Abstract
The fruit fly Drosophila melanogaster has been a valuable model to investigate the genetic mechanisms of innate immunity. Initially focused on the resistance to bacteria and fungi, these studies have been extended to include antiviral immunity over the last decade. Like all living organisms, insects are continually exposed to viruses and have developed efficient defense mechanisms. We review here our current understanding on antiviral host defense in fruit flies. A major antiviral defense in Drosophila is RNA interference, in particular the small interfering (si) RNA pathway. In addition, complex inducible responses and restriction factors contribute to the control of infections. Some of the genes involved in these pathways have been conserved through evolution, highlighting loci that may account for susceptibility to viral infections in humans. Other genes are not conserved and represent species-specific innovations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The fruit fly Drosophila melanogaster has been a favorite model organism for biologists since the beginning of the twentieth century. Their rapid generation, small size and the genetic tools created by the pioneering fly geneticists in the 1910s provided a unique model to investigate animal biology during the pre-genome editing era. As a result, studies on Drosophila led to many critical advances in developmental biology and neuroscience [1]. Since the 1990s, Drosophila has also been used as a model to investigate the innate immunity, which contributed greatly to renewed interest in this field [2]. Innate immunity is the first line of defense that multicellular organisms deploy to limit pathogen infections. In vertebrates, the innate response also regulates the production of cytokines and co-stimulatory molecules, which shape the subsequent adaptive immune response [3].
Studies on innate immunity in Drosophila initially focused on bacterial and fungal infections, and revealed that the production of antimicrobial peptides (AMPs) plays an important role in host defense [4]. Expression of AMPs is controlled by the evolutionarily conserved signaling pathways Toll and IMD (immune deficiency), which regulate the activity of transcription factors of the NF-κB family [5, 6]. Proteolytic cascades involving sequential activation of serine proteases participate in the activation of the Toll pathway, and in the clotting and melanization responses to wounding [7, 8]. Cellular responses involving both circulating and sessile hemocytes also participate in antimicrobial host defense in flies, in particular via phagocytosis of bacteria by macrophage-like plasmatocytes and encapsulation of parasitic wasp eggs in larvae by lamellocytes [9–11].
Viruses pose major threats to all organisms, including humans, as illustrated by epidemics such as influenza or HIV. Viruses also have a very significant economic impact through their effect on crops and livestock. Of note, several arboviruses (arthropod-borne viruses) are transmitted to mammalian hosts by hematophagous insect vectors. Recent epidemics associated with these viruses (e.g., Zika virus) are driving interest in the interactions between insect hosts and viruses, which remain poorly understood. As obligate intracellular pathogens, viruses present few targets for sensing or neutralization by the immune system. In addition, viruses evolve rapidly, which makes their control by the immune system a never-ending arms race. Investigating virus–host interactions in a wide set of hosts, including insects, can therefore provide interesting insights into fundamental antiviral strategies [12]. Over the last 12 years, a number of groups have started to investigate the genetic basis of antiviral resistance in Drosophila. It is now well established that the cell intrinsic mechanism of RNA interference (RNAi) plays a central role in the control of viral infections in flies, as it does in plants and other invertebrates. In addition, inducible responses and restriction factors also contribute to resistance to viral infections. Here, we review the current state of knowledge of these different mechanisms, focusing mainly on results published within the last 3 years. Excellent reviews on the earlier stages of the field can be found elsewhere [13–18].
RNA interference and nucleic acid-based immunity
RNA interference pathways in Drosophila
The term RNA interference (RNAi) includes an array of pathways, in which small non-coding (nc)RNAs (~20- to ~30-nt long) are used to regulate gene expression. First discovered in plants and subsequently in the nematode Caenorhabditis elegans as a way to silence genes with double stranded (ds)RNA, RNAi is conserved across the plant and animal kingdoms [19, 20]. RNAi involves the formation of an active RNA-induced silencing complex (RISC), which is formed by a small ncRNA (non-coding RNA) and a protein of the Argonaute (Ago) family. There are three major classes of small silencing RNAs in animals : microRNAs (miRNAs), small interfering RNAs (siRNAs) and piwi-interacting RNAs (piRNAs). In Drosophila, they interact with five AGO proteins: Ago1-3, Piwi and Aubergine [17].
miRNAs are involved in the post-transcriptional regulation of gene expression and participate in both developmental pathways and homeostasis. In flies, there are hundreds of miRNAs, which might provide regulation of up to half of the coding genome [21, 22]. miRNA biogenesis is a sequential process initiated in the nucleus and continuing in the cytoplasm with the help of several enzymes. Primary miRNAs (pri-miRNAs) are transcribed by RNA polymerase II and form a hairpin structure, which is processed by the nuclear RNase III enzyme Drosha and its dsRNA binding co-factor—Pasha [23]. This initial processing step leads to the formation of precursor miRNAs (pre-miRNAs), which are transported into the cytoplasm. A second RNase III enzyme, Dicer-1, then processes pre-miRNAs into mature 22-nt-long RNA duplexes. Again, this processing event is assisted by a dsRNA binding co-factor, the longest isoform of the Loquacious protein, Loqs-PB [24, 25]. Of note, the recent solving of the three-dimensional structure of Drosha points to a common ancestry with Dicer enzymes [26]. The 22-nt duplex miRNA is loaded onto the Ago1 protein, to form the miRNA programmed RNA-induced silencing complex (miRISC) [27]. Binding of miRISC to target mRNAs results in translation inhibition and mRNA degradation through slicing or decay. Several DNA viruses express miRNAs to regulate the expression of their genome, or the host cell genome [28]. Conversely, some viruses are targeted by host-encoded miRNAs [28]. The impact of miRNAs on Drosophila viruses remains poorly characterized [29]. The recent discovery of Kallithea virus, a fruit fly DNA virus of the Nudivirus family, which expresses a highly abundant miRNA, provides an opportunity to address its function in the Drosophila model [30].
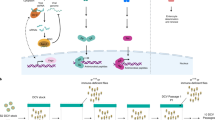
Double-stranded RNA (dsRNA) from exogenous and endogenous sources can trigger a second RNAi mechanism—the siRNA pathway. dsRNA is sensed by the second member of the Dicer family in insects, Dicer-2, which processes it into 21-nt duplex siRNAs (Fig. 1). These are loaded onto the AGO2 protein to form a pre-RISC complex, with the help of R2D2, a dsRNA binding co-factor of Dicer-2. One strand of the duplex, the passenger strand, is then ejected, leading to the formation of a mature RISC complex. The remaining strand, known as the guide strand, is stabilized by 2′-O-methylation of the 3′ nucleotide by the protein Hen1, and then targets the RNAs containing complementary sequence, which will be cleaved by the AGO2 slicer activity [17]. There is strong genetic evidence that the siRNA pathway, and in particular its three core components, Dcr-2, R2D2 and AGO2, play a major role in antiviral immunity in Drosophila. This is also supported by the production of Dcr-2-dependent virus-derived siRNAs in flies or cells infected by a range of different viruses, irrespective of their type of genome (RNA or DNA) (reviewed in [13, 14]). Virus-derived small RNAs provide a footprint of the action of the fly immune system, giving useful information on the sensing of viral nucleic acids. For example, immunostaining with antibodies recognizing dsRNA initially failed to detect dsRNA in cells infected by negative-strand RNA viruses [31]. However, the profile of siRNAs derived from vesicular stomatitis virus (VSV), a rhabdovirus containing a single-stranded RNA genome of negative polarity, is consistent with the processing of a viral dsRNA template [32, 33]. Indeed, a recent study with more sensitive antibodies has confirmed the presence of dsRNA in VSV-infected cells [34]. In the case of the DNA virus IIV6 (invertebrate iridescent virus 6), high-throughput sequencing pinpoints the genomic region whence the dsRNA is produced [35, 36]. That many insect and plant viruses express suppressors of the siRNA pathway (see below) also attests to the importance of this pathway for the control of viral infection. A major remaining question is the sensing of dsRNA by Dicer-2. Indeed, this mechanism appears to differ depending on the source of the trigger. For example, the short isoform of Loqs, Loqs-PD, is required for the processing of endo-siRNAs or exo-siRNAs by Dicer-2, but, intringuingly, is dispensable in the case of viral RNA [24, 37] (Fig. 1).
