other CD36 family members (Woodcock et al. 2015). Therefore, it is an open possibility that CRQ is indirectly involved in phagocytosis, as lipid uptake may affect the phagocytic process. Another class B receptor, Peste, is required for phagocytosis of Mycobacterium fortuitum and Listeria monocytogenes, but not E. coli or S. aureus, as shown in RNAi screens made in S2 cells. These results remain to be confirmed in flies (Agaisse et al. 2005; Philips et al. 2005).
Four receptors belong to the class C scavenger receptor family in Drosophila, Sr-CI, Sr-CII, Sr-CIII, and Sr-CIV. Sr-CI is specifically expressed on the surface of plasmatocytes and can recognize a large variety of ligands like the mammalian class A scavenger receptor (Pearson et al. 1995). In vitro studies delineated its capacity to bind to Gram-negative and Gram-positive bacteria, but not yeasts (Pearson et al. 1995; Ramet et al. 2002). Its transcription is upregulated after bacterial challenge (Irving et al. 2005). The Sr-CI locus shows a high degree of polymorphism, likely due to positive selection (Lazzaro 2005). However, the function of Sr-CI in bacterial recognition has never been confirmed in vivo by using null mutants.
Integrins
In addition to their function in adhesion, integrins have been shown to mediate phagocytosis in various organisms. The Drosophila genome encodes five α- and two β- integrin subunits. The βν integrin was shown to be involved in the phagocytosis of both apoptotic cells and S. aureus. PGN is likely to be the integrin ligand as shown using cell wall–deficient bacteria (Shiratsuchi et al. 2012). Furthermore, βν was shown to form a heterodimer with αPS3 for the phagocytosis of both apoptotic cells and S. aureus (Nonaka et al. 2013).
Opsonins
The complement factor C3b and antibodies are classical opsonins in mammals. In Drosophila, six genes encode proteins that carry a domain structurally related to the mammalian complement factor C3 family, named the thioester-containing proteins (TEPs).
The Tep1, Tep2, Tep3, and Tep4 genes contain a signal peptide, indicating that they are secreted proteins and could function as opsonins. It was shown that these four genes are upregulated in adults upon bacterial challenge (Bou Aoun et al. 2011; Irving et al. 2005; Lagueux et al. 2000). Tep5 is thought to be a pseudogene (Bou Aoun et al. 2011; De Gregorio et al. 2002b), while Tep6 (also called Mcr) plays an unexpected role in the establishment of septate junctions in the intestinal epithelium (Batz et al. 2014; Hall et al. 2014). TEP6 is therefore unlikely to play any role in phagocytosis, at least in this context. The expression of Tep genes had been studied by in situ hybridization and by using reporter gene constructs (Bou Aoun et al. 2011; Dostalova et al. 2017). Thus, it was shown that TEP1, TEP2, TEP3, TEP4, and TEP6 are all expressed in plasmatocytes. Tep1, Tep2, and Tep4 transcripts are also detected at low levels in the fat body and some barrier epithelia. They show a strong upregulation after bacterial challenge, since they are regulated by the stress-responsive JAK-STAT pathway (Bou Aoun et al. 2011; Lagueux et al. 2000). In addition, Tep2 and Tep4 appear to be regulated by the systemic immune response (De Gregorio et al. 2002b).
The importance of the TEPs in phagocytosis of bacteria was first demonstrated in the mosquito Anopheles gambiae (Levashina et al. 2001). A RNAi study in Drosophila S2 cells suggested that TEPs can bind and enhance phagocytosis of E. coli (Tep2), S. aureus (Tep3), and Candida albicans (Tep6) (Stroschein-Stevenson et al. 2006). The specificity of these associations remains to be established in vivo. Rather, additional evidence in support of opsonization in insects comes from work using a compound mutant lacking Tep1, Tep2, Tep3 and Tep4 (Dostalova et al. 2017). These four TEPs operate in both the humoral and cellular response of Drosophila immunity, by promoting Toll pathway activation and phagocytosis of Gram-positive bacteria (Dostalova et al. 2017). It was recently shown that TEP4 acts as an opsonin that favors the phagocytosis of P. aeruginosa in an oral infection assay (Haller et al. 2018). In conclusion, some TEPs may indeed bind to microbes and promote phagocytosis, although the possibility is open that they present additional functions. The receptors mediating the uptake of TEPs remain unidentified.
Another class of potential opsonins are the Nimrod B–type proteins, the function of which remains poorly characterized. The five members, NimB1-B5, contain multiple NIM domains that are conserved in other insect species (Estevez-Lao and Hillyer, 2014; Ju et al. 2006; Matsunaga and Fujiwara 2002). Drosophila NimB1 and NimB2 can bind bacteria in vitro (Zsámboki et al. 2013). A recent study has shown that NimB5 is not involved in phagocytosis, but it is produced by the fat body to regulate plasmatocyte adhesion and proliferation rate. NimB5 is induced upon starvation and adjusts plasmatocyte numbers to the metabolic state of the host (Ramond et al. 2020).
Finally, PGRP-SC1A was reported to act as an opsonin for S. aureus and Toll pathway activation (Garver et al. 2006). However, null mutations of PGRP-SC1 did not reveal any role in Toll activation, although an effect modest in comparison to that detected for PGRP-SC2 mutants has been reported after a challenge with E. faecalis (Paredes et al. 2011). This was further corroborated by exposing S2 cells overexpressing either of these two PGRP-SC genes to killed E. faecalis, which led to the stimulation of Drosomycin expression (Costechareyre et al. 2016). As the Toll pathway can be activated in S2 cells solely by providing the mature Spätzle ligand, the expression of Drosomycin likely results from IMD pathway activation. As PGRP-SC degrades peptidoglycan, it might affect bacterial cell wall structure and indirectly promote phagocytosis.
DSCAM1
Down syndrome cell adhesion molecule 1 (Dscam1) belongs to the immunoglobulin family, and its gene organization consists in clusters of variable exons, flanked by constant exons, leading to about 38,000 potential isoforms by alternative splicing (Schmucker et al. 2000). Dscam1 has initially been implicated in the development of the nervous system and neuron wiring, where Dscam1 isoform interactions shape the dendritic pattern (Wojtowicz et al. 2004; Zhan et al. 2004). In 2000, it was shown that infection leads to the production of specific secreted isoforms that can be detected in the Drosophila hemolymph (Watson et al. 2005). However, a study did not reveal any change of Dscam1 splicing upon infection (Armitage et al. 2014). A knockdown experiment in S2 cells showed that Dscam1 is involved in binding and uptake of E. coli by hemocytes and suggested that this protein might act as an opsonin (Watson et al. 2005), a function not yet demonstrated in vivo.
PGRP-LC
RNAi experiments initially showed that PGRP-LC is involved in the phagocytosis of Gram-negative but not Gram-positive bacteria (Ramet et al. 2002). However, it was shown later that PGRP-LC hardly contributes to Gram-negative bacteria phagocytosis in S2 cells (Kocks et al. 2005).
Links between the cellular immune response and the activation of the systemic immune response
As regards adults, the genetic ablation of plasmatocytes using hemocyte-specific drivers failed to reveal any impact on the inducibility of the tested AMP genes (Charroux and Royet 2009; Defaye et al. 2009). However, a later study revealed an unexpected requirement for hemocytes in the case of Drosocin expression in the fat body upon an immune challenge. The IMD pathway is activated through PGRP-LC in hemocytes and leads to the release of the Unpaired3 cytokine, which in turn triggers the expression of Drosocin in the neighboring fat body tissue and the tracheal epithelium (Sanchez Bosch et al. 2019). Thus, hemocytes appear to function as sentinels not only to phagocytose microorganisms that have escaped from the digestive tract but also in the case of systemic infections. Why this system functions solely to induce Drosocin and not other AMP genes remains to be addressed.
With respect to larvae, we have already described the requirement for hemocytes to trigger a systemic activation of the IMD pathway upon the ingestion of Ecc15 (Basset et al. 2000; Yang et al. 2019). The genetic ablation of hemocytes in larvae led to a decreased activation of the systemic immune responses mediated by the Toll and the IMD pathways (Shia et al. 2009). In the case of Toll, it has been proposed that hemocytes contribute to systemic activation of its pathway by providing the Spätzle cytokine whereas the case of the IMD pathway has not been further investigated (Shia et al. 2009). However, the domino mutant, which lacks hemocytes among other defects, nevertheless exhibited a normal induction of the Toll and IMD-mediated systemic immune responses (Braun et al. 1998).
Sensing viral infections
Viral nucleic acids elicit the innate immune response in both protostomes and deuterostomes (Fig. 4). In insects, the sensing of virus-derived dsRNA elicits several antiviral responses.
Sensing viral infections in Drosophila. Double-stranded (ds) RNAs are sensed by Dicer-2, that together with R2D2 process them into siRNAs, one strand of which is loaded on AGO2. The involvement of AGO2 allows the formation of the RISC complex that targets the mRNA corresponding to the complementary siRNA sequence. In parallel, siRNAs induce the expression of Vago. dsRNAs are also sensed by the cGAS-like receptor 1 (cGLR1) that synthesizes 3’2’cGAMP cyclic dinucleotides. Another sensor, cGLR2, is activated by an unidentified ligand, and synthesizes 3’2’cGAMP and 2’3’cGAMP. These two cGAMPs then activates Drosophila melanogaster STING. The latter activates the terminal part of the IMD pathway by interacting with IKKß independently of IKKγ. This STING-Relish axis controls the production of specific genes referred to as STING-regulated genes. The JAK-STAT pathway is likely induced indirectly by cytosolic components such as α-actinin that are released from infected cells lysed by viral infections (not shown)
The DEAD-box RNA helicase DCR-2 is the major sensor of the vsiRNA pathway that acts upon long dsRNAs and is related to mammalian RIG-I (Guo et al. 2019; Schneider and Imler 2021). It contains two domains potentially involved in sensing dsRNAs, a platform-PAZ domain and a N-terminal helicase domain that belongs to the mammalian RIG-I family of cytosolic RNA sensors. RIG-I discriminates between capped cellular transcripts and the phosphorylated termini of viral RNAs. Likewise, DCR-2 does not use cellular transcripts as templates. It uses its helicase domain to initiate the processive cleavage of blunt termini viral dsRNAs, in keeping with a potential evolutionarily ancient role in the detection of non-self RNA. Interestingly, there is a second mode of cleavage, this time of RNAs with 3′OH overhang termini that involves the platform-PAZ domain and is required for processing RNAs of the endo-siRNA pathway, an activity that may involve accessory dsRNA-binding proteins such as the PD isoform of Loquacious (Marques et al. 2013; Sinha et al. 2015, 2018).
As noted earlier, JAK-STAT pathway activation may result from the production of the Unpaired cytokines by damaged cells. In the case of IIV-6, the production of Upd3 depends on the production of ROS by NOX that in turn activates the p38b pathway. It has been proposed that the activation of NOX is somehow mediated by cellular debris of lysed cells as actin-containing fractions trigger a rather similar intracellular signaling pathway in “receiver” cells (see also Box 2) (West and Silverman 2018).
The noncanonical IMD pathway that partially mediates host defense against DCV, CrPV, vesicular stomatitis virus, and Kallithea virus is evocative of the mammalian cGAS-STING pathway that triggers the interferon response upon sensing DNAs in the cytosol, a compartment in which DNA is normally absent (Ablasser and Chen 2019; Goto et al. 2020; Wu and Chen 2014). cGAS senses dsDNA and generates a cyclic dinucleotide known as 2′3′cyclic GMP-AMP (cGAMP). cGAMP then activates STING that in turn acts through TBK1 to stimulate interferon regulatory factors. Interestingly, STING can also be activated by cyclic dinucleotides produce by invasive bacteria such as Listeria (Hansen et al. 2014). In contrast, DmelSTING activates the terminal part of the IMD pathway by interacting with IKKß independently of IKKγ that appears to be solely required for IMD canonical signaling. This STING-Relish axis controls the production of specific genes referred to as STING-regulated genes (Cai et al. 2020). The hunt for Drosophila cGAS homologs has been fruitful. Two cGAS-like receptors (cGLR) required for host defense against viral infections have been identified (Holleufer et al. 2021; Slavik et al. 2021). Insect cGLRs present a distinct nucleic binding site that lacks a Zn-finger motif as well as a secondary binding site. Accordingly, the substrate of cGLR1 is dsRNA longer than 30 bp and not DNA. cGLR2 is activated by an unidentified ligand. Single cGLR mutants are more sensitive than wild-type flies but are less susceptible than the cGLR1/cGLR2 double mutant to viral infections (DCV, Kallithea virus but not VSV nor IIV-6). Surprisingly, cGLR1 predominantly synthesizes 3′2′cGAMP instead of 2′3′cGAMP, a phenomenon that appears to be widespread in Diptera. This may reflect a selective pressure exerted by viruses able to counteract 2′3′cGAMP signaling. Indeed, a family of viral enzymes, poxins made by poxviruses, and some Lepidopteran viruses specifically cleave 2′3′cGAMP and thereby allow the virus to escape detection (Eaglesham et al. 2019). The selection of 3′2′cGAMP signaling would have allowed Dipterans to elude the action of these virulence factors.
A recent study based on a comparative affinity proteomics approach on human, mouse, and Drosophila cells led to the identification of 128 proteins with an impact on viral growth. TAO kinases were identified to bind specifically to dsRNA and to display antiviral activity (Pennemann et al., Nature communications, in press). In mammals, TAO kinases appear to be required for full induction of IRF3-dependent cytokines such as Interferon-ß and IP-10. In flies, the sole TAO kinase is required for the induction of Sting-regulated gene 1.
Our present understanding of the host defense against viral infections is that it relies on sensing viral dsRNAs or potentially on detecting host molecules released from cells lysed by the virus. Future studies will reveal whether evolution has also selected for ETI against specific viral virulence factors, the synthesis of 3′2′cGAMP to potentially avoid poxin action being a step in that direction. Another example is provided by a long noncoding RNA (lncRNA), the VSR-interacting RNA, VINR, which accumulates in the nucleus of DCV-infected S2 cells. This accumulation also occurs upon the ectopic expression of the DCV viral suppressor of RNAi (VSR) protein 1A (DCV-1A). DCV-1A suppresses RNAi by binding to dsRNAs and masking them from the innate immunity sensing system. It likely stabilizes VINR steady-state expression by protecting this lncRNA from degradation. VINR in turn activates a noncanonical IMD pathway involved in host defense against DCV and against bacteria. Thus, VINR is part of a host counter-defense strategy against a viral effector that suppresses the canonical RNAi antiviral pathway, a counter counter-defense (Zhang et al. 2020).