The siRNA pathway in Drosophila. The three core proteins of the RNAi machinery (Dicer-2, R2D2 and AGO2) associate with cofactors to sense RNAs and form the RNA-induced silencing complex (RISC). The siRNA pathway can be activated by the delivery of exogenous dsRNA in the cytosol or by viruses, but also by the transcription in the nucleus of natural antisense transcripts (NATs) or structured RNA. Virus-encoded suppressor molecules (in red triangles) inhibit the pathway at different steps (see the text for details)
The third RNAi mechanism in flies, the piRNA pathway, acts in germ cells and in some somatic tissues such as the follicular cells surrounding the ovaries [38]. piRNAs are small, 24- to 27-nt-long ncRNAs, which originate from specific genomic loci, mainly containing defective transposable elements [39]. As implied by their name, piRNAs associate with Piwi and two related proteins named Aubergine and AGO3, which form a separate clade of AGO proteins. Like siRNA, their 3′ end is protected by 2′-O-methylation [40]. The synthesis of piRNAs does not depend on Dicer proteins, but rather involves the exonuclease Zucchini and the AGO proteins from the Piwi clade themselves. piRNAs derived from viruses have been observed in mosquito cell lines and, for some viruses, in infected Aedes mosquitoes [41–46]. This suggests that the piRNA pathway may participate in antiviral immunity in Drosophila, in addition to its well-established role in genomic protection against transposable elements. However, in the case of Drosophila, virus-derived piRNAs have only been reported in a cell line derived from ovaries, and the piRNA pathway does not appear to participate in antiviral response in flies [47, 48].
Cell biology and regulation of the antiviral siRNA pathway
Besides the core components of the siRNA pathway, namely Dicer-2, R2D2 and AGO2, it is becoming apparent that other factors participate in this central antiviral pathway (Fig. 1).
Indeed, loading of the siRNA duplex onto AGO2 to form a pre-RISC complex cannot occur solely in the presence of the Dicer-2/R2D2 complex. Three chaperone proteins, Hsc70, Hsp90 and Hop, are essential for pre-RISC formation, whereas two others (Droj2 and p23) further improve the efficiency of AGO2–RISC assembly [49]. Overall, the in vitro addition of the five chaperone proteins to the core siRNA pathway components reconstituted a fully active RISC complex. The function of the chaperone machinery is ATP dependent, and ATP hydrolysis provides the energy required to accommodate the siRNA duplex onto AGO2. Hsc70 and Hsp90 also extend the dwell time of the Dicer-2/R2D2/siRNA complex on AGO2, therefore enabling the efficient loading of the siRNA duplex [49].
A further essential component of the siRNA pathway was recently identified in an unbiased EMS mutagenesis screen. The screen monitored silencing of the gene white, which encodes a transporter for a precursor of an eye color pigment, by a long hairpin RNA expressed from an inverted repeat transgene. In TAF11 mutant flies, silencing of white is significantly reduced, as observed in r2d2 mutant flies [50]. In agreement with these findings, GFP expression from a recombinant viral replicon derived from flock house virus (FHV), which is efficiently silenced by the siRNA pathway in cells from the S2 line, is derepressed after CRISPR/Cas-mediated knockout of the TAF11 gene. TAF11 interacts with Dicer-2, R2D2 and AGO2 and co-localizes in the cytoplasm with Dicer-2/R2D2 in cytoplasmic foci known as D2 bodies [51]. Although TAF11 does not bind siRNAs itself, it enhances the loading of radiolabeled duplex siRNA on AGO2. Altogether, these results are consistent with a scaffold model in which four TAF11 molecules bind to and facilitate the formation of Dicer-2/R2D2 heterotetramers, which are necessary to bind duplex siRNA [50]. The resulting RISC loading complex is localized to cytoplasmic D2 bodies.
The siRNA pathway is also regulated at the transcriptional level. The Drosophila forkhead box O (dFOXO) protein is the single member of the highly conserved family of FOXO transcriptional regulators present in flies [52]. This transcription factor binds the AGO2 and Dicer-2 promoters and regulates their activity. Accordingly, dFOXO null flies are more susceptible to cricket paralysis virus (CrPV) and FHV infection and this defect can be rescued by ectopic expression of Dicer-2.
Other genes can have an impact on the siRNA pathway, albeit indirectly. One function of Dicer-2 is to degrade stress-induced tRNA fragments [53]. The RNA methyl transferase, DNMT2, limits tRNAs cleavage. In flies mutant for this enzyme, tRNA fragments accumulate and divert Dicer-2 from other substrates, e.g., dsRNA. This might account for the increased sensitivity of DNMT2 mutant flies to infections by RNA viruses [54].
Together, the components of the siRNA pathway exert strong selective pressure on viruses, which have developed countermeasures in the form of suppressor proteins.
Insect viruses express suppressors of RNAi
Many insect viruses, including those of Drosophila, encode viral suppressors of RNAi (VSRs). Some of them bind to long viral dsRNAs and prevent binding of Dcr-2. This is the case for the proteins 1 A and 340R from Drosophila C virus (DCV) and IIV6, respectively, which contain canonical dsRNA binding domains, dsRBDs [55–57]. The proteins B2 of FHV and VP3 of the birnavirus Drosophila X virus (DXV) also bind dsRNA, through a noncanonical domain [58]. In addition to long dsRNA, 340R and B2 also bind 21-nt-long siRNA duplexes, thus also inhibiting efficient loading onto AGO2. Two other VSRs, 1 A and VP1 from the CrPV and Nora viruses, respectively, bind to AGO2 and interfere with its enzymatic activity (Fig. 1).
The importance of the VSRs has been particularly well illustrated in the case of FHV. Nodaviruses have small bipartite RNA genomes that are easy to manipulate genetically. RNA1 encodes the replicase, whereas RNA2 encodes the capsid proteins. A third RNA transcript, RNA3, is also produced from RNA1 and encodes the VSR B2. Whereas the wild-type FHV is highly pathogenic upon injection into the body cavity of the fly, viral mutants unable to express B2 are completely attenuated. As expected, the virus regains virulence when injected into Dcr-2 or AGO2 mutant flies [59, 60]. Interestingly however, even in the absence of a functional siRNA pathway, the B2-deficient virus exhibits reduced virulence compared to wild-type FHV. Indeed, a second function of B2 is to bind double-stranded regions of RNA2 and prevent this RNA being recruited into subcellular structures where its translation would be repressed. As a result, translation of the capsid protein is impaired in the absence of B2, even in RNAi-deficient cells [60].
The intimate relationship between some viral suppressors and components of the siRNA pathway (e.g., the VP1 suppressor of Nora virus and AGO2) exerts a strong selective pressure on the host genes to escape targeting. As a consequence, AGO2, r2d2 and Dcr-2 are among the fastest evolving genes in Drosophila [61]. Thus, viral suppressors can have host-specific activities. For example, the VP1 protein from a divergent Nora virus isolated from D. immigrans interacts with and suppresses D. immigrans AGO2, but not D. melanogaster AGO2 [62]. This provides an excellent example for the co-evolution of the host RNAi machinery and viral supression mechanisms.
From a cell-intrinsic resistance mechanism to systemic immunity?
The host defense against infection engages both cell-intrinsic mechanisms and systemic responses. The latter involve communication of immune signals, which alert uninfected cells and amplify the host response. In plants and the worm Caenorhabditis elegans, antiviral RNAi includes a systemic component that relies on the spreading of siRNAs [17]. This spreading can initiate the production of secondary siRNAs in non-infected cells, upon synthesis of dsRNA by host-encoded RNA-dependent RNA polymerases (RdRPs). In C. elegans, transmembrane transporters also participate in a systemic RNAi response. Genes homologous to plant and nematodes RdRP that could trigger systemic RNAi responses have not been identified in insects. Accordingly, RNAi is cell autonomous in Drosophila [63], and exposing flies to a low dose of DCV, which will trigger RNAi in infected cells, is not sufficient to induce protection against a challenge with a higher dose of virus [64]. However, dsRNA released by infected cells may trigger RNAi in distant cells, upon internalization by the dsRNA uptake pathway [65, 66]. In addition, viral RNA can be reverse transcribed into DNA in Drosophila and transcripts produced by this DNA can be processed by the siRNA pathway to inhibit viral replication [67]. Nanotubes-like structures may also be used to transfer dsRNA and components of the RNAi machinery between cells [68].
Inducible responses to viral infection in Drosophila
Evolutionarily conserved innate immune pathways and antiviral immunity
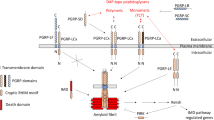
In Drosophila, bacterial and fungal infections activate the Toll and IMD pathways, which contribute to the resolution of the infection. Fungi and most Gram-positive bacteria activate the Toll pathway, in which the kinase Pelle phosphorylates the IκB protein Cactus, triggering its polyubiquitination and degradation by the proteasome. This allows the NF-κB proteins DIF and Dorsal to translocate to the nucleus and activate transcription of AMPs such as Drosomycin. Toll is activated by the neurotrophin-like cytokine Spaetzle. This molecule circulates in the hemolymph as an inactive precursor and is activated by proteolytic processing. The proteolytic cascade acting upstream of Spaetzle is triggered by circulating pattern recognition receptors (PRR) sensing lysine-type peptidoglycan (e.g., PGRP-SA) or β-glucans (e.g., GNBP3). Alternatively, the pathway can be induced by abnormal proteolytic activity in the hemolymph sensed by the serine-protease persephone (reviewed in [69]). Gram-negative bacteria activate the IMD pathway via their cell wall components. In particular, di-aminopimelic acid is sensed by the transmembrane PRR, PGRP-LC, which activates a cytoplasmic signaling pathway leading to phosphorylation and activation of another NF-κB protein called Relish. Upon nuclear translocation, Relish transcribes antimicrobial peptide genes (e.g., diptericin) (reviewed in [70]). The evolutionary ancient Toll and IMD pathways share several similarities with the inflammatory pathways regulated by Toll-like receptors, the interleukin-1 receptor and the TNF receptor in mammals. Activation of the Jak/STAT pathway represents another hallmark of the response to septic injury in Drosophila. The Drosophila genome encodes a single JAK kinase (Hopscotch) and a single STAT transcription factor (STAT92E), which respond to activation of the gp130-like cytokine receptor Domeless by three cytokines of the Unpaired (UPD) family [71]. As described below, the Toll, IMD and Jak/STAT pathways have been proposed to play a role in antiviral immunity (Fig. 2).
The Jak/STAT pathway participates in the control of infection by dicistroviruses
Some 130 genes are upregulated by a factor of at least two in response to DCV infection [72]. Characterization of the promoter of a strongly induced gene, vir-1, revealed the importance of DNA motifs corresponding to binding sites for the transcription factor STAT92E. Several DCV-induced genes share this property and, like vir-1, were no longer induced in flies either mutant for hopscotch or expressing a dominant-negative version of Domeless. Hopscotch mutant flies have increased viral load and succumb more rapidly than controls to DCV infection, indicating that at least some of the induced genes participate in the control of the infection [72]. Like inflammation in mammals, induction of the innate immunity pathways is associated with toxicity. This is best illustrated by the phenotype of flies mutant for the histone H3 lysine 9 (H3K9) methyltransferase G9a. These flies are highly susceptible to infection by RNA viruses, including DCV, and lethality is associated with hyperactivation of the Jak/STAT pathway [73]. Thus, the epigenetic regulator G9a mediates tolerance to virus infection in Drosophila by modulating activation of the Jak/STAT pathway. Importantly, even though vir-1 and other Jak/STAT regulated genes such as members of the Turandot (Tot) family are induced by several RNA viruses, hopscotch mutant flies appear to be uniquely susceptible to viruses of the Dicistroviridae family (e.g., DCV, CrPV) [35]. Probably, some aspects of the Jak/STAT-dependent inducible response may be redundant with additional defenses for viruses other than Dicistroviridae in flies.
Contribution of the NF-κB family transcription factors Dorsal, DIF and Relish to antiviral immunity
The Toll pathway has been proposed to play a role in antiviral immunity. An initial report indicated that two mutants of the Toll pathway, affecting the genes Dif and Toll, die more rapidly than controls following infection by Drosophila X virus (DXV), which belongs to the Birnaviridae family [74]. Intriguingly, the mutation in Dif was a loss of function, whereas the mutation in Toll was a gain of function, resulting in a constitutively activated pathway. A possible explanation for this finding could be that, as described above, immunopathology caused by activation of innate immunity pathways in Drosophila contributes to the lethality. Indeed, Dif 1 mutant flies exhibited a significant increase in DXV titer, in agreement with a role of DIF in the activation of an antiviral program, whereas the viral titer was slightly reduced in Toll 10b mutant flies. Of note, loss of function mutations in other genes of the pathway (spaetzle, Toll, pelle, tube) did not show a phenotype following DXV infection, suggesting that DIF regulates antiviral activity by a non-classical mechanism in this context. An independent study recently reported that the genes spaetzle, Toll, pelle and dorsal are required for resistance to oral infection by several RNA viruses (DCV, CrPV, FHV and Nora virus) [75]. Interestingly, although the Toll pathway is activated in the fat body upon oral infection or by direct injection of DCV into the hemocele, only the oral infection route gave a phenotype. This suggests that the antiviral action of the Toll pathway targets a step of the viral cycle specific to the oral infection route, which is bypassed when the virus is directly injected in the body cavity. Of note, polydnaviruses, which are mutualists of parasitic wasps, express vankyrin (vank) genes that encode inhibitors of Dorsal and Dif. Two vankyrins from Microplitis demolitor Bracovirus (MdBV) also bind to and inhibit Relish [76, 77].
Two studies initially reported that flies mutant for several genes encoding components of the IMD pathway were more susceptible than controls to infection by CrPV or SINV [78, 79]. A follow-up study by Hardy and co-workers identified 95 genes differentially expressed in transgenic flies expressing an SINV replicon [80]. Analysis of the upstream regions of these genes revealed the presence of Rel and STAT binding sites in most of them and the presence of these binding sites correlated with a decreased expression in flies heterozygotes for Relish and STAT92E. Among the genes tested, silencing of diptericin B (Relish dependent) and attacin C (STAT dependent) resulted in increased viral load. Thus, these two antimicrobial peptides appear to participate in the control of SINV infection in Drosophila. A completely independent set of experiments recently confirmed that the IMD pathway is involved in antiviral immunity [81]. These experiments investigated the function of the gene Diedel, which is strongly upregulated by some viruses, including SINV. Diedel mutant flies have reduced viability and succumb more rapidly than controls when infected by SINV, but not the unrelated vesicular stomatitis virus (VSV). This increased lethality does not result from uncontrolled viral replication, but rather from overactivation of the IMD pathway. Indeed, a large number of IMD regulated genes are upregulated in die mutant flies, and viability is rescued in flies double mutant for die and imd or ikkγ, two genes encoding key components of the IMD pathway [81]. Thus, die, which encodes a circulating 12 kDa protein, mediates tolerance to SINV infection by downregulating activation of the IMD pathway. Most interestingly, homologues of die are found in the genome of several insect DNA viruses [e.g., Spodoptera frugiperda ascovirus 1a (SfAV1a)], and one of them can partially rescue the phenotype of die mutant flies. The hijacking of a negative regulator of the IMD pathway by DNA viruses emphasizes the relevance of this pathway for the control of viral infections [81].
In summary, the genetic data now at hand point to an involvement of the three main evolutionarily conserved “inflammatory” pathways in the control of viral infections in flies. In addition, profiling of the transcriptome of infected cells and RNAi screens revealed that the heat shock pathway as well as transcriptional pausing participates in a rapid response to viral infection in Drosophila [82, 83]. In the intestinal epithelium, the cytokine Pvf2 can also trigger an antiviral program upon activating the PVR receptor and the ERK pathway [84, 85]. Altogether, these findings open interesting questions on the nature of the antiviral molecules these pathways upregulate, and on the mechanisms by which they are activated in the context of viral infections.
Sensing viral infection to trigger inducible responses
How viral infections are sensed in Drosophila remains largely unknown. In mammals, several innate immunity receptors sense viral nucleic acids and trigger the interferon response (reviewed in [86]). The only known sensor for viral RNA in flies is Dicer-2, which triggers RNA interference. Interestingly, Dicer-2 can also trigger an inducible response. Indeed, induction of the gene Vago is abolished in flies mutant for Dicer-2 or expressing FHV-B2, a VSR that competes with Dicer-2 for the binding to dsRNA [87]. The induction of an ortholog of Vago in Culex mosquitoes following infection by West Nile virus also depends on Dicer-2 [88]. Thus, this cytosolic sensor for viral RNA, which shares an evolutionarily conserved duplex RNA-activated ATPase domain with RIG-I like receptors, can trigger two types of responses in Drosophila, RNAi and induced expression of molecules associated with antiviral immunity [89]. The signaling pathway connecting Dicer-2 to induction of Vago has not yet been characterized in flies. In Culex however, the Relish ortholog REL2 and a TRAF factor have been associated with the upregulation of CxVago [90]. Thus, at least one component of the IMD pathway, the transcription factor REL2, can be activated by a pathway activated upon sensing dsRNA by Dicer-2 in Culex mosquitoes.
Excess DNA in the cytosol of Drosophila cells can also activate an immune response, suggesting that the sensing of viral DNA could contribute to antiviral immunity. Indeed, mutation of the gene encoding the lysosomal enzyme DNAseII results in the constitutive expression of the IMD-regulated AMPs Diptericin and Attacin A, whereas the Toll-regulated AMP drosomycin is not affected [91]. This suggests that the IMD pathway can be activated upon sensing cytosolic DNA. Although the sensor for DNA remains unknown, this pathway involves the serine phosphatase Eya, which can associate with the kinase IKKβ and the transcription factor Relish [92]. The possible involvement of this pathway in the resistance to DNA virus infection remains to be investigated.
Besides nucleic acids, other components of viral particles may be sensed by the immune system of the fly. For example, the gene diedel (die) is strongly upregulated by the enveloped viruses SINV and VSV, but the non-enveloped viruses DCV, CrPV and FHV induce little or no response. Furthermore, UV inactivation of both SINV and VSV does not impair induction of die. Altogether, these findings suggest that sensing of molecules from the viral envelope may trigger induction of this host cytokine [81]. Interestingly, induction of die does not involve Relish, but DIF. However, the signal transducer MyD88 is not required for die induction, suggesting that in the context of these viral infections, DIF is not activated by the canonical Toll pathway.