References
Ablasser A, Chen ZJ (2019) cGAS in action: expanding roles in immunity and inflammation. Science 363:eaat8657
Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF et al (2000) The genome sequence of Drosophila melanogaster. Science 287:2185–2195
Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE (2005) Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309:1248–1251
Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N (2003) Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell 5:441–450
Aggarwal K, Rus F, Vriesema-Magnuson C, Erturk-Hasdemir D, Paquette N, Silverman N (2008) Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog 4:e1000120
Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV et al (2009) Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121
Armitage SA, Sun W, You X, Kurtz J, Schmucker D, Chen W (2014) Quantitative profiling of Drosophila melanogaster Dscam1 isoforms reveals no changes in splicing after bacterial exposure. PLoS One 9:e108660
Atilano ML, Pereira PM, Vaz F, Catalao MJ, Reed P, Grilo IR, Sobral RG, Ligoxygakis P, Pinho MG, Filipe SR (2014) Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. eLife 3:e02277
Atilano ML, Yates J, Glittenberg M, Filipe SR, Ligoxygakis P (2011) Wall teichoic acids of Staphylococcus aureus limit recognition by the drosophila peptidoglycan recognition protein-SA to promote pathogenicity. PLoS Pathog 7:e1002421
Attieh Z, Kallassy Awad M, Rejasse A, Courtin P, Gomperts Boneca I, Chapot-Chartier MP, Sanchis Borja V, El Chamy L (2019. D-alanylation of teichoic acids in Bacilli impedes the immune sensing of peptidoglycan in Drosophila. bioRxiv:631523
Attieh Z, Mouawad C, Rejasse A, Jehanno I, Perchat S, Hegna IK, Okstad OA, Kallassy Awad M, Sanchis-Borja V, El Chamy L (2020) The fliK gene is required for the resistance of Bacillus thuringiensis to antimicrobial peptides and virulence in Drosophila melanogaster. Frontiers in microbiology 11:611220
Barajas-Azpeleta R, Wu J, Gill J, Welte R, Seidel C, McKinney S, Dissel S, S, K (2018) Antimicrobial peptides modulate long-term memory. PLoS Genet 14:e1007440
Basbous N, Coste F, Leone P, Vincentelli R, Royet J, Kellenberger C, Roussel A (2011) The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway. EMBO Rep 12:327–333
Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, Lemaitre B (2000) The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci U S A 97:3376–3381
Batz T, Forster D, Luschnig S (2014) The transmembrane protein Macroglobulin complement-related is essential for septate junction formation and epithelial barrier function in Drosophila. Development 141:899–908
Bellen HJ, Tong C, Tsuda H (2010) 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci 11:514–522
Benmimoun B, Papastefanaki F, Perichon B, Segklia K, Roby N, Miriagou V, Schmitt C, Dramsi S, Matsas R, Speder P (2020) An original infection model identifies host lipoprotein import as a route for blood-brain barrier crossing. Nat Commun 11:6106
Bergeret E, Perrin J, Williams M, Grunwald D, Engel E, Thevenon D, Taillebourg E, Bruckert F, Cosson P, Fauvarque MO (2008) TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J Cell Sci 121:3325–3334
Bergner A, Muta T, Iwanaga S, Beisel HG, Delotto R, Bode W (1997) Horseshoe crab coagulogen is an invertebrate protein with a nerve growth factor-like domain. Biol Chem 378:283–287
Bettencourt R, Asha H, Dearolf C, Ip YT (2004) Hemolymph-dependent and -independent responses in Drosophila immune tissue. J Cell Biochem 92:849–863
Bidla G, Dushay MS, Theopold U (2007) Crystal cell rupture after injury in Drosophila requires the JNK pathway, small GTPases and the TNF homolog Eiger. J Cell Sci 120:1209–1215
Binggeli O, Neyen C, Poidevin M, Lemaitre B (2014. Prophenoloxidase activation is required for survival to microbial infections in Drosophila. PLoS Pathog 10:e1004067
Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J (2004) Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol 5:1175–1180
Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J (2006) Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog 2:e14
Blum ID, Keles MF, Baz ES, Han E, Park K, Luu S, Issa H, Brown M, Ho MCW, Tabuchi M, et al (2021) Astroglial calcium signaling encodes sleep need in Drosophila. Curr Biol 31:150–162 e157
Bosco-Drayon V, Poidevin M, Boneca IG, Narbonne-Reveau K, Royet J, Charroux B (2012) Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 12:153–165
Bou Aoun R, Hetru C, Troxler L, Doucet D, Ferrandon D, Matt N (2011) Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster. J Innate Immun 3:52–64
Boulet M, Renaud Y, Lapraz F, Benmimoun B, Vandel L, Waltzer L (2021) Characterization of the Drosophila adult hematopoietic system reveals a rare cell population with differentiation and proliferation potential. Front Cell Dev Biol 9:739357
Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, Charriere GM, Ip WK, Fracchia S, Hennessy E et al (2011) Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity 35:536–549
Boyer L, Paquette N, Silverman N, Stuart LM (2012) Bacterial effectors: learning on the fly. Adv Exp Med Biol 710:29–36
Braun A, Hoffmann JA, Meister M (1998) Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc Natl Acad Sci U S A 95:14337–14342
Brennan CA, Delaney JR, Schneider DS, Anderson KV (2007) Psidin is required in Drosophila blood cells for both phagocytic degradation and immune activation of the fat body. Curr Biol 17:67–72
Bretscher AJ, Honti V, Binggeli O, Burri O, Poidevin M, Kurucz E, Zsamboki J, Ando I, Lemaitre B (2015) The Nimrod transmembrane receptor Eater is required for hemocyte attachment to the sessile compartment in Drosophila melanogaster. Biology Open 4:355–363
Broderick NA, Lemaitre B (2012) Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3:307–321
Brown MS, Goldstein JL (1983) Lipoprotein metabolism in the macrophage: implications for cholesterol deposition in atherosclerosis. Annu Rev Biochem 52:223–261
Brun S, Vidal S, Spellman P, Takahashi K, Tricoire H, Lemaitre B (2006) The MAPKKK Mekk1 regulates the expression of Turandot stress genes in response to septic injury in Drosophila. Genes Cells 11:397–407
Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, Lemaitre B (2009) A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci U S A 106:12442–12447
Buchon N, Silverman N, Cherry S (2014) Immunity in Drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat Rev Immunol 14:796–810
Bulet P, Stocklin R (2005) Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett 12:3–11
Cai H, Holleufer A, Simonsen B, Schneider J, Lemoine A, Gad HH, Huang J, Huang J, Chen D, Peng T et al (2020) 2'3'-cGAMP triggers a STING- and NF-kappaB-dependent broad antiviral response in Drosophila. Sci Signal 13
Cai XT, Li H, Borch Jensen M, Maksoud E, Borneo J, Liang Y, Quake SR, Luo L, Haghighi P, Jasper H (2021) Gut cytokines modulate olfaction through metabolic reprogramming of glia. Nature 596:97–102
Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B (2013) Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc Natl Acad Sci U S A 110:E1752-1760
Capilla A, Karachentsev D, Patterson RA, Hermann A, Juarez MT, McGinnis W (2017) Toll pathway is required for wound-induced expression of barrier repair genes in the Drosophila epidermis. Proc Natl Acad Sci U S A 114:E2682–E2688
Capo F, Chaduli D, Viallat-Lieutaud A, Charroux B, Royet J (2017) Oligopeptide transporters of the SLC15 family are dispensable for peptidoglycan sensing and transport in Drosophila. J Innate Immun 9:483–492
Chakrabarti S, Dudzic JP, Li X, Collas EJ, Boquete JP, Lemaitre B (2016) Remote control of intestinal stem cell activity by haemocytes in Drosophila. PLoS Genet 12:e1006089
Chakrabarti S, Visweswariah SS (2020) Intramacrophage ROS primes the innate immune system via JAK/STAT and toll activation. Cell reports 33:108368
Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J (2006) Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 311:1761–1764
Chang CI, Ihara K, Chelliah Y, Mengin-Lecreulx D, Wakatsuki S, Deisenhofer J (2005) Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc Natl Acad Sci U S A 102:10279–10284
Chang CI, Pili-Floury SS, Herve M, Parquet C, Chelliah Y, Lemaitre B, Mengin-Lecreulx D, Deisenhofer J (2004) A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l, d-carboxypeptidase activity. PLoS Biol 2:E277
Charriere GM, Ip WE, Dejardin S, Boyer L, Sokolovska A, Cappillino MP, Cherayil BJ, Podolsky DK, Kobayashi KS, Silverman N et al (2010) Identification of Drosophila Yin and PEPT2 as evolutionarily conserved phagosome-associated muramyl dipeptide transporters. J Biol Chem 285:20147–20154
Charroux B, Capo F, Kurz CL, Peslier S, Chaduli D, Viallat-Lieutaud A, Royet J (2018) Cytosolic and secreted peptidoglycan-degrading enzymes in Drosophila respectively control local and systemic immune responses to microbiota. Cell Host Microbe 23:215–228 e214
Charroux B, Royet J (2009) Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc Natl Acad Sci U S A 106:9797–9802
Choe KM, Lee H, Anderson KV (2005) Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A 102:1122–1126
Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV (2002) Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in Drosophila. Science 296:359–362
Chung YS, Kocks C (2011) Recognition of pathogenic microbes by the Drosophila phagocytic pattern recognition receptor eater. J Biol Chem 286:26524–26532
Clark RI, Woodcock KJ, Geissmann F, Trouillet C, Dionne MS (2011) Multiple TGF-beta superfamily signals modulate the adult Drosophila immune response. Curr Biol 21:1672–1677
Clemmons AW, Lindsay SA, Wasserman SA (2015) An effector peptide family required for Drosophila toll-mediated immunity. PLoS Pathog 11:e1004876
Costechareyre D, Capo F, Fabre A, Chaduli D, Kellenberger C, Roussel A, Charroux B, Royet J (2016) Tissue-specific regulation of Drosophila NF-kappaB pathway activation by peptidoglycan recognition protein SC. J Innate Immun 8:67–80
Cuttell L, Vaughan A, Silva E, Escaron CJ, Lavine M, Van Goethem E, Eid JP, Quirin M, Franc NC (2008) Undertaker, a Drosophila junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell 135:524–534
Davis MM, Engstrom Y (2012) Immune response in the barrier epithelia: lessons from the fruit fly Drosophila melanogaster. J Innate Immun 4:273–283
De Gregorio E, Han SJ, Lee WJ, Baek MJ, Osaki T, Kawabata S, Lee BL, Iwanaga S, Lemaitre B, Brey PT (2002a) An immune-responsive Serpin regulates the melanization cascade in Drosophila. Dev Cell 3:581–592
De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B (2002b) The Toll and Imd pathways are the major regulators of the immune response in Drosophila. Embo J 21:2568–2579
Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, Dostert C, Antoniewski C, Hoffmann JA, Imler JL (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol 9:1425–1432
Defaye A, Evans I, Crozatier M, Wood W, Lemaitre B, Leulier F (2009) Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infections. J Innate Immun 1:322–334
Dostalova A, Rommelaere S, Poidevin M, Lemaitre B (2017) Thioester-containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps. BMC Biol 15:79
Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL (2005) The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol 6:946–953
Dudzic JP, Hanson MA, Iatsenko I, Kondo S, Lemaitre B (2019) More than black or white: melanization and toll share regulatory serine proteases in Drosophila. Cell reports 27:1050–1061 e1053
Dudzic JP, Kondo S, Ueda R, Bergman CM, Lemaitre B (2015) Drosophila innate immunity: regional and functional specialization of prophenoloxidases. BMC Biol 13:81
Eaglesham JB, Pan Y, Kupper TS, Kranzusch PJ (2019) Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature 566:259–263
Ekengren S, Tryselius Y, Dushay MS, Liu G, Steiner H, Hultmark D (2001) A humoral stress response in Drosophila. Curr Biol 11:714–718
El Chamy L, Leclerc V, Caldelari I, Reichhart JM (2008) Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat Immunol 9:1165–1170
Elrod-Erickson M, Mishra S, Schneider D (2000) Interactions between the cellular and humoral immune responses in Drosophila. Curr Biol 10:781–784
Engström Y, Kadalayil L, Sun S-C, Samakovlis C, Hultmark D, Faye I (1993) KappaB-like motifs regulate the induction of immune genes in Drosophila. J Mol Biol 232:327–333
Estevez-Lao TY, Hillyer JF (2014) Involvement of the Anopheles gambiae Nimrod gene family in mosquito immune responses. Insect Biochem Mol Biol 44:12–22
Evans IR, Rodrigues FS, Armitage EL, Wood W (2015) Draper/CED-1 mediates an ancient damage response to control inflammatory blood cell migration in vivo. Curr Biol 25:1606–1612
Fast D, Duggal A, Foley E (2018) Monoassociation with Lactobacillus plantarum disrupts intestinal homeostasis in adult Drosophila melanogaster. mBio 9:e01114–01118
Ferguson M, Foley E (2021) Microbial recognition regulates intestinal epithelial growth in homeostasis and disease. Febs J
Ferrandon D (2013) The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: from resistance to resilience. Curr Opin Immunol 25:59–70
Ferrandon D, Imler JL, Hetru C, Hoffmann JA (2007) The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol 7:862–874
Ferrandon D, Jung AC, Criqui MC, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart JM, Hoffmann JA (1998) A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J 17:1217–1227
Ferreira AG, Naylor H, Esteves SS, Pais IS, Martins NE, Teixeira L (2014) The Toll-dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog 10:e1004507
Filipe SR, Tomasz A, Ligoxygakis P (2005) Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep 6:327–333
Foley E, O’Farrell PH (2003) Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev 17:115–125
Franc NC, Dimarcq J-L, Lagueux M, Hoffmann J, Ezekowitz AB (1996) Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4:431–443
Franc NC, Heitzler P, Ezekowitz RA, White K (1999) Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science 284:1991–1994
Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ (2003) Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron 38:567–580
Gaidt MM, Morrow A, Fairgrieve MR, Karr JP, Yosef N, Vance RE (2021). Self-guarding of MORC3 enables virulence factor-triggered immunity. Nature in press
Ganesan S, Aggarwal K, Paquette N, Silverman N (2011) NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster. Curr Top Microbiol Immunol 349:25–60
Garver LS, Wu J, Wu LP (2006) The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proc Natl Acad Sci U S A 103:660–665
Gay NJ, Keith FJ (1991) Drosophila Toll and IL-1 receptor. Nature 351:355–356
Gendrin M, Welchman DP, Poidevin M, Herve M, Lemaitre B (2009) Long-range activation of systemic immunity through peptidoglycan diffusion in Drosophila. PLoS Pathog 5:e1000694
Gendrin M, Zaidman-Remy A, Broderick NA, Paredes J, Poidevi M, Roussel A, Lemaitre B (2013) Functional analysis of PGRP-LA in Drosophila immunity. PLoS One 8:e69742
Glaser RW (1918) On the existence of immunity principles in insects. Psyche 25:39–46
Gobert V, Gottar M, Matskevich A, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D (2003) Dual activation of the Drosophila Toll pathway by two pattern recognition receptors. Science 302:2126–2130
Goic B, Vodovar N, Mondotte JA, Monot C, Frangeul L, Blanc H, Gausson V, Vera-Otarola J, Cristofari G, Saleh MC (2013) RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol 14:396–403
Gordon O, Henry CM, Srinivasan N, Ahrens S, Franz A, Deddouche S, Chakravarty P, Phillips D, George R, Kjaer S et al (2018) Alpha-actinin accounts for the bioactivity of actin preparations in inducing STAT target genes in Drosophila melanogaster. eLife 7:e19662
Goto A, Okado K, Martins N, Cai H, Barbier V, Lamiable O, Troxler L, Santiago E, Kuhn L, Paik D et al (2020) The kinase IKKbeta regulates a STING-and NF-kappaB- dependent antiviral response pathway in Drosophila. Immunity 52:200
Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, Belvin M, Hoffmann JA, Ferrandon D (2006) Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell 127:1425–1437
Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J (2002) The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416:640–644
Grootjans S, Vanden Berghe T, Vandenabeele P (2017) Initiation and execution mechanisms of necroptosis: an overview. Cell Death Differ 24:1184–1195
Guillou A, Troha K, Wang H, Franc NC, Buchon N (2016) The Drosophila CD36 Homologue croquemort is required to maintain immune and gut homeostasis during development and aging. PLoS Pathog 12:e1005961
Guo L, Karpac J, Tran SL, Jasper H (2014) PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 156:109–122
Guo Z, Li Y, Ding SW (2019) Small RNA-based antimicrobial immunity. Nat Rev Immunol 19:31–44
Ha EM, Lee KA, Park SH, Kim SH, Nam HJ, Lee HY, Kang D, Lee WJ (2009a) Regulation of DUOX by the Galphaq-phospholipase Cbeta-Ca2+ pathway in Drosophila gut immunity. Dev Cell 16:386–397
Ha EM, Lee KA, Seo YY, Kim SH, Lim JH, Oh BH, Kim J, Lee WJ (2009b) Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in drosophila gut. Nat Immunol 10:949–957
Ha EM, Oh CT, Bae YS, Lee WJ (2005a) A direct role for dual oxidase in Drosophila gut immunity. Science 310:847–850
Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, Brey PT, Lee WJ (2005b) An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell 8:125–132
Hall S, Bone C, Oshima K, Zhang L, McGraw M, Lucas B, Fehon RG, Ward, R.E.t. (2014) Macroglobulin complement-related encodes a protein required for septate junction organization and paracellular barrier function in Drosophila. Development 141:889–898
Haller S, Franchet A, Hakkim A, Chen J, Drenkard E, Yu S, Schirmeier S, Li Z, Martins N, Ausubel FM et al (2018) Quorum-sensing regulator RhlR but not its autoinducer RhlI enables Pseudomonas to evade opsonization. EMBO Rep 19:e44880
Hamon Y, Trompier D, Ma Z, Venegas V, Pophillat M, Mignotte V, Zhou Z, Chimini G (2006) Cooperation between engulfment receptors: the case of ABCA1 and MEGF10. PLoS One 1:e120
Han C, Song Y, Xiao H, Wang D, Franc NC, Jan LY, Jan YN (2014) Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron 81:544–560
Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA et al (2014) Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J 33:1654–1666
Hao Y, Yu S, Luo F, Jin LH (2018) Jumu is required for circulating hemocyte differentiation and phagocytosis in Drosophila. Cell Commun Signal 16:95
Harnish JM, Link N, Yamamoto S (2021) Drosophila as a model for infectious diseases. Int J Mol Sci 22:2724
Hashimoto Y, Tabuchi Y, Sakurai K, Kutsuna M, Kurokawa K, Awasaki T, Sekimizu K, Nakanishi Y, Shiratsuchi A (2009) Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. J Immunol 183:7451–7460
Holleufer A, Winther KG, Gad HH, Ai X, Chen Y, Li L, Wei Z, Deng H, Liu J, Frederiksen NA et al (2021) Two cGAS-like receptors induce antiviral immunity in Drosophila. Nature 597:114–118
Hultmark D (1993) Immune-reactions in Drosophila and other insects - a model for innate immunity. Trends Genet 9:178–183
Hultmark D, Steiner H, Rasmuson T, Boman HG (1980) Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem 106:7–16
Hunter MV, Willoughby PM, Bruce AEE, Fernandez-Gonzalez R (2018) Oxidative stress orchestrates cell polarity to promote embryonic wound healing. Dev Cell 47:377–387 e374
Iatsenko I, Boquete JP, Lemaitre B (2018) Microbiota-derived lactate activates production of reactive oxygen species by the intestinal NADPH oxidase Nox and shortens Drosophila lifespan. Immunity 49:929–942 e925
Iatsenko I, Kondo S, Mengin-Lecreulx D, Lemaitre B (2016) PGRP-SD, an extracellular pattern-recognition receptor, enhances peptidoglycan-mediated activation of the Drosophila Imd pathway. Immunity 45:1013–1023
Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, Hoffmann JA, Hetru C, Meister M (2005) New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol 7:335–350
Issa N, Guillaumot N, Lauret E, Matt, N, Schaeffer-Reiss C, Van Dorsselaer A, Reichhart JM, Veillard F (2018) The circulating protease persephone is an immune sensor for microbial proteolytic activities upstream of the Drosophila toll pathway. Mol Cell 69:539–550 e536
Janeway CA (1989) Approaching the asymptote: evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54:1–13
Janeway CA Jr, Medzhitov R (2002) Innate immune recognition. Annu Rev Immunol 20:197–216
Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, Kambris Z, Brun S, Hashimoto C, Ashida M et al (2006) A Spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell 10:45–55
Jones JD, Dangl JL (2006) The plant immune system. Nature 444:323–329
Jones RM, Desai C, Darby TM, Luo L, Wolfarth AA, Scharer CD, Ardita CS, Reedy AR, Keebaugh ES, Neish AS (2015) Lactobacilli modulate epithelial cytoprotection through the Nrf2 pathway. Cell Rep 12:1217–1225
Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA et al (2013) Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J 32:3017–3028
Jones RM, Luo L, Moberg KH (2012) Aeromonas salmonicida-secreted protein AopP is a potent inducer of apoptosis in a mammalian and a Drosophila model. Cell Microbiol 14:274–285
Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS (2008) Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe 3:233–244
Ju JS, Cho MH, Brade L, Kim JH, Park JW, Ha NC, Soderhall I, Soderhall K, Brade H, Lee BL (2006) A novel 40-kDa protein containing six repeats of an epidermal growth factor-like domain functions as a pattern recognition protein for lipopolysaccharide. J Immunol 177:1838–1845
Jugder BE, Kamareddine L, Watnick PI (2021) Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex. Immunity 54:1683–1697 e1683
Kamar R, Rejasse A, Jehanno I, Attieh Z, Courtin P, Chapot-Chartier MP, Nielsen-Leroux C, Lereclus D, El Chamy L, Kallassy M et al (2017) DltX of Bacillus thuringiensis is essential for D-Alanylation of teichoic acids and resistance to antimicrobial response in insects. Front Microbiol 8:1437
Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, Watnick PI (2018) The Drosophila immune deficiency pathway modulates enteroendocrine function and host metabolism. Cell Metab 28:449–462 e445
Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N (2004) Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20:637–649
Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH et al (2006) PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol 7:715–723
Kiehart DP, Crawford JM, Aristotelous A, Venakides S, Edwards GS (2017) Cell sheet morphogenesis: dorsal closure in Drosophila melanogaster as a model system. Annu Rev Cell Dev Biol 33:169–202
Kim BY, Wang JR, Miller DE, Barmina O, Delaney E, Thompson A, Comeault AA, Peede D, D’Agostino ER, Pelaez J et al (2021) Highly contiguous assemblies of 101 drosophilid genomes. eLife 10:e66405
Kim MS, Byun M, Oh BH (2003) Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol 4:787–793
Kim SH, Lee WJ (2014) Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Front Cell Infect Microbiol 3:116
Kim YS, Ryu JH, Han SJ, Choi KH, Nam KB, Jang IH, Lemaitre B, Brey PT, Lee WJ (2000) Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J Biol Chem 275:32721–32727
Kleino A, Ramia NF, Bozkurt G, Shen Y, Nailwal H, Huang J, Napetschnig J, Gangloff M, Chan FK, Wu H, et al (2017). Peptidoglycan-sensing receptors trigger the formation of functional amyloids of the adaptor protein imd to initiate Drosophila NF-kappaB signaling. Immunity 47:635–647 e636
Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM et al (2005) Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123:335–346
Kurant E, Axelrod S, Leaman D, Gaul U (2008) Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell 133:498–509
Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E et al (2007) Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol 17:649–654
Kurz CL, Charroux B, Chaduli D, Viallat-Lieutaud A, Royet J (2017) Peptidoglycan sensing by octopaminergic neurons modulates Drosophila oviposition. eLife 6:e50559