Finally, some genes may be induced by the stress associated with viral infection, or in response to alterations in the host physiology. For example, infection by DCV induces a heat shock response [82]. This response may be activated by the accumulation of unfolded viral proteins in the cytosol of infected cells. Activation of the Toll pathway in apoptosis-deficient flies may provide another mechanism for induction of immunity genes [93]. As detailed below, apoptosis is an evolutionarily conserved antiviral mechanism and many DNA viruses have evolved suppressors to escape it [94]. Virus-induced necrotic death, when apoptosis is inhibited, could result in the release of damage-associated molecular patterns activating the cytokine Spaetzle and the Toll pathway [93]. A third example is provided by viruses such as CrPV, which inhibit the cap-dependent translation of cellular mRNAs and may thus promote the synthesis of cap-independent mRNAs involved in stress response [95]. Finally, altered physiology of the infected host may contribute to the upregulation of immune genes. For example, DCV infects the smooth muscle cells of the crop in the anterior midgut, resulting in intestinal obstruction and a depletion of energy stores [96]. This starvation-like condition can lead to activation of the transcription factor FOXO, which participates in the regulation of the expression of AMP genes [97] and, as mentioned above, Dicer-2 and AGO2 [52].
Toward the functional characterization of virus-induced Drosophila genes
Only a handful of the genes identified as induced or upregulated by viral infection have been characterized functionally. Some of them do not act directly on the virus, but rather participate in the consolidation of the induced response or the homeostasis of this response. For example, the cytokines Upd2 and Upd3, which activate the Jak/STAT pathway, are induced by DCV infection [35]. The nucleoporin Nup98, which is upregulated by SINV and VSV, participates in the induced expression of a subset of putative antiviral genes, together with the transcription factor FoxK [98, 99]. The gene Vago encodes a 18 kDa cysteine-rich peptide containing a single von Willebrand factor type C domain, which probably represents another cytokine participating in the amplification of the immune response [87]. Indeed, Vago restricts DCV infection in the Drosophila fat body. Although Vago has not been characterized further in Drosophila, in Culex mosquitoes it restricts WNV infection by activating the Jak/STAT pathway, suggesting an antiviral cytokine function [88]. The last example is the cytokine Die, which is strongly induced by SINV and VSV and represses activation of the IMD pathway in the context of SINV infection [81]. Overactivation of the IMD pathway is associated with deleterious effects on the host. Accordingly, die mutant flies succumb more rapidly than wild-type controls following SINV infection. These mutants contain viral loads similar to that of controls, but exhibit an exacerbated induction of IMD-regulated genes, which reduces viability.
Other virus-induced genes encode antiviral effectors. As mentioned above, silencing the expression of two antimicrobial peptides induced by SINV infection, Diptericin B and Attacin C, results in increased SINV replication [80]. Antimicrobial peptides have also been associated with antiviral activity in vector mosquitoes and mammals [100, 101]. Activation of the heat shock response in flies improves the control of DCV infection and survival of the flies, indicating that this response is a constituent of antiviral innate immunity in Drosophila and that some of the molecules induced are associated with antiviral activity. Indeed, overexpression of the heat shock factor Hsp70 is sufficient to increase survival following DCV infection, suggesting that this factor has antiviral activity [82].
In summary, we are still largely ignorant of the function of most of the host factors induced by viral immune challenge. Even for the few candidate antiviral molecules that have been identified, we have limited understanding of their mode of action. The identification of pathways associated with antiviral resistance (e.g., Toll, IMD, Jak/STAT, transcriptional pausing) by genetic screens opens the way to the characterization of the transcriptome of infected flies mutant for these pathways. This will narrow down the number of candidate antiviral molecules to test functionally. Such approach was successfully used in Aedes mosquitoes, where the investigation of 18 genes regulated by the Jak/STAT pathway and induced by dengue virus infection led to the identification of two anti-dengue factors [102].
Induced apoptosis and phagocytosis
Apoptosis is a conserved mechanism of programmed cell death restricting viral replication and dissemination in insects. Caspases, the proteases that trigger apoptosis, are tightly regulated by members of the IAP (inhibitor of apoptosis protein) family (e.g., dIAP1 in Drosophila), which are themselves controlled by antagonist proteins (encoded in Drosophila by the RHG genes: reaper, hid, grim and sickle). Apoptosis can be triggered as a result of depletion of the labile protein dIAP1 when host translation shuts down as a result of viral infection [103]. Alternatively, viral infection can trigger the expression of pro-apoptotic RHG genes, following activation of the transcription factor p53 [104]. Programmed cell death can stop the infection before viral replication is completed. Additionally, apoptosis may promote clearance of infected cells by phagocytes, thus preventing dissemination. Clearly, both the presence of hemocytes (blood cells) and active phagocytosis are required to control FHV, DCV and CrPV in infected flies [79, 105, 106].
Intrinsic antiviral immunity and restriction factors in Drosophila
The first line of innate immune defense encountered by infectious viruses is constituted by restriction factors. These proteins are constitutively expressed in host cells already before the infection and target one or more steps of the viral replication cycle, resulting in a severe drop in the virus titer. Host proteins must fulfill several criteria to be considered as restriction factors. They are frequently encoded in the germline, and antiviral activity is often their main function. Their basal expression level can be upregulated by a viral infection, although their function is often antagonized by viral polypeptides. In addition, the direct interaction between rapidly evolving viral proteins and restriction factors exerts a constant pressure on the latter, resulting in a positive selection of these host genes [107, 108].
Multiple mechanisms of viral restriction have been described, with some factors specifically targeting one virus or virus family, while others have a broad spectrum of activity across several viral families. In the last two decades, many restriction factors and their mechanisms of action have been discovered in mammals [107–114]. Restriction factors also exist in Drosophila, although they remain poorly characterized. Most of them were identified while studying natural viral pathogens of Drosophila including the Sigma virus (DMelSV), a rhabdovirus, and the DCV dicistrovirus [115].
The p62 ortholog Ref(2)P restricts the sigma rhabdovirus (DMelSV)
Five loci involved in resistance to DMelSV infections referred to as ref(1)H, ref(2)M, ref(2)P, ref(3)D and ref(3)O were roughly mapped nearly 40 years ago by genetic analysis on the Drosophila melanogaster genome [116]. DMelSV is transmitted vertically and infects natural fly populations. The infection appears relatively benign, but can be easily detected since infected flies succumb following an exposure to CO2 [117]. The first and so far best characterized locus at which genetic variation affects virus multiplication in flies is ref(2)P, which is located on the left arm of the second chromosome. The gene was cloned by P-element tagging and common permissive ref(2)P o and restrictive ref(2)P p alleles were identified [118]. In fact, Ref(2)P is strongly polymorphic, as revealed by 14 different protein sequences obtained from 14 sequenced fly haplotypes [119]. Three polymorphisms located in the N-terminal PB1 (Phox and Bem 1) domain contribute to the sensitive or restrictive phenotype [119, 120]. Both restrictive and permissive forms of Ref(2)P can be co-immunoprecipitated with the N and P proteins from DMelSV [121], indicating a direct interaction. Interestingly, some mutations in the virus can overcome the restriction [122, 123], eventually resulting in the invasion of natural populations by variant viruses. Thus, Ref(2)P fulfills most of the criteria of a restriction factor. Surprisingly, however, flies with permissive alleles are more susceptible to DMelSV infection than those carrying a null allele. This suggests that the protein encoded by the permissive allele functions as a dominant-negative, with the virus having co-opted this protein for its replication, and the restrictive allele arisen a posteriori [124].
How does Ref(2)P affect DMelSV replication cycle? This protein forms a complex with the Drosophila atypical protein kinase C (daPKC), which positively regulates the Toll-signalling pathway and induces the synthesis of AMPs [125, 126]. Activation of the Toll pathway may therefore contribute to the inhibitory effect of Ref(2)P on DMelSV. Intruigingly, Ref(2)P is the Drosophila ortholog of the mammalian polyubiquitin-binding scaffold protein P62, (aka sequestosome 1). P62 recognizes autophagic cargos and allows their engulfment into autophagosomes through binding to members of the Atg8/LC3 family [127]. Accordingly, Ref(2)P co-localizes with cytoplasmic protein aggregates induced by aging, reduced proteasomal or autophagic activity, and neurodegenerative diseases in humans [128]. Autophagy, which has been associated with antiviral activity in flies [105, 129], may therefore contribute to the restrictive activity of Ref(2)P against DMelSV. In this regard, the pro- and antiviral action of the permissive and restrictive alleles of Ref(2)P may reflect its autophagic functions (e.g., [130–133]). Clearly, it would be interesting to investigate further the role of autophagy in the control of Sigma virus infections as well as the contribution of Ref(2)P in the resistance to other rhabdoviruses (e.g., VSV).
Two other restriction factors control susceptibility to DMelSV in Drosophila
Recently, two additional restriction factors for DMelSV, CHKov1 and Ge-1 have been characterized in Drosophila melanogaster. The gene Ge-1 is located on the left arm of the second chromosome. It was identified through high-resolution genetic mapping in the DMelSV refractory locus called ref(2)M [116, 134]. The Ge-1 protein is composed of an N-terminal WD40 domain and a C-terminal region separated by a serine-rich linker region. A rare polymorphism in Ge-1 consisting of a deletion of 26 amino acids from the linker region is associated with increased resistance to DMelSV infections in Drosophila [134]. No cross-resistance to DCV or Drosophila A virus (DAV) could be observed. Such specificity may be expected for bona fide restriction factors, although proof of an interaction between Ge-1 and a rhabdovirus factor/process is still missing. Of note, silencing of the common sensitive allele of Ge-1 increases susceptibility of flies to the rhabdovirus, indicating that even this allele is endowed with some restrictive activities against DMelSV. Ge-1 is a central component of processing bodies (P-bodies). These cytoplasmic foci are composed of RNA and proteins and are involved in mRNA degradation and posttranscriptional gene regulation. In these cytoplasmic structures, Ge-1 bridges Decapping protein 1 (Dcp1) and Decapping protein 2 (Dcp2), which together remove the 5′ cap from mRNAs (decapping) leading to their exonuclease-dependent degradation [135, 136]. AGO2, a key effector of the antiviral siRNA pathway, has also been localized in P-bodies. However, a link between AGO2 and the Ge-1 resistance to DMelSV could not be experimentally established [134]. Alternatively, the antiviral effect of Ge-1 may be mediated by the decapping complex. Indeed, the core component of the decapping complex, Dcp2, restricts the bunyavirus RVFV in Drosophila cells [137]. RNAi-mediated silencing of Dcp1 led to increased DMelSV loads in flies. However, silencing Dcp2 had no effect on DMelSV replication, suggesting that the canonical decapping complex is not involved [134].
The gene CHKov1 was identified by linkage mapping as the locus previously called ref(3)D [116]. The insertion of a transposable element (Doc) into the coding sequence of CHKov1 leads to the expression of shorter CHKov1 proteins that are endowed with a higher resistance capacity against DMelSV infections [138]. This genotype is the most common in natural populations of Drosophila. In few lines, another rearrangement, associated with even stronger resistance against this rhabdovirus, was discovered in this region. This complex rearrangement results in flies containing one partial and two full copies of CHKov2, a paralog of CHKov1, together with three copies of the CHKov1 allele containing the Doc insertion [138, 139]. A strong decrease in neutral genetic variation was also found in the regions surrounding the Doc insertion, probably reflecting a recent selection pressure exerted by the rhabdovirus on the CHKov1 gene [139]. In summary, there is strong genetic evidence indicating that CHKov1 is a third restriction factor for DMelSV, although some criteria, such as an interaction of the polypeptide with a viral factor/process, have not yet been fulfilled. Identifying such interaction would shed light on the mechanism of action of CHKov1.
The gene Pastrel restricts DCV infections in Drosophila
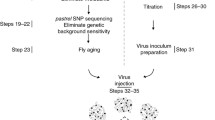
Wild Drosophila populations were found to differ in their sensitivity to infection by another natural viral pathogen, DCV. A genome-wide association study (GWAS) identified a cluster of polymorphisms in and around a gene called pastrel (pst), associated with resistance or susceptibility to DCV (Fig. 3) [139]. Pst is localized on the left arm of the third chromosome. Its restrictive activity seems to be virus specific, since no significant associations with susceptibility to another positive-strand RNA virus, the nodavirus FHV, or the rhabdovirus Drosophila affinis sigma virus (DAffSV), could be observed in flies. Notably, Pst was also found to restrict infection by another dicistrovirus, namely CrPV [140]. This implies that the restriction factor may target a conserved step of the replication cycle of these picorna-like viruses. The resistant allele of pst codes for alanine at position 598, whereas the susceptible variant encodes a threonine (Fig. 3). In this study, the key role of pst in DCV restriction was validated by functional RNAi-based analysis. These experiments revealed in addition that the pst-sensitive allele is also endowed with antiviral effects, although to a lesser extent than its restrictive counterpart [139]. Interestingly, an independent study assessing the genetic and phenotypic changes underlying the adaptation of D. melanogaster to DCV infections also identified the polymorphism affecting position 598 of Pst as a key determinant of resistance to this virus [140]. A second locus involved in the resistance to DCV was identified by genome-wide sequencing in this study. Ubc-2EH is located on the X chromosome and two polymorphisms associated with resistance to DCV were identified in introns of the gene. Involvement of Ubc-2EH was confirmed by RNAi, and, like pst, it was observed to also restrict CrPV. However, Ubc-2EH had no impact on FHV, which belongs to a different virus family [140].
Adaptation of Drosophila to DCV highlights the importance of restriction factors. a Flies grown for 20 generations under DCV infection show increased survival upon infection with the virus compared to controls. b Comparison of allele frequencies between control and virus-selected populations at generation 20 identified by genome-wide sequencing. The arrowheads indicate the localization of causal polymorphisms on the left arm of chromosome 3 and on chromosome X. CMH Cochran–Mantel–Haenszel test. c Schematic representation of the Pst and Ubc-E2H genes, with coding regions in green, and the location of polymorphism indicated in the exon 7 of Pst and in the intron of Ubc-E2H. a and b were redrawn and adapted from [140]
Other factors participating in intrinsic immunity in flies
Several other host factors were recently identified as limiting or restricting the multiplication of viruses in Drosophila. However, the viruses used do not naturally infect flies, which prevents these host factors being classified as bona fide restriction factors. We will refer to them as resistance factors, as proposed by Doyle et al. [141], for factors that do not fulfill all the criteria of restriction factors. One example of such factor is the protein Rm62, the fly ortholog of the mammalian DEAD-box helicase DDX17 [142]. This peptide was identified by an RNAi screen as restricting the replication of bunyaviruses including Rift Valley fever virus (RVFV) and La Crosse virus (LACV), in vivo and in vitro. In contrast, Rm62 did not impact the replication of VSV, SINV or DCV. Similar results were obtained in infected human cell cultures silenced for DDX17, the human homolog of Rm62. DDX17 is a nuclear protein which participates in the processing of host pri-miRNA via interactions with stem loop structures. In infected cells, it translocates to the cytoplasm where it can interact with viral RNA. Cross-linking immunoprecipitation followed by high-throughput sequencing (CLIP-seq) revealed that DDX17 binds an essential stem loop in bunyaviral RNA, explaining its antiviral action [142]. RVFV can also be restricted in Drosophila cells by dXPO1, an export receptor, and dRUVBL1, a component of the Tip60 histone acetylase complex [143].
The dsRNA-specific endoribonuclease Drosha that processes pri-miRNAs in the nucleus is another example of resistance factor. This enzyme relocalizes to the cytoplasm in Drosophila DL1 cells following infection by SINV, as mentioned above for Rm62 and RVFV. Drosha depletion in both Drosophila and mammalian cells results in increased viral RNA expression, demonstrating that the protein functions as an intrinsic antiviral factor upon relocalization to the cytoplasm [144]. Two other nuclear proteins, dMtr4 and dZcch7, were also reported to relocalise to the cytosol upon viral infection. These proteins belong to the exosome cofactor TRAMP complex and participate in viral RNA degradation after exiting the nucleus [145].
In summary, only a few proteins have been identified in D. melanogaster as fulfilling all the criteria of classical restriction factors. This is probably due to the relative lack of interest in Drosophila viral diseases and the low number of natural viral pathogens identified for this species to date. The recent discovery of at least 20 new RNA and DNA viruses infecting wild Drosophila using metagenomics will provide opportunities to identify additional restriction factors in flies [30]. Whether these will have mammalian orthologs, or not (like Pst), their functional characterization will provide powerful insights into critical steps of viral replication cycles, or components of the virus that can be targeted for intervention.
Concluding remarks
The choice of the fly D. melanogaster as a model to investigate innate immunity in the late 1980s, when the field was largely ignored by the majority of immunologists, was insightful. Indeed, the large range of genetic tools available for this model organism has provided important information on the functioning and regulation of innate immunity. One of the lessons from these studies is that innate immunity is not as unspecific as was formally thought. Indeed, insects and among them Drosophila can discriminate between different types of infectious microorganisms and mount somewhat directed responses. The study of antiviral immunity provides another striking example of the degree of specificity that can be achieved by innate immune responses. Indeed, the antiviral siRNA pathway, which relies on base pairing between 21 nucleotide-long siRNAs produced by the host and viral RNAs provides a striking example of extreme specificity and evolutionary adaptability.
Even though the role of the siRNA pathway in the control of viral infections is well established, important questions remain. They include the mechanism of the sensing of viral RNAs, which appears to differ from the sensing of non-viral dsRNA. Identification and characterization of novel regulators of the core components of the siRNA pathway provide useful information. The exploitation of profiles of virus-derived siRNAs, identified by high-throughput sequencing, will certainly also provide insights. Questions on the cell biology of antiviral RNAi and on cell to cell communication and amplification of responses are also fascinating issues that can be addressed with this model organism.