Lafont M, Vergnes A, Vidal-Dupiol J, de Lorgeril J, Gueguen Y, Haffner P, Petton B, Chaparro C, Barrachina C, Destoumieux-Garzon D et al (2020) A sustained immune response supports long-term antiviral immune priming in the Pacific oyster. Crassostrea Gigas Mbio 11:e02777-e12719
Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA (2000) Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci U S A 97:11427–11432
Lanot R, Zachary D, Holder F, Meister M (2001) Postembryonic hematopoiesis in Drosophila. Dev Biol 230:243–257
Lau GW, Goumnerov BC, Walendziewicz CL, Hewitson J, Xiao W, Mahajan-Miklos S, Tompkins RG, Perkins LA, Rahme LG (2003) The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect Immun 71:4059–4066
Lazzaro BP (2005) Elevated polymorphism and divergence in the class C scavenger receptors of Drosophila melanogaster and D. simulans. Genetics 169:2023–2034
Lee KA, Kim B, Bhin J, Kim DH, You H, Kim EK, Kim SH, Ryu JH, Hwang D, Lee WJ (2015) Bacterial uracil modulates Drosophila DUOX-dependent gut immunity via Hedgehog-induced signaling endosomes. Cell Host Microbe 17:191–204
Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, Lee WJ (2013) Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153:797–811
Lee WJ, Lee JD, Kravchenko VV, Ulevitch RJ, Brey PT (1996) Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm, Bombyx mori. Proc Natl Acad Sci U S A 93:7888–7893
Lee WJ, Miura M (2014) Mechanisms of systemic wound response in Drosophila. Curr Top Dev Biol 108:153–183
Lemaitre B, Hoffmann J (2007) The host defense of Drosophila melanogaster. Annu Rev Immunol 25:697–743
Lemaitre B, Miguel-Aliaga I (2013) The digestive tract of Drosophila melanogaster. Annu Rev Genet 47:377–404
Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA (1996) The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973–983
Lemaitre B, Reichhart JM, Hoffmann JA (1997) Drosophila host defense: differential display of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA 94:14614–14619
Leone P, Bischoff V, Kellenberger C, Hetru C, Royet J, Roussel A (2008) Crystal structure of Drosophila PGRP-SD suggests binding to DAP-type but not lysine-type peptidoglycan. Mol Immunol 45:2521–2530
Lesperance DN, Broderick NA (2020) Microbiomes as modulators of Drosophila melanogaster homeostasis and disease. Curr Opin Insect Sci 39:84–90
Leulier F, Lemaitre B (2008) Toll-like receptors–taking an evolutionary approach. Nat Rev Genet 9:165–178
Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B (2003) The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol 4:478–484
Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM (1999) Constitutive activation of Toll-mediated antifungal defense in serpin-deficient Drosophila. Science 285:1917–1919
Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC (2001) Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104:709–718
Levy F, Bulet P, Ehret-Sabatier L (2004) Proteomic analysis of the systemic immune response of Drosophila. Mol Cell Proteomics 3:156–166
Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM (2002) Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297:114–116
Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH (2006) Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem 281:8286–8295
Limmer S, Haller S, Drenkard E, Lee J, Yu S, Kocks C, Ausubel FM, Ferrandon D (2011) Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc Natl Acad Sci U S A 108:17378–17383
Lin SJH, Cohen LB, Wasserman SA (2020) Effector specificity and function in Drosophila innate immunity: getting AMPed and dropping Boms. PLoS Pathog 16:e1008480
Liu S, Liu H, Johnston A, Hanna-Addams S, Reynoso E, Xiang Y, Wang Z (2017) MLKL forms disulfide bond-dependent amyloid-like polymers to induce necroptosis. Proc Natl Acad Sci U S A 114:E7450-e7459
Liu X, Sano T, Guan Y, Nagata S, Hoffmann JA, Fukuyama H (2012) Drosophila EYA regulates the immune response against DNA through an evolutionarily conserved threonine phosphatase motif. PLoS One 7:e42725
Loof TG, Morgelin M, Johansson L, Oehmcke S, Olin AI, Dickneite G, Norrby-Teglund A, Theopold U, Herwald H (2011) Coagulation, an ancestral serine protease cascade, exerts a novel function in early immune defense. Blood 118:2589–2598
Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J (2008) The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe 3:293–303
Mangahas PM, Zhou Z (2005) Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol 16:295–306
Marques JT, Wang JP, Wang X, de Oliveira KP, Gao C, Aguiar ER, Jafari N, Carthew RW (2013) Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog 9:e1003579
Matos RC, Schwarzer M, Gervais H, Courtin P, Joncour P, Gillet B, Ma D, Bulteau AL, Martino ME, Hughes S et al (2017) D-Alanylation of teichoic acids contributes to Lactobacillus plantarum-mediated Drosophila growth during chronic undernutrition. Nat Microbiol 2:1635–1647
Matskevich AA, Quintin J, Ferrandon D (2010) The Drosophila PRR GNBP3 assembles effector complexes involved in antifungal defenses independently of its Toll-pathway activation function. Eur J Immunol 40:1244–1254
Matsunaga TM, Fujiwara H (2002) Identification and characterization of genes abnormally expressed in wing-deficient mutant (flugellos) of the silkworm, Bombyx mori. Insect Biochem Mol Biol 32:691–699
Meehan TL, Joudi TF, Timmons AK, Taylor JD, Habib CS, Peterson JS, Emmanuel S, Franc NC, McCall K (2016).Components of the engulfment machinery have distinct roles in corpse processing. PLoS One 11:e0158217
Meister M, Braun A, Kappler C, Reichhart J-M, Hoffmann JA (1994) Insect immunity. A transgenic analysis in Drosophila defines several functional domains in the diptericin promoter. EMBO J 13:5958–5966
Melcarne C, Lemaitre B, Kurant E (2019a) Phagocytosis in Drosophila: from molecules and cellular machinery to physiology. Insect Biochem Mol Biol 109:1–12
Melcarne C, Ramond E, Dudzic J, Bretscher AJ, Kurucz E, Ando I, Lemaitre B (2019b) Two Nimrod receptors, NimC1 and Eater, synergistically contribute to bacterial phagocytosis in Drosophila melanogaster. FEBS J 286:2670–2691
Mellroth P, Karlsson J, Steiner H (2003) A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem 278:7059–7064
Metchnikoff E (1884) Über eine Sprosspilzkrankheit der Daphnien; Beitrag zur lehre über Kampf der Phagocyten gegen Krankheitserreger. Archiv Für Pathologische Anatomie Und Physiologie Und Für Klinische Medecin 96:177–195
Michel T, Reichhart J, Hoffmann JA, Royet J (2001) Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414:756–759
Ming M, Obata F, Kuranaga E, Miura M (2014) Persephone/Spatzle pathogen sensors mediate the activation of Toll receptor signaling in response to endogenous danger signals in apoptosis-deficient Drosophila. J Biol Chem 289:7558–7568
Mishima Y, Quintin J, Aimanianda V, Kellenberger C, Coste F, Clavaud C, Hetru C, Hoffmann JA, Latge JP, Ferrandon D et al (2009) The N-terminal domain of drosophila gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors. J Biol Chem 284:28687–28697
Mondotte JA, Gausson V, Frangeul L, Blanc H, Lambrechts L, Saleh MC (2018) Immune priming and clearance of orally acquired RNA viruses in Drosophila. Nat Microbiol 3:1394–1403
Mondotte JA, Gausson V, Frangeul L, Suzuki Y, Vazeille M, Mongelli V, Blanc H, Failloux AB, Saleh MC (2020) Evidence for long-lasting transgenerational antiviral immunity in insects. Cell reports 33:108506
Montanari M, Royet J (2021) Impact of microorganisms and parasites on neuronally controlled Drosophila behaviours. Cells 10:2350
Mukae N, Yokoyama H, Yokokura T, Sakoyama Y, Nagata S (2002) Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev 16:2662–2671
Muta T, Iwanaga S (1996) The role of hemolymph coagulation in innate immunity. Curr Opin Immunol 8:41–47
Nam HJ, Jang IH, You H, Lee KA, Lee WJ (2012) Genetic evidence of a redox-dependent systemic wound response via Hayan protease-phenoloxidase system in Drosophila. Embo J 31:1253–1265
Nappi AJ, Vass E, Frey F, Carton Y (1995) Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur J Cell Biol 68:450–456