In addition to antiviral RNAi, several important questions remain regarding the induced antiviral response of Drosophila. We are still largely ignorant of the receptors and effector mechanisms involved, but the tools (e.g., mutant strains, differentially expressed transgenic strains, together with markers and reporters of the antiviral response) are now at hand to analyze these interactions. Restriction factors represent a third topic worthy of interest, and one can predict that the structural and functional characterization of the products of the genes that have been identified will provide important insight. Finally, the mechanism by which the intracellular endosymbiont bacterium Wolbachia potently interferes with viral infection in flies represent another exciting question [146, 147].
In conclusion, the studies of the past 10 years in the field of Drosophila antiviral immunity have revealed an array of mechanisms. Some of them have been conserved through evolution, while others have not. Until now, the D. melanogaster model has largely benefitted from the interest raised by the identification and characterization of evolutionarily conserved molecules and pathways. Arguably, the specificities of insect immunity are as interesting as the evolutionarily conserved aspects. Indeed, they represent species-specific innovations in host defense. In view of the rapid evolution of microorganisms, these innovations could be enlightening for the design of new therapeutic strategies for infectious diseases.
References
Bellen HJ, Yamamoto S (2015) Morgan’s legacy: fruit flies and the functional annotation of conserved genes. Cell 163:12–14. doi:10.1016/j.cell.2015.09.009
Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA (1999) Phylogenetic perspectives in innate immunity. Science 284:1313–1318
Medzhitov R, Janeway CAJ (2002) Decoding the patterns of self and nonself by the innate immune system. Science 296:298–300
Imler J-L, Bulet P (2005) Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy 86:1–21. doi:10.1159/000086648
Hoffmann JA (2003) The immune response of Drosophila. Nature 426:33–38
Hultmark D (2003) Drosophila immunity: paths and patterns. Curr Opin Immunol 15:12–19
Veillard F, Troxler L, Reichhart J-M (2016) Drosophila melanogaster clip-domain serine proteases: structure, function and regulation. Biochimie 122:255–269. doi:10.1016/j.biochi.2015.10.007
Theopold U, Krautz R, Dushay MS (2014) The Drosophila clotting system and its messages for mammals. Dev Comp Immunol 42:42–46. doi:10.1016/j.dci.2013.03.014
Weavers H, Evans IR, Martin P, Wood W (2016) Corpse engulfment generates a molecular memory that primes the macrophage inflammatory response. Cell 165:1658–1671. doi:10.1016/j.cell.2016.04.049
Gold KS, Brückner K (2015) Macrophages and cellular immunity in Drosophila melanogaster. Semin Immunol 27:357–368. doi:10.1016/j.smim.2016.03.010
Letourneau M, Lapraz F, Sharma A et al (2016) Drosophilahematopoiesis under normal conditions and in response to immune stress. FEBS Lett. doi:10.1002/1873-3468.12327
Marques JT, Imler J-L (2016) The diversity of insect antiviral immunity: insights from viruses. Curr Opin Microbiol 32:71–76. doi:10.1016/j.mib.2016.05.002
Bronkhorst AW, van Rij RP (2014) The long and short of antiviral defense: small RNA-based immunity in insects. Current Opin Virol 7:19–28. doi:10.1016/j.coviro.2014.03.010
Karlikow M, Goic B, Saleh M-C (2014) RNAi and antiviral defense in Drosophila: setting up a systemic immune response. Dev Comp Immunol 42:85–92. doi:10.1016/j.dci.2013.05.004
Xu J, Cherry S (2014) Viruses and antiviral immunity in Drosophila. Dev Comp Immunol 42:67–84. doi:10.1016/j.dci.2013.05.002
Kingsolver MB, Huang Z, Hardy RW (2013) Insect antiviral innate immunity: pathways, effectors, and connections. J Mol Biol 425:4921–4936. doi:10.1016/j.jmb.2013.10.006
Ding SW, Ding S-W (2010) RNA-based antiviral immunity. Nat Rev Immunol 10:632–644. doi:10.1038/nri2824
Lamiable O, Imler J-L (2014) Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol 20:62–68. doi:10.1016/j.mib.2014.05.006
Ratcliff F (1997) A similarity between viral defense and gene silencing in plants. Science 276:1558–1560. doi:10.1126/science.276.5318.1558
Fire A, Fire A, Xu S et al (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. doi:10.1038/35888
Berezikov E, Robine N, Samsonova A et al (2011) Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21:203–215. doi:10.1101/gr.116657.110
Carthew RW, Agbu P, Giri R (2016) MicroRNA function in Drosophila melanogaster. Semin Cell Dev Biol. doi:10.1016/j.semcdb.2016.03.015
Landthaler M, Yalcin A, Tuschl T (2004) The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14:2162–2167. doi:10.1016/j.cub.2004.11.001
Marques JT, Kim K, Wu P-H et al (2009) Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol 17:24–30. doi:10.1038/nsmb.1735
Saito K, Ishizuka A, Siomi H, Siomi MC (2005) Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol 3:e235. doi:10.1371/journal.pbio.0030235
Kwon SC, Nguyen TA, Choi Y-G et al (2016) Structure of human DROSHA. Cell 164:81–90. doi:10.1016/j.cell.2015.12.019
Förstemann K, Horwich MD, Wee L et al (2007) Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell 130:287–297. doi:10.1016/j.cell.2007.05.056
Skalsky RL, Cullen BR (2010) Viruses, microRNAs, and host interactions. Annu Rev Microbiol 64:123–141. doi:10.1146/annurev.micro.112408.134243
Hussain M, Asgari S (2014) MicroRNAs as mediators of insect host–pathogen interactions and immunity. J Insect Physiol 70:151–158. doi:10.1016/j.jinsphys.2014.08.003
Webster CL, Waldron FM, Robertson S et al (2015) The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol 13:e1002210. doi:10.1371/journal.pbio.1002210
Weber F, Wagner V, Rasmussen SB et al (2006) Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80:5059–5064
Mueller S, Gausson V, Vodovar N et al (2010) RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci USA 107:19390–19395. doi:10.1073/pnas.1014378107
Sabin LR, Zheng Q, Thekkat P et al (2013) Dicer-2 processes diverse viral RNA species. PLoS One 8:e55458. doi:10.1371/journal.pone.0055458
Son K-N, Liang Z, Lipton HL (2015) Double-stranded rna is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA viruses. J Virol 89:9383–9392. doi:10.1128/JVI.01299-15
Kemp C, Mueller S, Goto A et al (2013) Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol 190:650–658. doi:10.4049/jimmunol.1102486
Bronkhorst AW, van Cleef KWR, Vodovar N et al (2012) The DNA virus invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci USA 109:E3604–E3613. doi:10.1073/pnas.1207213109
Marques JT, Wang J-P, Wang X et al (2013) Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog 9:e1003579. doi:10.1371/journal.ppat.1003579
Malone CD, Malone CD, Brennecke J et al (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137:522–535. doi:10.1016/j.cell.2009.03.040
Brennecke J, Aravin AA, Stark A et al (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103
Wang H, Ma Z, Niu K, et al (2016) Antagonistic roles of Nibbler and Hen1 in modulating piRNA 3′ ends in Drosophila. Development 143:530–539. doi:10.1242/dev.128116
Vodovar N, Bronkhorst AW, van Cleef KWR et al (2012) Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 7:e30861. doi:10.1371/journal.pone.0030861
Morazzani EM, Wiley MR, Murreddu MG et al (2012) Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog 8:e1002470. doi:10.1371/journal.ppat.1002470
Léger P, Lara E, Jagla B et al (2013) Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J Virol 87:1631–1648. doi:10.1128/JVI.02795-12
Miesen P, Girardi E, van Rij RP (2015) Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res 43:6545–6556. doi:10.1093/nar/gkv590
Schnettler E, Donald CL, Human S et al (2013) Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J Gen Virol 94:1680–1689. doi:10.1099/vir.0.053850-0
Aguiar ERGR, Olmo RP, Paro S et al (2015) Sequence-independent characterization of viruses based on the pattern of viral small RNAs produced by the host. Nucleic Acids Res 43(13):6191–6206
Wu Q, Luo Y, Lu R et al (2010) Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci 107:1606–1611. doi:10.1073/pnas.0911353107
Petit M, Mongelli V, Frangeul L et al (2016) piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc Natl Acad Sci USA 113:E4218–E4227. doi:10.1073/pnas.1607952113
Iwasaki S, Sasaki HM, Sakaguchi Y et al (2015) Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature 521:533–536. doi:10.1038/nature14254
Liang C, Wang Y, Murota Y et al (2015) TAF11 assembles the RISC loading complex to enhance RNAi efficiency. Mol Cell 59:807–818. doi:10.1016/j.molcel.2015.07.006
Nishida KM, Miyoshi K, Ogino A et al (2013) Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol Cell 49:680–691. doi:10.1016/j.molcel.2012.12.024
Spellberg MJ, Marr MT, Marr MT II (2015) FOXO regulates RNA interference in Drosophilaand protects from RNA virus infection. Proc Natl Acad Sci 112:14587–14592. doi:10.1073/pnas.1517124112
Durdevic Z, Mobin MB, Hanna K, et al (2013) The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep 4:931–937. doi:10.1016/j.celrep.2013.07.046
Durdevic Z, Hanna K, Gold B et al (2013) Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep 14:269–275. doi:10.1038/embor.2013.3
van Rij RP, Saleh M-C, Berry B et al (2006) The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20:2985–2995. doi:10.1101/gad.1482006
van Cleef KWR, van Mierlo JT, Miesen P et al (2014) Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res 42:8732–8744. doi:10.1093/nar/gku528
Bronkhorst AW, van Cleef KWR, Venselaar H, van Rij RP (2014) A dsRNA-binding protein of a complex invertebrate DNA virus suppresses the Drosophila RNAi response. Nucleic Acids Res 42:12237–12248. doi:10.1093/nar/gku910
Chao JA, Lee JH, Chapados BR et al (2005) Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12:952–957
Han YH, Luo YJ, Wu Q et al (2011) RNA-based immunity terminates viral infection in adult Drosophila in the absence of viral suppression of RNA interference: characterization of viral small interfering RNA populations in wild-type and mutant flies. J Virol 85:13153–13163. doi:10.1128/JVI.05518-11
Petrillo JE, Petrillo JE, Venter PA et al (2013) Cytoplasmic granule formation and translational inhibition of nodaviral RNAs in the absence of the double-stranded RNA binding protein B2. J Virol 87:13409–13421. doi:10.1128/JVI.02362-13
Obbard DJ, Jiggins FM, Halligan DL, Little TJ (2006) Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol 16:580–585
van Mierlo JT, Overheul GJ, Obadia B et al (2014) Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog 10:e1004256. doi:10.1371/journal.ppat.1004256
Roignant J-Y, Carré C, Mugat B et al (2003) Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9:299–308. doi:10.1261/rna.2154103
Longdon B, Cao C, Martinez J, Jiggins FM (2013) Previous exposure to an RNA virus does not protect against subsequent infection in Drosophila melanogaster. PLoS One 8:e73833. doi:10.1371/journal.pone.0073833
Saleh M-C, Tassetto M, van Rij RP et al (2009) Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458:346–350. doi:10.1038/nature07712
Saleh M-C, van Rij RP, Hekele A et al (2006) The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol 8:793–802. doi:10.1038/ncb1439
Goic B, Vodovar N, Mondotte JA et al (2013) RNA-mediated interference and reverse transcription control the persistence of RNA viruses in the insect model Drosophila. Nat Immunol 14:396–403. doi:10.1038/ni.2542
Karlikow M, Goic B, Mongelli V, et al (2016) Drosophila cells use nanotube-like structures to transfer dsRNA and RNAi machinery between cells. Sci Rep 6:27085. doi:10.1038/srep27085
Valanne S, Wang J-H, Rämet M (2011) The Drosophila Toll signaling pathway. J Immunol 186:649–656. doi:10.4049/jimmunol.1002302
Kleino A, Silverman N (2014) The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol 42:25–35. doi:10.1016/j.dci.2013.05.014
Zeidler MP, Bausek N (2013) The Drosophila JAK-STAT pathway. JAKSTAT 2:e25353. doi:10.4161/jkst.25353
Dostert C, Jouanguy E, Irving P et al (2005) The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol 6:946–953. doi:10.1038/ni1237
Merkling SH, Bronkhorst AW, Kramer JM et al (2015) The epigenetic regulator G9a mediates tolerance to RNA virus infection in Drosophila. PLoS Pathog 11:e1004692. doi:10.1371/journal.ppat.1004692
Zambon RA, Nandakumar M, Vakharia VN, Wu LP (2005) The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci 102:7257–7262. doi:10.1073/pnas.0409181102
Ferreira ÁG, Naylor H, Esteves SS et al (2014) The Toll-dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog 10:e1004507. doi:10.1371/journal.ppat.1004507
Bitra K, Suderman RJ, Strand MR (2012) Polydnavirus Ank proteins bind NF-κB homodimers and inhibit processing of Relish. PLoS Pathog 8:e1002722. doi:10.1371/journal.ppat.1002722
Gueguen G, Kalamarz ME, Ramroop J et al (2013) Polydnaviral ankyrin proteins aid parasitic wasp survival by coordinate and selective inhibition of hematopoietic and immune NF-kappa B signaling in insect hosts. PLoS Pathog 9:e1003580. doi:10.1371/journal.ppat.1003580
Avadhanula V, Weasner BP, Hardy GG et al (2009) A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog 5:e1000582. doi:10.1371/journal.ppat.1000582
Costa A, Jan E, Sarnow P, Schneider D (2009) The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE 4:e7436. doi:10.1371/journal.pone.0007436
Huang Z, Kingsolver MB, Avadhanula V, Hardy RW (2013) An antiviral role for antimicrobial peptides during the arthropod response to alphavirus replication. J Virol 87:4272–4280. doi:10.1128/JVI.03360-12
Lamiable O, Kellenberger C, Kemp C et al (2016) Cytokine Diedel and a viral homologue suppress the IMD pathway in Drosophila. Proc Natl Acad Sci USA 113:698–703. doi:10.1073/pnas.1516122113
Merkling SH, Overheul GJ, van Mierlo JT, et al (2015) The heat shock response restricts virus infection in Drosophila. Sci Rep 5:12758. doi:10.1038/srep12758
Xu J, Grant G, Sabin LR et al (2012) Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila. Cell Host Microbe 12:531–543. doi:10.1016/j.chom.2012.08.011
Xu J, Hopkins K, Sabin L et al (2013) ERK signaling couples nutrient status to antiviral defense in the insect gut. Proc Natl Acad Sci USA 110:15025–15030. doi:10.1073/pnas.1303193110
Sansone CL, Cohen J, Yasunaga A et al (2015) Microbiota-dependent priming of antiviral intestinal immunity in Drosophila. Cell Host Microbe 18:571–581. doi:10.1016/j.chom.2015.10.010
Goubau D, Deddouche S, Reis e Sousa C (2013) Cytosolic sensing of viruses. Immunity 38:855–869. doi:10.1016/j.immuni.2013.05.007
Deddouche S, Matt N, Budd A et al (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol 9:1425–1432. doi:10.1038/ni 0.1664
Paradkar PN, Trinidad L, Voysey R et al (2012) Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc Natl Acad Sci USA 109:18915–18920. doi:10.1073/pnas.1205231109
Paro S, Imler J-L, Meignin C (2015) Sensing viral RNAs by Dicer/RIG-I like ATPases across species. Curr Opin Immunol 32:106–113. doi:10.1016/j.coi.2015.01.009
Paradkar PN, Duchemin J-B, Voysey R, Walker PJ (2014) Dicer-2-dependent activation of Culex Vago occurs via the TRAF-Rel2 signaling pathway. PLoS Negl Trop Dis 8:e2823. doi:10.1371/journal.pntd.0002823
Mukae N, Yokoyama H, Yokokura T et al (2002) Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev 16:2662–2671. doi:10.1101/gad.1022802
Liu X, Sano T, Guan Y et al (2012) Drosophila EYA regulates the immune response against DNA through an evolutionarily conserved threonine phosphatase motif. PLoS One 7:e42725. doi:10.1371/journal.pone.0042725
Ming M, Obata F, Kuranaga E, Miura M (2014) Persephone/Spätzle pathogen sensors mediate the activation of Toll receptor signaling in response to endogenous danger signals in apoptosis-deficient Drosophila. J Biol Chem 289:7558–7568. doi:10.1074/jbc.M113.543884
Clem RJ (2015) Viral IAPs, then and now. Semin Cell Dev Biol 39:72–79. doi:10.1016/j.semcdb.2015.01.011
Garrey JL, Lee Y-Y, Au HHT et al (2010) Host and viral translational mechanisms during cricket paralysis virus infection. J Virol 84:1124–1138. doi:10.1128/JVI.02006-09
Chtarbanova S, Lamiable O, Lee K-Z et al (2014) Drosophila C virus systemic infection leads to intestinal obstruction. J Virol 88:14057–14069. doi:10.1128/JVI.02320-14
Becker T, Loch G, Beyer M et al (2010) FOXO-dependent regulation of innate immune homeostasis. Nature 463:369–373. doi:10.1038/nature08698
Panda D, Pascual-Garcia P, Dunagin M et al (2014) Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila. Proc Natl Acad Sci USA 111:E3890–E3899. doi:10.1073/pnas.1410087111
Panda D, Gold B, Tartell MA, et al (2015) The transcription factor FoxK participates with Nup98 to regulate antiviral gene expression. MBio 6:e02509–14. doi:10.1128/mBio.02509-14
Klotman ME, Chang TL (2006) Defensins in innate antiviral immunity. Nat Rev Immunol 6:447–456. doi:10.1038/nri1860