Nehme NT, Liegeois S, Kele B, Giammarinaro P, Pradel E, Hoffmann JA, Ewbank JJ Ferrandon D (2007) A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog 3:e173
Nehme NT, Quintin J, Cho JH, Lee J, Lafarge MC, Kocks C, Ferrandon D (2011) Relative roles of the cellular and humoral responses in the Drosophila host defense against three Gram-positive bacterial infections. PLoS One 6:e14743
Neyen C, Binggeli O, Roversi P, Bertin L, Sleiman MB, Lemaitre B (2015) The Black cells phenotype is caused by a point mutation in the Drosophila pro-phenoloxidase 1 gene that triggers melanization and hematopoietic defects. Dev Comp Immunol 50:166–174
Neyen C, Poidevin M, Roussel A, Lemaitre B (2012) Tissue- and ligand-specific sensing of gram-negative infection in drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol 189:1886–1897
Neyen C, Runchel C, Schupfer F, Meier P, Lemaitre B (2016) The regulatory isoform rPGRP-LC induces immune resolution via endosomal degradation of receptors. Nat Immunol 17:1150–1158
Niethammer P (2016) The early wound signals. Curr Opin Genet Dev 40:17–22
Nonaka S, Nagaosa K, Mori T, Shiratsuchi A, Nakanishi Y (2013) Integrin alphaPS3/betanu-mediated phagocytosis of apoptotic cells and bacteria in Drosophila. J Biol Chem 288:10374–10380
Nüsslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287:795–801
O'Connor JT, Stevens AC, Shannon EK, Akbar FB, LaFever KS, Narayanan NP, Gailey CD, Hutson MS, Page-McCaw A (2021) Proteolytic activation of growth-blocking peptides triggers calcium responses through the GPCR Mthl10 during epithelial wound detection. Dev Cell 56:2160–2175 e2165
Ochiai M, Ashida M (1988) Purification of a beta-1,3-glucan recognition protein in the prophenoloxidase activating system from hemolymph of the silkworm, Bombyx mori. J Biol Chem 263:12056–12062
Onfelt Tingvall T, Roos E, Engstrom Y (2001) The imd gene is required for local Cecropin expression in Drosophila barrier epithelia. EMBO Rep 2:239–243
Pahl HL, Krauss B, Schulze-Osthoff K, Decker T, Traenckner EB, Vogt M, Myers C, Parks T, Warring P, Muhlbacher A et al (1996) The immunosuppressive fungal metabolite gliotoxin specifically inhibits transcription factor NF-kappaB. J Exp Med 183:1829–1840
Paik D, Monahan A, Caffrey DR, Elling R, Goldman WE, Silverman N (2017) SLC46 Family transporters facilitate cytosolic innate immune recognition of monomeric peptidoglycans. J Immunol 199:263–270
Pais IS, Valente RS, Sporniak M, Teixeira L (2018) Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol 16:e2005710
Palmer WH, Joosten J, Overheul GJ, Jansen PW, Vermeulen M, Obbard DJ, Van Rij RP (2019) Induction and suppression of NF-kappaB signalling by a DNA virus of Drosophila. J Virol 93:e01443-e11418
Paquette N, Conlon J, Sweet C, Rus F, Wilson L, Pereira A, Rosadini CV, Goutagny N, Weber AN, Lane WS et al (2012) Serine/threonine acetylation of TGFbeta-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci U S A 109:12710–12715
Paradkar PN, Trinidad L, Voysey R, Duchemin JB, Walker PJ (2012) Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc Natl Acad Sci U S A 109:18915–18920
Paredes JC, Welchman DP, Poidevin M, Lemaitre B (2011) Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35:770–779
Park JW, Kim CH, Kim JH, Je BR, Roh KB, Kim SJ, Lee HH, Ryu JH, Lim JH, Oh BH et al (2007) Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci U S A 104:6602–6607
Pearson A, Lux A, Krieger M (1995) Expression cloning of dSR-CI, a class C macrophage-specific scavenger receptor from Drosophila melanogaster. Proc Natl Acad Sci U S A 92:4056–4060
Perrin J, Mortier M, Jacomin AC, Viargues P, Thevenon D, Fauvarque MO (2015) The nonaspanins TM9SF2 and TM9SF4 regulate the plasma membrane localization and signalling activity of the peptidoglycan recognition protein PGRP-LC in Drosophila. J Innate Immun 7:37–46
Philips JA, Rubin EJ, Perrimon N (2005) Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science 309:1251–1253
Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, Ueda R, Lemaitre B (2004) In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem 279:12848–12853
Poirier EZ, Goic B, Tome-Poderti L, Frangeul L, Boussier J, Gausson V, Blanc H, Vallet T, Loyd H, Levi LI et al (2018) Dicer-2-dependent generation of viral DNA from defective genomes of RNA viruses modulates antiviral immunity in insects. Cell Host Microbe 23:353–365 e358
Qiao X, Wang L, Song L (2021) The primitive interferon-like system and its antiviral function in molluscs. Dev Comp Immunol 118:103997
Quintin J (2009) Études de la famille des GNBP/ßGRP dans la réponse immunitaire de la mouche du vinaigre Drosophila melanogaster et des relations entre cet hôte et les champignons opportunistes du genre Candida. (Université de Strasbourg)
Quintin J, Asmar J, Matskevich AA, Lafarge MC, Ferrandon D (2013) The Drosophila toll pathway controls but does not clear Candida glabrata infections. J Immunol 190:2818–2827
Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA (2002) Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416:644–648
Ramond E, Petrignani B, Dudzic JP, Boquete JP, Poidevin M, Kondo S, Lemaitre B (2020) The adipokine NimrodB5 regulates peripheral hematopoiesis in Drosophila. FEBS J 287:3399–3426
Razzell W, Evans IR, Martin P, Wood W (2013) Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr Biol 23:424–429
Reiser JB, Teyton L, Wilson IA (2004) Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution. J Mol Biol 340:909–917
Roh KB, Kim CH, Lee H, Kwon HM, Park JW, Ryu JH, Kurokawa K, Ha NC, Lee WJ, Lemaitre B et al (2009) Proteolytic cascade for the activation of the insect toll pathway induced by the fungal cell wall component. J Biol Chem 284:19474–19481
Royet J, Dziarski R (2007) Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol 5:264–277
Rubin GM, Spradling AC (1982) Genetic transformation of Drosophila with transposable elements vectors. Science 218:348–353
Rutschmann S, Jung AC, Rui Z, Silverman N, Hoffmann JA, Ferrandon D (2000) Role of Drosophila IKKg in a Toll-independent antibacterial immune response. Nat Immunology 1:342–347
Ryu JH, Ha EM, Oh CT, Seol J-H, Brey P, Jin I, Lee DL, Kim J, Lee D, Lee WJ (2006) An essential complementary role of NF-kappaB pathway to microbicidal oxidants in Drosophila gut immunity. Embo J 25:3693–3701
Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ (2008) Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319:777–782
Sanchez Bosch P, Makhijani K, Herboso L, Gold KS, Baginsky R, Woodcock KJ, Alexander B, Kukar K, Corcoran S, Jacobs T et al (2019) Adult Drosophila lack hematopoiesis but rely on a blood cell reservoir at the respiratory epithelia to relay infection signals to surrounding tissues. Dev Cell 51:787–803 e785
Savill J, Hogg N, Ren Y, Haslett C (1992) Thrombospondin cooperates with CD36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J Clin Invest 90:1513–1522
Schmidt RL, Rinaldo FM, Hesse SE, Hamada M, Ortiz Z, Beleford DT, Page-McCaw A, Platt JL, Tang AH (2011) Cleavage of PGRP-LC receptor in the Drosophila IMD pathway in response to live bacterial infection in S2 cells. Self Nonself 2:125–141
Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL (2000) Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101:671–684
Schneider J, Imler JL (2021) Sensing and signalling viral infection in Drosophila. Dev Comp Immunol 117:103985
Shia AK, Glittenberg M, Thompson G, Weber AN, Reichhart JM, Ligoxygakis P (2009) Toll-dependent antimicrobial responses in Drosophila larval fat body require Spatzle secreted by haemocytes. J Cell Sci 122:4505–4515
Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ (2011) Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674
Shiratsuchi A, Mori T, Sakurai K, Nagaosa K, Sekimizu K, Lee BL, Nakanishi Y (2012) Independent recognition of Staphylococcus aureus by two receptors for phagocytosis in Drosophila. J Biol Chem 287:21663–21672
Sinha NK, Iwasa J, Shen PS, Bass BL (2018) Dicer uses distinct modules for recognizing dsRNA termini. Science 359:329–334