Luplertlop N, Surasombatpattana P, Patramool S et al (2011) Induction of a peptide with activity against a broad spectrum of pathogens in the Aedes aegypti salivary gland, following infection with dengue virus. PLoS Pathog 7:e1001252. doi:10.1371/journal.ppat.1001252
Souza-Neto JA, Sim S, Dimopoulos G (2009) An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci USA 106:17841–17846. doi:10.1073/pnas.0905006106
Settles EW, Friesen PD (2008) Flock house virus induces apoptosis by depletion of Drosophila inhibitor-of-apoptosis protein DIAP1. J Virol 82:1378–1388
Liu B, Behura SK, Clem RJ et al (2013) P53-mediated rapid induction of apoptosis conveys resistance to viral infection in Drosophila melanogaster. PLoS Pathog 9:e1003137. doi:10.1371/journal.ppat.1003137
Lamiable O, Arnold J, de Faria IJDS et al (2016) Analysis of the contribution of hemocytes and autophagy to Drosophila antiviral immunity. J Virol 90:5415–5426. doi:10.1128/JVI.00238-16
Nainu F, Nainu F, Tanaka Y et al (2015) Protection of insects against viral infection by apoptosis-dependent phagocytosis. J Immunol 195:5696–5706. doi:10.4049/jimmunol.1500613
Kluge SF, Sauter D, Kirchhoff F (2015) SnapShot: antiviral restriction factors. Cell 163(774–774):e1. doi:10.1016/j.cell.2015.10.019
Duggal NK, Emerman M (2012) Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12:687–695. doi:10.1038/nri3295
Harris RS, Hultquist JF, Evans DT (2012) The restriction factors of human immunodeficiency virus. J Biol Chem 287:40875–40883. doi:10.1074/jbc.R112.416925
van Montfoort N, Olagnier D, Hiscott J (2014) Unmasking immune sensing of retroviruses: Interplay between innate sensors and host effectors. Cytokine Growth Factor Rev 25:657–668. doi:10.1016/j.cytogfr.2014.08.006
Smith RM, Pernstich C, Halford SE (2014) TstI, a Type II restriction-modification protein with DNA recognition, cleavage and methylation functions in a single polypeptide. Nucleic Acids Res 42:5809–5822. doi:10.1093/nar/gku187
Simon V, Bloch N, Landau NR (2015) Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi:10.1038/ni.3156
Jia X, Zhao Q, Xiong Y (2015) HIV suppression by host restriction factors and viral immune evasion. Curr Opin Struct Biol 31:106–114. doi:10.1016/j.sbi.2015.04.004
Zhou L-Y (2016) Host restriction factors for hepatitis C virus. World J Gastroenterol 22:1477. doi:10.3748/wjg.v22.i4.1477
Cogni R, Cao C, Day JP et al (2016) The genetic architecture of resistance to virus infection in Drosophila. Mol Ecol. doi:10.1111/mec.13769
Gay P (1978) Drosophila genes which intervene in multiplication of sigma virus. Mol Gen Genet 159:269–283. doi:10.1007/BF00268263
Lhéritier P (1958) The hereditary virus of Drosophila. In: Advances in virus research, vol 5. Elsevier, Amsterdam, pp 195–245
Contamine D, Petitjean AM, Ashburner M (1989) Genetic resistance to viral infection: the molecular cloning of a Drosophila gene that restricts infection by the rhabdovirus sigma. Genetics 123:525–533
Wayne ML, Contamine D, Kreitman M (1996) Molecular population genetics of ref(2)P, a locus which confers viral resistance in Drosophila. Mol Biol Evol 13:191–199
Dru P, Bras F, Dezélée S et al (1993) Unusual variability of the Drosophila melanogaster ref(2)P protein which controls the multiplication of sigma rhabdovirus. Genetics 133:943–954
Wyers F, Dru P, Simonet B, Contamine D (1993) Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J Virol 67:3208–3216
Contamine D (1981) Role of the Drosophila genome in Sigma virus multiplication. I. Role of the ret(2)P gene; selection of host-adapted mutants at the nonpermissive allele Pp. Virology 114:474–488
Fleuriet A, Periquet G (1993) Evolution of the Drosophila melanogaster-sigma virus system in natural populations from Languedoc (southern France). Arch Virol 129:131–143
Carré-Mlouka A, Gaumer S, Gay P et al (2007) Control of sigma virus multiplication by the ref(2)P gene of Drosophila melanogaster: an in vivo study of the PB1 domain of Ref(2)P. Genetics 176:409–419. doi:10.1534/genetics.106.063826
Avila A, Silverman N, Diaz-Meco MT, Moscat J (2002) The Drosophila atypical protein kinase C-ref(2)p complex constitutes a conserved module for signaling in the toll pathway. Mol Cell Biol 22:8787–8795. doi:10.1128/MCB.22.24.8787-8795.2002
Goto A, Blandin S, Royet J et al (2003) Silencing of Toll pathway components by direct injection of double-stranded RNA into Drosophila adult flies. Nucleic Acids Res 31:6619–6623. doi:10.1093/nar/gkg852
Ktistakis NT, Tooze SA (2016) Digesting the expanding mechanisms of autophagy. Trends Cell Biol 26:624–635. doi:10.1016/j.tcb.2016.03.006
Nezis IP, Simonsen A, Sagona AP et al (2008) Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol 180:1065–1071. doi:10.1083/jcb.200711108
Shelly S, Lukinova N, Bambina S et al (2009) Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30:588–598. doi:10.1016/j.immuni.2009.02.009
Joubert P-E, Meiffren G, Grégoire IP et al (2009) Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 6:354–366. doi:10.1016/j.chom.2009.09.006
Richetta C, Faure M (2013) Autophagy in antiviral innate immunity. Cell Microbiol 15:368–376. doi:10.1111/cmi.12043
Richetta C, Grégoire IP, Verlhac P et al (2013) Sustained autophagy contributes to measles virus infectivity. PLoS Pathog 9:e1003599. doi:10.1371/journal.ppat.1003599
Mauthe M, Langereis M, Jung J et al (2016) An siRNA screen for ATG protein depletion reveals the extent of the unconventional functions of the autophagy proteome in virus replication. J Cell Biol 214:619–635. doi:10.1083/jcb.201602046
Cao C, Magwire MM, Bayer F, Jiggins FM (2016) A polymorphism in the processing body component Ge-1 controls resistance to a naturally occurring rhabdovirus in Drosophila. PLoS Pathog 12:e1005387. doi:10.1371/journal.ppat.1005387
Yu JH, Yang W-H, Gulick T et al (2005) Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11:1795–1802. doi:10.1261/rna.2142405
Xu J, Yang J-Y, Niu Q-W, Chua N-H (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18:3386–3398. doi:10.1105/tpc.106.047605
Hopkins KC, McLane LM, Maqbool T et al (2013) A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev 27:1511–1525. doi:10.1101/gad.215384.113
Magwire MM, Bayer F, Webster CL et al (2011) Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet 7:e1002337. doi:10.1371/journal.pgen.1002337
Magwire MM, Fabian DK, Schweyen H et al (2012) Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet 8:e1003057. doi:10.1371/journal.pgen.1003057
Martins NE, Faria VG, Nolte V et al (2014) Host adaptation to viruses relies on few genes with different cross-resistance properties. Proc Natl Acad Sci USA 111:5938–5943. doi:10.1073/pnas.1400378111
Doyle T, Goujon C, Malim MH (2015) HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol 13:403–413. doi:10.1038/nrmicro3449
Moy RH, Cole BS, Yasunaga A et al (2014) Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell 158:764–777. doi:10.1016/j.cell.2014.06.023
Yasunaga A, Hanna SL, Li J et al (2014) Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog 10:e1003914. doi:10.1371/journal.ppat.1003914
Shapiro JS, Schmid S, Aguado LC et al (2014) Drosha as an interferon-independent antiviral factor. Proc Natl Acad Sci USA 111:7108–7113. doi:10.1073/pnas.1319635111
Molleston JM, Sabin LR, Moy RH et al (2016) A conserved virus-induced cytoplasmic TRAMP-like complex recruits the exosome to target viral RNA for degradation. Genes Dev 30:1658–1670. doi:10.1101/gad.284604.116
Faria VG, Martins NE, Magalhães S et al (2016) Drosophila adaptation to viral infection through defensive symbiont evolution. PLoS Genet 12:e1006297. doi:10.1371/journal.pgen.1006297
Wong ZS, Brownlie JC, Johnson KN (2016) Impact of ERK activation on fly survival and Wolbachia-mediated protection during virus infection. J Gen Virol 97:1446–1452. doi:10.1099/jgv.0.000456
Acknowledgements
We thank Dr. Nelson Martins and Dr. David Gubb for critical reading and comments on the manuscript and Dr. Carine Meignin for help with the figures. Work in our laboratory was supported by CNRS and grants from NIH (PO1 AI070167), ANR (ANR-13-BSV3-009), Infect-ERA (ANR-14-IFEC-0005), and Investissements d’Avenir Programs (ANR-10-LABX-36 ; ANR-11-EQPX-0022).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mussabekova, A., Daeffler, L. & Imler, JL. Innate and intrinsic antiviral immunity in Drosophila. Cell. Mol. Life Sci. 74, 2039–2054 (2017). https://doi.org/10.1007/s00018-017-2453-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2453-9