Sinha NK, Trettin KD, Aruscavage PJ, Bass BL (2015) Drosophila dicer-2 cleavage is mediated by helicase- and dsRNA termini-dependent states that are modulated by Loquacious-PD. Mol Cell 58:406–417
Slavik KM, Morehouse BR, Ragucci AE, Zhou W, Ai X, Chen Y, Li L, Wei Z, Bahre H, Konig M et al (2021) cGAS-like receptors sense RNA and control 3’2’-cGAMP signalling in Drosophila. Nature 597:109–113
Smith-Bolton R (2016) Drosophila imaginal discs as a model of epithelial wound repair and regeneration. Adv Wound Care (new Rochelle) 5:251–261
Soderhall K, Cerenius L (1998) Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol 10:23–28
Soldano A, Alpizar YA, Boonen B, Franco L, Lopez-Requena A, Liu G, Mora N, Yaksi E, Voets T, Vennekens R et al (2016) Gustatory-mediated avoidance of bacterial lipopolysaccharides via TRPA1 activation in Drosophila. eLife 5:e13133
Somogyi K, Sipos B, Penzes Z, Kurucz E, Zsamboki J, Hultmark D, Ando I (2008) Evolution of genes and repeats in the Nimrod superfamily. Mol Biol Evol 25:2337–2347
Song W, Veenstra JA, Perrimon N (2014) Control of lipid metabolism by tachykinin in Drosophila. Cell Rep 9:40–47
Srinivasan N, Gordon O, Ahrens S, Franz A, Deddouche S, Chakravarty P, Phillips D, Yunus AA, Rosen MK, Valente RS et al (2016) Actin is an evolutionarily-conserved damage-associated molecular pattern that signals tissue injury in Drosophila melanogaster. eLife 5:e19662
Steiner H, Hultmark D, Engström A, Bennich H, Boman HG (1981) Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292:246–248
Stensmyr MC, Dweck HK, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S et al (2012) A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151:1345–1357
Stramer BM, Dionne MS (2014) Unraveling tissue repair immune responses in flies. Semin Immunol 26:310–314
Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD (2006) Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol 4:e4
Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RA, Moore KJ (2005) Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol 170:477–485
Stuart LM, Ezekowitz RA (2008) Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol 8:131–141
Sugumaran M, Barek H (2016) Critical analysis of the melanogenic pathway in insects and higher animals. Int J Mol Sci 17:1753
Tabuchi Y, Shiratsuchi A, Kurokawa K, Gong JH, Sekimizu K, Lee BL, Nakanishi Y (2010) Inhibitory role for D-alanylation of wall teichoic acid in activation of insect Toll pathway by peptidoglycan of Staphylococcus aureus. J Immunol 185:2424–2431
Takahasi K, Ochiai M, Horiuchi M, Kumeta H, Ogura K, Ashida M, Inagaki F (2009) Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition. Proc Natl Acad Sci U S A 106:11679–11684
Takehana A, Katsuyama T, Yano T, Oshima Y, Takada H, Aigaki T, Kurata S (2002) Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc Natl Acad Sci U S A 99:13705–13710
Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S (2004) Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. Embo J 23:4690–4700
Tanji T, Hu X, Weber AN, Ip YT (2007) Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol 27:4578–4588
Tanji T, Yun EY, Ip YT (2010) Heterodimers of NF-{kappa}B transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proc Natl Acad Sci U S A 107:14715–14720
Tassetto M, Kunitomi M, Andino R (2017) Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 169:314–325 e313
Theopold U, Schmidt O, Soderhall K, Dushay MS (2004) Coagulation in arthropods: defence, wound closure and healing. Trends Immunol 25:289–294
Tsai CR, Wang Y, Galko MJ (2018) Crawling wounded: molecular genetic insights into wound healing from Drosophila larvae. Int J Dev Biol 62:479–489
Tsarouhas V, Yao L, Samakovlis C (2014) Src kinases and ERK activate distinct responses to Stitcher receptor tyrosine kinase signaling during wound healing in Drosophila. J Cell Sci 127:1829–1839
Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL (2000) Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13:737–748
Veillard F, Troxler L, Reichhart JM (2016) Drosophila melanogaster clip-domain serine proteases: structure, function and regulation. Biochimie 122:255–269
Wang L, Gilbert RJ, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P (2008) Peptidoglycan recognition protein-SD provides versatility of receptor formation in Drosophila immunity. Proc Natl Acad Sci U S A 105:11881–11886
Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P (2006) Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. Embo J 25:5005–5014
Wang S, Tsarouhas V, Xylourgidis N, Sabri N, Tiklova K, Nautiyal N, Gallio M, Samakovlis C (2009) The tyrosine kinase Stitcher activates Grainy head and epidermal wound healing in Drosophila. Nat Cell Biol 11:890–895
Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D (2005) Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309:1874–1878
Weavers H, Martin P (2020) The cell biology of inflammation: from common traits to remarkable immunological adaptations. J Cell Biol 219:e202004003
Weavers H, Wood W, Martin P (2019) Injury activates a dynamic cytoprotective network to confer stress resilience and drive repair. Curr Biol 29:3851–3862 e3854
Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelievre E, Gascan H, Ray KP, Morse MA, Imler JL, Gay NJ (2003) Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat Immunol 4:794–800
Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D (2000) A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci U S A 97:13772–13777
West C, Silverman N (2018) p38b and JAK-STAT signaling protect against Invertebrate iridescent virus 6 infection in Drosophila. PLoS Pathog 14:e1007020
Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC (2004) Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell 118:619–633
Woodcock KJ, Kierdorf K, Pouchelon CA, Vivancos V, Dionne MS, Geissmann F (2015) Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity 42:133–144
Wu J, Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488
Wu SC, Liao CW, Pan RL, Juang JL (2012) Infection-induced intestinal oxidative stress triggers organ-to-organ immunological communication in Drosophila. Cell Host Microbe 11:410–417
Yakubovich N, Silva EA, O’Farrell PH (2010) Nitric oxide synthase is not essential for Drosophila development. Curr Biol 20:R141-142
Yang S, Zhao Y, Yu J, Fan Z, Gong ST, Tang H, Pan L (2019) Sugar alcohols of polyol pathway serve as alarmins to mediate local-systemic innate immune communication in Drosophila. Cell Host Microbe 26:240–251 e248
Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N et al (2008) Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol 9:908–916
Yoshida H, Kinoshita K, Ashida M (1996) Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem 271:13854–13860
Yoshida H, Ochiai M, Ashida M (1986) Beta-1,3-glucan receptor and peptidoglycan receptor are present as separate entities within insect prophenoloxidase activating system. Biochem Biophys Res Commun 141:1177–1184
Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B (2006) The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24:463–473
Zaidman-Remy, A, Poidevin M, Herve M, Welchman DP, Paredes JC, Fahlander C, Steiner H, Mengin-Lecreulx D, Lemaitre B (2011) Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS One 6:e17231
Zhan XL, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL (2004) Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron 43:673–686
Zhang L, Xu W, Gao X, Li W, Qi S, Guo D, Ajayi OE, Ding SW, Wu Q (2020) lncRNA sensing of a viral suppressor of RNAi activates non-canonical innate immune signaling in Drosophila. Cell Host Microbe 27:115-128.e118
Zsámboki J, Csordás G, Honti V, Pintér L, Bajusz I, Galgóczy L, Andó I, Kurucz É (2013) Drosophila Nimrod proteins bind bacteria. Open Life Sciences 8:633–645
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liegeois, S., Ferrandon, D. Sensing microbial infections in the Drosophila melanogaster genetic model organism. Immunogenetics 74, 35–62 (2022). https://doi.org/10.1007/s00251-021-01239-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-021-01239-0